Regulated responses to extracellular signals depend on cell-surface proteins that are internalized and recycled back to the plasma membrane. Two novel WD40 domain proteins, Ere1 and Ere2 (endosomal recycling proteins), are found to mediate cargo-specific recognition by the retromer pathway.
Abstract
Regulated secretion, nutrient uptake, and responses to extracellular signals depend on cell-surface proteins that are internalized and recycled back to the plasma membrane. However, the underlying mechanisms that govern membrane protein recycling to the cell surface are not fully known. Using a chemical-genetic screen in yeast, we show that the arginine transporter Can1 is recycled back to the cell surface via two independent pathways mediated by the sorting nexins Snx4/41/42 and the retromer complex, respectively. In addition, we identify two novel WD40-domain endosomal recycling proteins, Ere1 and Ere2, that function in the retromer pathway. Ere1 is required for Can1 recycling via the retromer-mediated pathway, but it is not required for the transport of other retromer cargoes, such as Vps10 and Ftr1. Biochemical studies reveal that Ere1 physically interacts with internalized Can1. Ere2 is present in a complex containing Ere1 on endosomes and functions as a regulator of Ere1. Taken together, our results suggest that Snx4/41/42 and the retromer comprise two independent pathways for the recycling of internalized cell-surface proteins. Moreover, a complex containing the two novel proteins Ere1 and Ere2 mediates cargo-specific recognition by the retromer pathway.
INTRODUCTION
The balance between down-regulation and recycling pathways controls the cell-surface expression and function of plasma membrane (PM) proteins (Saksena et al., 2007; Hanyaloglu and von Zastrow, 2008; Mudd and Kass, 2008; Grant and Donaldson, 2009). The plasma membrane continually undergoes remodeling in response to environmental cues, by the delivery and removal of lipids and proteins via the secretory and endocytic pathways. The fate of an internalized cell-surface protein is determined at endosomes by either the ESCRT–mediated (endosomal complexes required for transport) multivesicular body (MVB) sorting pathway for subsequent degradation in lysosomes (equivalent to the yeast vacuole) or by recycling pathways that target PM proteins back to the cell surface. Thus the degradation and the recycling of internalized PM proteins are essential for the proper control of cell signaling and quality control pathways. Defects in the function of the ESCRT machinery have been associated with several diseases (Saksena et al., 2007). Likewise, recycling pathways for targeting internalized transmembrane proteins to the cell surface are critical for nutrient uptake and responses to extracellular signals. The transferrin receptor and low-density-lipoprotein receptor are two classic examples of PM proteins in mammalian cells that are efficiently recycled to the cell surface following their internalization and sorting at endosomes (Brown et al., 1983; Grant and Donaldson, 2009). In addition, prolonged or continued responses to G protein–coupled receptor (GPCR) agonists (e.g., resensitization of β-adrenergic receptors; Hanyaloglu and von Zastrow, 2008), the recycling of synaptic vesicle proteins necessary for neurotransmitter release at the synapse (Dittman and Ryan, 2009), and the delivery of glucose transporters (e.g., GLUT4) from intracellular stores in response to insulin signaling (Hoffman and Elmendorf, 2011) provide examples for the importance of recycling pathways in cell signaling. In yeast, several plasma membrane proteins have been shown to undergo recycling following endocytic internalization, including the soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) protein Snc1, the chitin synthase Chs3, and the iron transporter Ftr1 (Bonifacino and Rojas, 2006; Grant and Donaldson, 2009). Of interest, distinct pathways for the recycling of these proteins have been implicated in yeast.
However, the pathways that orchestrate the recycling of internalized cell-surface proteins are not completely understood. For recycling, sorting at endosomes seems to be the first and the most crucial step. Protein sorting occurs on different types of endosomes (early and late) and can be carried out by different protein complexes, such as clathrin and its adaptors, the sorting nexins, and the retromer complex (Bonifacino and Rojas, 2006; Johannes and Popoff, 2008; Grant and Donaldson, 2009). Each of these protein complexes is evolutionarily conserved. In yeast and mammalian cells, clathrin and its AP-1 adaptor complex are localized at the trans-Golgi network (TGN) and endosomes. The function of clathrin and the AP-1 adaptor complex in sorting proteins between the TGN and endosomes has been shown for cargoes such as Chs3 and Kex2 in yeast (Seeger and Payne, 1992; Valdivia et al., 2002) and Shiga toxin and mannose 6-phosphate receptors in mammalian cells (Johannes and Popoff, 2008).
The yeast sorting nexins, Snx4, Snx41, and Snx42, function in a distinct pathway that mediates the sorting of proteins out of endosomes (Bonifacino and Rojas, 2006). The best-known cargo for Snx4, Snx41, and Snx42 is the vesicle-SNARE protein Snc1, which is transported from early endosomes to the TGN. Snx4 can be cross-linked to Snc1 and binds to Snx41 and Snx42, indicating that these sorting nexins constitute a complex that directly mediates the sorting of Snc1 from endosomes to the TGN (Hettema et al., 2003). The mammalian homologues of Snx4, Snx41, and Snx42 are SNX4, SNX7, and SNX30, respectively (Teasdale et al., 2001). Mammalian SNX4 also functions in the recycling of cell-surface proteins (Traer et al., 2007). In addition to mediating endosome-to-TGN transport, mammalian SNX4 also directs transport from the endosome to the perinuclear endosomal recycling compartment (ERC; Skanland et al., 2007). Little is known about the function of SNX7 and SNX30 in mammalian cells.
Another conserved protein complex involved in endosomal sorting is the retromer complex (Seaman, 2005; Bonifacino and Rojas, 2006). A role for the retromer complex was first described in yeast for mediating the retrograde transport of the vacuolar protein sorting receptor Vps10 from endosomes to the TGN. The retromer complex has five components and can be divided into two subcomplexes: a cargo recognition subcomplex containing Vps26, Vps29, and Vps35, and a membrane deformation subcomplex consisting of two PX and BAR domain–containing sorting nexin proteins, Vps5 and Vps17. Recently another sorting nexin, Snx3, was found to function with the retromer for the recycling of the iron permease proteins, Fet3 and Ftr1, in yeast. This study indicated that Snx3 physically interacted with Ftr1 and the cargo recognition complex (Strochlic et al., 2007). In higher eukaryotes, the retromer has been shown to recycle the sorting receptor Wntless that is required for the Wnt signaling and developmental pathway (Belenkaya et al., 2008; Franch-Marro et al., 2008; Pan et al., 2008; Port et al., 2008; Yang et al., 2008).
Although clathrin, Snx4/Snx41/Snx42, and retromer all mediate the sorting of proteins out of endosomes, the relative roles of these proteins in recycling remain largely unknown. Moreover, even less is understood about how specific cargoes are recognized for recycling by each of these pathways. We performed a chemical-genetic screen designed to identify mutants that are defective in cell-surface targeting and stability of the arginine permease Can1. In this screen, single-deletion mutants of genes encoding the Snx4/41/42 complex components, the Snx3 sorting nexin, and the retromer components were impaired in Can1 cell-surface stability. In addition, we identified two uncharacterized WD40 domain–containing proteins, Ybr246w and Ypl183c (which we named Ere1 and Ere2, for endosomal recycling), as necessary for efficient Can1 cell-surface targeting. Our data further support that Snx4/41/42 and the retromer act in two independent pathways that mediate the recycling of cell-surface proteins. Moreover, our data indicate that a complex containing the two novel proteins, Ere1 and Ere2, mediates cargo-specific recognition by the retromer pathway.
RESULTS
Snx4/41/42, retromer, and the novel proteins Ere1 and Ere2 are involved in the recycling of Can1
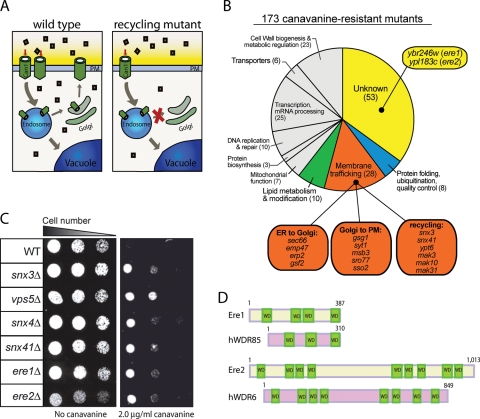
To identify factors involved in cell-surface protein targeting and recycling, we performed a screen of the yeast nonessential gene library containing ∼4700 single-deletion mutants. We used the arginine permease Can1 as a model PM protein, reasoning that mutant cells with reduced Can1 cell-surface expression levels would display resistance to the toxic arginine analogue canavanine. In particular, we predicted that canavanine resistance (cvr) would result from reduced canavanine uptake due to defects in cell-surface targeting or recycling of the Can1 permease (Figure 1A). We identified 173 cvr mutant strains that displayed a canavanine-resistant phenotype (Figure 1B; see Supplemental Table for the full list). Genes identified in the canavanine resistance screen have been implicated in numerous cellular processes, including membrane trafficking (28 genes), lipid metabolism (10 genes), transcription and RNA metabolism (25 genes), protein folding and quality control (8 genes), and metabolic regulation (23 genes; Figure 1B and Supplemental Table).
FIGURE 1:
Cells lacking Snx4/41/42, retromer components, Ere1, or Ere2 are resistant to canavanine. (A) Model for Can1 trafficking and the screen for recycling mutants. In wild-type cells, the lifetime of Can1 at the PM is regulated by endocytosis, recycling, and degradation (left). In mutant cells deficient for Can1 recycling, Can1 cell-surface levels are reduced, resulting in reduced uptake of the toxic arginine analogue canavanine (squares). (B) Classification of cvr mutants identified in the canavanine resistance screen. (C) Cells impaired in endosomal recycling are resistant to canavanine. Serial dilutions (fourfold) of wild-type (WT) and deletion mutant cells (as indicated) were grown on synthetic media plates with or without 2 μg/ml canavanine for 2 d at 26°C. (D) Schematic diagrams for Ere1, Ere2, and their human orthologues WDR85 and WDR6, respectively.
As expected, we identified several canavanine-resistant mutants lacking factors with known roles in endosomal recycling. These included deletions in SNX3 and SNX41 that encode PX domain–containing sorting nexin family member proteins (Figure 1, B and C, and Table 1), as well asYPT6, which encodes a Rab GTPase, and genes (MAK3, MAK10, and MAK31) that encode subunits of the NatC acetylase complex previously suggested to regulate the Arf-like GTPase Arl3 in recycling from endosomes (Figure 1B and Supplemental Table; Setty et al., 2004). In addition, we found that loss of the sorting nexins Snx4/41/42 or the retromer subunits (Vps5, Vps17, Vps26, Vps29, Vps35, and Snx3) resulted in canavanine-resistant phenotypes (Figure 1C and Table 1), implying that both the Snx4/41/42 and retromer complexes are involved in Can1 recycling.
TABLE 1:
Canavanine-resistant phenotypes of recycling mutants.
| Strain | Score | Strain | Score | Strain | Score |
|---|---|---|---|---|---|
| Wild type | − | snx3Δ vps5Δ | ++ | snx41Δ snx3Δ | ++++ |
| snx4Δ | ++ | snx4Δ snx41Δ snx4Δ | ++ | snx4Δ snx3Δ | ++++ |
| snx41Δ | ++ | vps5Δ ere1Δ | ++ | snx4Δ vps35Δ | ++++ |
| snx42Δ | + | snx3Δ ere1Δ | ++ | snx4Δ ere1Δ | ++++ |
| snx3Δ | ++ | snx3Δ ere2Δ | ++ | snx41Δ ere1Δ | ++++ |
| vps5Δ | ++ | ere1Δ ere2Δ | ++ | snx41Δ ere2Δ | ++++ |
| vps17Δ | ++ | apl2Δ | − | ||
| vps26Δ | + | gga2Δ | − | ||
| vps29Δ | ++ | ent3Δ | − | ||
| vps35Δ | ++ | ent5Δ | − | ||
| ere1Δ | ++ | ||||
| ere2Δ | ++ |
The Ere1 and Ere2 proteins function in the retromer recycling pathway. Serial dilutions (fourfold) of wild-type and different mutant cells (as indicated) were grown on synthetic media plates with or without 2.0 μg/ml canavanine for 2 d at 26°C. The canavanine-resistant phenotype of each strain was scored based on the relative growth of serial dilutions on canavanine plates. Relative growth: no growth at 0.1 OD600/ml was scored as −; growth at 0.1 OD600/ml was scored as +; growth at 2.5 × 10−2 OD600/ml was scored as ++; growth at 1.56 × 10−3 OD600/ml was scored as ++++.
Examination of other known membrane trafficking and lipid metabolism mutants identified in the cvr screen revealed multiple steps involved in Can1 cell-surface targeting and stability (Figure 1B and Supplemental Table). A subset of the membrane trafficking mutants has previously been implicated in transport of cargo proteins from the ER (Figure 1B and Supplemental Table). These include Erp2 and Emp47, involved in COP-II vesicle sorting and formation, and Gsf2, which has been implicated in sorting integral membrane proteins, such as hexose transporters, into COP-II–coated vesicles that bud from the endoplasmic reticulum (Figure 1B; Marzioch et al., 1999; Sherwood and Carlson, 1999; Sato and Nakano, 2002). The cvr screen also identified components implicated in Golgi transport, such as Gsg1, a component of the TRAPP tether complex, and an Arf GTPase guanine nucleotide exchange factor, Syt1 (Figure 1B; Sacher et al., 2000; Chen et al., 2010). In addition, factors that act at a late step in the secretory pathway involved in fusion of secretory vesicles with the plasma membrane were identified in the cvr screen. These included the SNARE protein Sso2 and the Sec4 GTPase-activating protein Msb3 (Figure 1B; Aalto et al., 1993; Gao et al., 2003). In addition, several factors involved in ergosterol and sphingolipid lipid metabolism pathways, including Osh1, Osh2, Ipt1, Sur2, and Sur4, were identified in the cvr screen (Supplemental Table). Previous work suggested that sorting of Can1 into specific PM domains enriched in ergosterol and sphingolipids (termed PMC domains) regulates Can1 cell-surface stability (Grossmann et al., 2008).
In addition, we identified 53 genes with uncharacterized or not well-characterized functions in the cvr screen (Figure 1B), which we designated CVR1–53 (Supplemental Table). Of interest, 60% (32 of the 53) of the predicted gene products encoded by CVR1–53 have orthologues in other species (Supplemental Table). For example, two factors that we chose for further study and named Ere1 and Ere2 (for endosomal recycling; see later discussion) have human orthologues, hWDR85 and hWDR6, respectively (Figure 1D). Because these uncharacterized factors could be involved in various aspects of Can1 expression, activity, and targeting, we next performed tests to specifically identify factors involved in Can1 cell-surface recycling.
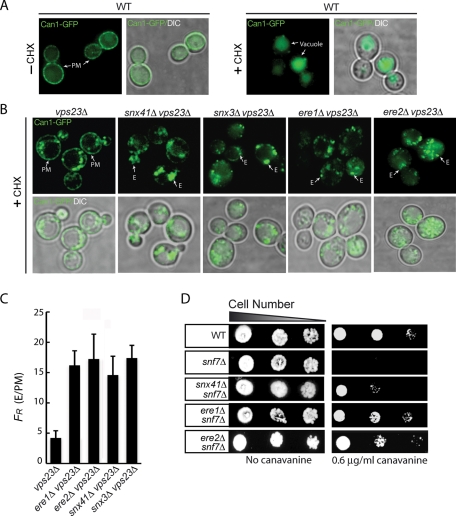
In exponentially growing wild-type cells, green fluorescent protein (GFP)–tagged Can1 localizes primarily to the plasma membrane (Figure 2A; Lin et al., 2008). Under stress or starvation conditions, such as in the presence of cycloheximide (Figure 2A; Lin et al., 2008), Can1-GFP is rapidly endocytosed and subsequently sorted by the ESCRT-mediated MVB pathway to the vacuole for degradation (Figure 2A; Lin et al., 2008; Teis et al., 2008). However, in ESCRT mutant cells, such as vps23Δ cells, Can1-GFP is not delivered to vacuoles and instead accumulates in abnormal endosomal compartments (class E compartments) and at the PM due to recycling from these aberrant endosomes (Teis et al., 2008). We reasoned that loss of factors involved in Can1 recycling should impair its return to the PM in ESCRT-mutant cells. Consistent with this idea, loss of Snx41 or Snx3 in ESCRT-mutant cells impaired Can1 recycling to the PM, as Can1-GFP localized only to intracellular puncta in the double-mutant cells (snx41Δ vps23Δ and snx3Δ vps23Δ cells; Figure 2B). Quantification of the fluorescence ratio of Can1-GFP at endosomes and the PM, FR, showed an increase of Can1-GFP accumulation at the class E compartments in snx41Δ vps23Δ and snx3Δ vps23Δ cells relative to vps23Δ cells (Figure 2C; see Materials and Methods). Of interest, deletion of the previously uncharacterized genes YBR246w or YPL183c in ESCRT-mutant (vps23Δ) cells greatly reduced Can1 recycling to the PM. YBR246w and YPL183c encode WD40 repeat domain–containing proteins that we named Ere1 and Ere2 (for endosomal recycling proteins; Figure 1D). Consistent with a role for Ere1 and Ere2 in recycling, Can1-GFP localized only to intracellular puncta in ere1Δ vps23Δ and ere2Δ vps23Δ double-mutant cells (Figure 2B). Accordingly, the fluorescence ratio of Can1-GFP at endosomes and the PM, FR, was increased in ere1Δ vps23Δ cells and ere2Δ vps23Δ cells relative to vps23Δ cells (Figure 2C).
FIGURE 2:
Snx4/41/42, the retromer, Ere1, and Ere2 are involved in the recycling of Can1. (A) Can1 localization in wild-type cells under steady conditions (left) and following treatment with cycloheximide (+ CHX, right). Wild-type cells expressing Can1-GFP were grown to mid-log phase, subjected to cycloheximide for 4 h to induce Can1 internalization, and analyzed by fluorescence microscopy. The plasma membrane (PM) and vacuoles are indicated. (B) The recycling of Can1 in ESCRT-mutant cells is mediated by Ere1, Ere2, Snx41, and the retromer complex. Mutant vps23Δ, snx41Δ vps23Δ, snx3Δ vps23Δ, ere1Δ vps23Δ, and ere2Δ vps23Δ cells expressing Can1-GFP were grown to mid-log phase, subjected to cycloheximide for 4 h to induce Can1 internalization, and analyzed by fluorescence microscopy. The PM and endosomes (E) are indicated. (C) The ratio of Can1-GFP fluorescence at endosomes and the PM in vps23Δ and ere1Δ vps23Δ, ere1Δ vps23Δ, snx41Δ vps23Δ, and snx3Δ vps23Δ double-mutant cells as described in Figure 1B; N = 25 for each cell type. See Materials and Methods for additional details. (D) Loss of Ere1 or Ere2 rescues the canavanine-hypersensitive phenotype in ESCRT-mutant cells. Serial dilutions of wild-type, snf7Δ, ere1Δ snf7Δ, ere2Δ snf7Δ, and snx41Δ snf7Δ cells were grown on synthetic media plates with or without 0.6 μg/ml canavanine for 2 d at 26°C.
Because we initially identified Ere1 and Ere2 in a screen for canavanine-resistant mutants (Figure 1, B and C, and Table 1), we reasoned that mutations that impair Can1 recycling would confer canavanine resistance in ESCRT-mutant cells, such as snf7Δ cells lacking ESCRT-III function. Consistent with this idea, deletion of SNX41 rescued the canavanine-sensitive phenotype of snf7Δ cells (snx41Δ snf7Δ double-mutant cells; Figure 2D; Teis et al., 2010). Of interest, we found that loss of Ere1 conferred a canavanine-resistant phenotype in snf7Δ-mutant cells (ere1Δ snf7Δ double-mutant cells; Figure 2D), consistent with a role for Ere1 in Can1 recycling. Similarly, loss of Ere2 conferred a canavanine-resistant phenotype in snf7Δ-mutant cells (ere2Δ snf7Δ double-mutant cells; Figure 2D).
As mentioned, Ere1 and Ere2 each contain WD40 repeat domains (four and nine WD40 repeats, respectively; Figure 1D). Both have predicted human orthologues (WD40 repeat–containing protein WRDR85, with 26% identity to Ere1; and WD40 repeat–containing protein WDR6, with 25% identity to Ere2; Figure 1D and unpublished data), suggesting that these proteins might have conserved functions. Because our initial results identified Ere1 and Ere2 as conserved factors involved in recycling, we chose these factors for further study.
The Ere1 and Ere2 proteins function in the retromer pathway
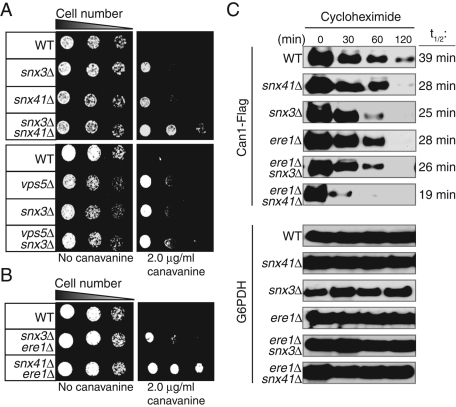
Previous reports demonstrated that the Snx4/41/42 and retromer complexes function in two separate pathways that mediate the recycling of different cargoes (such as Snc1 and Vps10, respectively) from endosomes. However, our initial data indicated that both the Snx4/41/42 and retromer complexes are involved in the recycling of the cell-surface protein Can1. So we next examined whether these two complexes function independently or together to recycle Can1. To establish whether the Snx4/41/42 and the retromer complexes function independently to recycle Can1, we created several double- and triple-deletion mutants of genes encoding the Snx4/41/42 and retromer complexes. The retromer complex consists of Vps5, Vps17, Vps35, Vps26, and Vps29. In addition, the Snx3 adaptor protein functions in the retromer pathway to mediate endosome-to-Golgi trafficking of the cell-surface iron transporter Ftr1 (Strochlic et al., 2007). We tested multiple combinations—vps5Δ snx3Δ, snx4Δ snx41Δ snx42Δ, snx4Δ vps35Δ, snx41Δ snx3Δ, snx4Δ snx3Δ, snx41Δ vps5Δ, and snx4Δ vps5Δ—to examine the consequence of loss of the retromer and Snx4/41/42 complexes. Consistent with previous findings that Snx3 and Vps5 function in the same pathway whereas Snx4, Snx41, and Snx42 function in another, vps5Δ snx3Δ double-mutant cells and snx4Δ snx41Δ snx42Δ triple-mutant cells did not display increased canavanine-resistant phenotypes as compared with their corresponding single-deletion mutants (Figure 3A and Table 1). However, snx41Δ snx3Δ, snx4Δ snx3Δ, and snx4Δ vps35Δ double-mutant cells show enhanced canavanine-resistant phenotypes due to loss of both the Snx4/41/42 and retromer pathways (Figure 3A and Table 1).
FIGURE 3:
The retromer and Snx4/41/42 pathways independently recycle Can1, and Ere1 functions in the retromer pathway. (A) Loss of both retromer and Snx41 function results in additive resistance to canavanine (top), whereas loss of multiple retromer subunits is not additive (bottom). (B) Loss of both Ere1 and Snx41 confers an additive canavanine resistance phenotype, but loss of both Ere1 and the retromer subunit Snx3 is not additive. (A, B) Serial dilutions of wild-type and various mutant cells (as indicated) were grown on synthetic media plates with or without 2 μg/ml canavanine for 2 d at 26°C. (C) Rates of Can1 degradation are increased in cells lacking Snx41, Snx3, or Ere1. Double deletion of ERE1 and SNX41 further increases the rate of Can1 turnover. Wild-type and mutant cells (as indicated) expressing FLAG-tagged Can1 were grown to mid-log phase and treated with cycloheximide to induce Can1 endocytosis. Cell lysates were prepared at the indicated time points and analyzed by immunoblotting to monitor Can1 degradation rates (top; t1/2 times are shown on the right). G6PDH was used as a protein loading control (bottom).
To check the pathway in which Ere1 and Ere2 function, we created ere1Δ snx3Δ, ere2Δ snx3Δ, ere1Δ vps5Δ, ere1Δ snx41Δ, ere2Δ snx41Δ, and ere1Δ snx4Δ double-mutant cells to test the functional relationships among Ere1, Ere2, the retromer, and the Snx4/41/42 proteins. The ere1Δ snx3Δ and ere1Δ vps5Δ double-mutant cells did not show increased canavanine resistance as compared with the corresponding ere1Δ, snx3Δ, and vps5Δ single-deletion mutants (Figure 3B and Table 1). Likewise, ere2Δ snx3Δ double-mutant cells showed a canavanine resistance phenotype similar to that of ere2Δ and snx3Δ single mutants (Table 1). In contrast, ere1Δ snx41Δ and ere1Δ snx4Δ double-mutant cells displayed increased canavanine resistance as compared with the single mutants (Figure 3B and Table 1). Likewise, ere2Δ snx41Δ double-mutant cells displayed increased canavanine resistance as compared with the single mutants (Table 1). These data indicated that Ere1 and Ere2 function in the retromer pathway for the recycling of Can1. We also determined whether the Ere1 and Ere2 proteins function together. Of interest, ere1Δ ere2Δ double-mutant cells did not display increased canavanine-resistant phenotypes as compared with ere1Δ and ere2Δ single-mutant cells (Table 1), further suggesting that these proteins act in common in the retromer-mediated pathway.
In wild-type cells, internalized Can1 can be either delivered to the vacuole for degradation or recycled back to the PM. Thus impaired recycling of Can1 would lead to increased rates of degradation. So we measured the lifetimes of Can1 in wild-type and various recycling-deficient cells under conditions that stimulate Can1 internalization. Consistent with our previous results, the degradation of Can1-FLAG in snx41Δ, snx3Δ, and ere1Δ mutant cells is slightly increased as compared with wild-type rates (Figure 3C). In addition, the lifetime of Can1 in snx3Δ ere1Δ double-deletion-mutant cells is similar to that of ere1Δ and snx3Δ single-mutant cells, consistent with the model that Ere1 and Snx3 function in a common pathway. In contrast, the degradation of Can1-FLAG in snx41Δ ere1Δ double-mutant cells is significantly increased as compared with ere1Δ and snx41Δ single-mutant cells (Figure 3C). Taken together, these data further suggest that the Ere-mediated retromer and Snx4/41/42 pathways function independently to recycle Can1.
Ere1 is necessary for recycling of specific cargoes via the retromer pathway
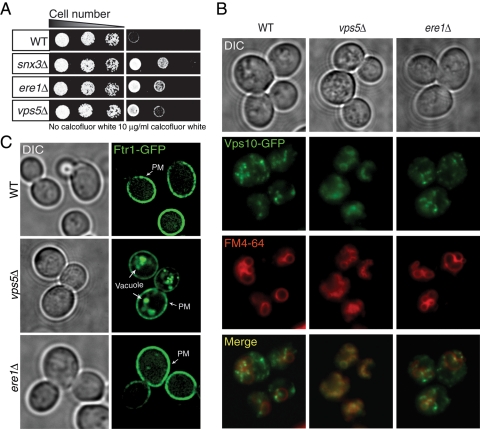
The retromer complex mediates the retrograde transport of many cargoes, such as the vacuolar hydrolase sorting receptor Vps10 (Seaman et al., 1998) and the iron transporter proteins Fet3 and Ftr1 (Strochlic et al., 2007). Because Ere1 functions with the retromer in the recycling of Can1, we further examined roles for Ere1 in recycling other cargoes. First, we examined additional plasma membrane proteins that undergo recycling to the cell surface following their internalization. Similar to the canavanine resistance phenotype, ere1Δ mutant cells show increased growth on media containing calcofluor white (Figure 4A). Calcofluor white binds chitin in the cell wall, and previous work showed that cells with reduced cell-surface expression levels of a chitin synthase (Chs3) display resistance to calcofluor white (Valdivia et al., 2002). Consistent with a role for recycling in chitin synthase cell-surface targeting, we also found that loss of the retromer subunits Snx3 and Vps5 resulted in calcofluor white–resistant phenotypes (Figure 4A). In addition, similar to the Can1-recycling defects in ere1Δ-mutant cells, the methionine transporter Mup1 was not efficiently recycled to the PM in ere1Δ snf7Δ double-mutant cells (Supplemental Figure S1A). Accordingly, the fluorescence ratio of Mup1-GFP at endosomes and the PM, FR, was increased in ere1Δ snf7Δ cells relative to snf7Δ cells (Supplemental Figure S1A). As a control, we also found that Mup1 recycling was reduced in snx41Δ snf7Δ double-mutant cells (Supplemental Figure S1A; Teis et al., 2010), consistent with our previous results indicating two distinct pathways for recycling Can1. In contrast, trafficking of the recycling SNARE protein Snc1 was unaffected in ere1Δ-mutant cells, as Snc1 localized normally to the PM and Golgi compartments in both wild-type and ere1Δ-mutant cells (Supplemental Figure S1B).
FIGURE 4:
Ere1 is not required for the transport of the retromer cargoes Vps10 and Ftr1. (A) Serial dilutions (fourfold) of wild-type, snx3Δ, ere1Δ, and vps5Δ cells were grown on synthetic media plates with or without 10 μg/ml calcofluor white for 2 d at 26°C. (B) Localization of Vps10-GFP in wild-type, vps5Δ, and ere1Δ cells. Cells were grown in yeast extract/peptone/dextrose (YPD), and vacuoles were labeled with FM4-64. Note: vps5Δ deletion-mutant cells show a fragmented-vacuole phenotype. (C) Localization of Ftr1-GFP in wild-type, vps5Δ, and ere1Δ mutant cells in the absence of exogenous iron.
Next we examined possible roles for Ere1 in trafficking between the Golgi and endosomes. In wild-type cells, the sorting receptor Vps10 traffics between the Golgi and endosomes. In cells with impaired retromer function, such as cells lacking Vps5, Vps10 is not recycled from endosomes and instead is delivered to the vacuole. As a result, cargoes normally destined for the vacuole, such as carboxypeptidase Y (CPY), are instead secreted (Marcusson et al., 1994). Although ere1Δ-mutant cells were identified in a genome-wide screen for mutants that secrete CPY (Bonangelino et al., 2002), we did not observe a strong defect in the transport of CPY to vacuoles by following CPY processing in the vacuole in pulse-chase labeling experiments (unpublished data). Consistent with this, the localization of Vps10-GFP was similar in wild-type and ere1Δ-mutant cells (Figure 4B). As a control, Vps10-GFP accumulated in fragmented vacuoles labeled with the marker FM4-64 in cells lacking Vps5 (Figure 4B). Thus Ere1 is not required for the efficient recycling of Vps10 from endosomes to the Golgi.
In iron-starved cells, Ftr1 is sorted at endosomes by the retromer complex to direct its recycling back to the PM (Strochlic et al., 2007). In cells lacking retromer function, such as vps5Δ-mutant cells, the recycling of internalized Ftr1 is impaired and Ftr1–GFP fusion proteins accumulate in the vacuole, where they are degraded (Figure 4C). In contrast, Ftr1–GFP is stably localized to the PM in both wild-type and ere1Δ-mutant cells (Figure 4C), suggesting that Ere1 provides cargo-specific functions in the retromer pathway.
Ere1 and Ere2 are cytoplasmic proteins that are recruited to endosomes
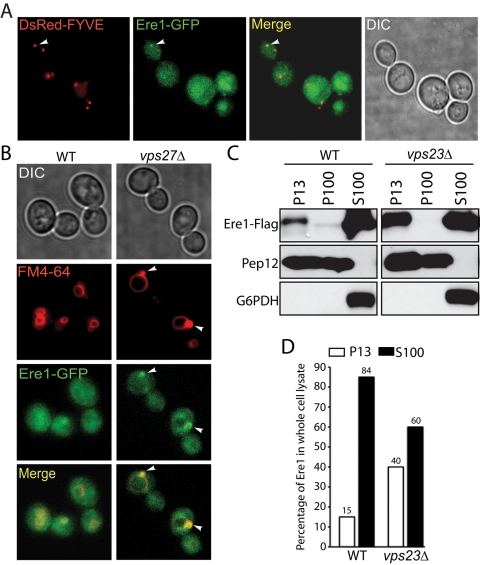
To examine Ere1 localization in vivo, we generated a functional Ere1-GFP fusion, as cells expressing Ere1-GFP alone do not exhibit a canavanine-resistant phenotype (unpublished data). In wild-type cells, Ere1-GFP was mostly cytoplasmic (Figure 5, A and B). However, in a small fraction of wild-type cells (∼10%), Ere1-GFP localized to intracellular punctate structures that colocalized with the PI3P-binding endosomal marker DsRed-FYVE (Figure 5A), suggesting that Ere1-GFP may be transiently recruited to endosomes. The endosomal localization of Ere1-GFP was more apparent in ESCRT-mutant cells (vps27Δ) that accumulate abnormal endosomal compartments (class E compartments) adjacent to vacuoles (Figure 5B). Consistent with the microscopy results, subcellular fractionation assays indicated that ∼15% of FLAG-tagged Ere1 was present in the membrane pellet fraction (P13) in wild-type cells (Figure 5C). Moreover, membrane association of Ere1 was increased more than twofold in ESCRT-mutant cells (40% of the total was present in the P13 fraction from vps23Δ cells; Figure 5, C and D). In addition, the endosomal localization of Ere1 was dependent on phosphatidylinositol 3-phosphate or its downstream effectors, as membrane association of Ere1 was not detected in lysates prepared from cells with impaired Vps34 phosphatidylinositol 3-kinase activity (Supplemental Figure S1C). Next we examined the localization and fractionation of a functional Ere2-GFP fusion in wild-type and ESCRT-mutant cells. Similar to Ere1-GFP, we found that endosomal localization and membrane association of Ere2-GFP were stabilized in ESCRT-mutant cells (Supplemental Figure S2). Taken together, these results suggested that both Ere1 and Ere2 are recruited to endosomes as needed, since the Ere proteins accumulate on endosomes in ESCRT-mutant cells in which increased recycling of internalized PM proteins occurs (Figure 2B; Teis et al., 2008).
FIGURE 5:
Ere1 localizes to endosomal compartments. (A) Representative fluorescence microscopy images showing wild-type cells coexpressing Ere1-GFP and the endosomal marker DsRed-FYVE. Ere1-GFP localized to intracellular puncta (corresponding to endosomes with arrow) in ∼10% of wild-type cells. (B) Ere1 is stabilized on endosomal compartments in ESCRT -mutant cells. Wild-type and vps27Δ mutant cells expressing Ere1-GFP were grown in YPD medium at 30°C. Vacuoles and E compartments (in vps27Δ cells) were labeled with FM4-64. (C) Subcellular fractionation of Ere1-FLAG in lysates from wild-type and vps23Δ mutant cells was analyzed by differential centrifugation and subsequent SDS–PAGE and immunoblotting. Pep12 was used as a marker for membrane fractions (P13 and P100), whereas glucose-6-phosphate dehydrogenase was used as a cytosolic marker (S100). (D) Quantification of Ere1-FLAG in P13 and S100 fractions from wild-type and vps23Δ cell lysates.
Ere1 interacts with the cargo protein Can1 and colocalizes with Vps5
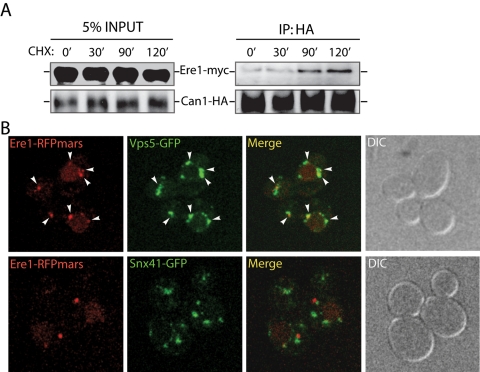
Our initial results suggested that Ere1 provides a cargo-specific function in retromer-mediated recycling from endosomes. To test whether Ere1 physically interacts with cargo proteins, such as Can1, we performed cross-linking experiments using lysates from ESCRT-mutant cells exposed to conditions that induce Can1 internalization. Notably, increasing amounts of Ere1-myc were cross-linked and coimmunopurified with Can1-hemagglutinin (HA) upon longer incubations with cycloheximide, which induces Can1 internalization (Figure 6A, 0- vs. 120-min time points). Next we examined whether Ere1 and the retromer subunit Vps5 colocalize on endosomes. Ere1-RFPmars colocalized with Vps5-GFP but not Snx41-GFP in ESCRT-mutant cells in which Ere1 is stabilized on membranes (Figure 6B). We did observe puncta with Vps5-GFP that lacked Ere1-RFPmars. However, all of the Ere1-RFPmars structures overlapped with Vps5-GFP. Thus these results further supported our model that Ere1 provides cargo-specific functions in the retromer-sorting pathway at endosomes.
FIGURE 6:
Ere1 associates with an internalized plasma membrane protein, Can1, and colocalizes with Vps5. (A) ESCRT-mutant (vps23Δ) cells coexpressing an integrated Ere1-13xmyc fusion and Can1-3xHA (from its own promoter on a centromeric plasmid) were treated with cycloheximide for the indicated times and then cross-linked with DSP and DTBP. Can1-3xHA was immunoprecipitated with antibodies specific for HA from solubilized cell lysates. Immunoprecipitates were subsequently analyzed by Western blotting using antibodies against the myc or HA epitopes to detect Ere1 and Can1, respectively. (B) Ere1 and Vps5 colocalize on endosomal compartments. ESCRT-mutant (vps23Δ) cells coexpressing Ere1-RFPmars and Vps5-GFP (top) or coexpressing Ere1-RFPmars and Snx41-GFP (bottom) were examined by confocal microscopy.
The WD40-repeat proteins Ere1 and Ere2 assemble into protein complexes on endosomes
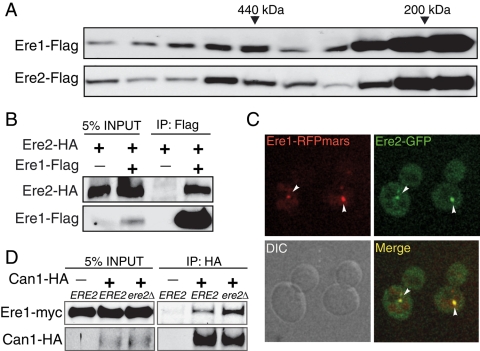
Our initial genetic results indicated that Ere1 and Ere2 function together in the retromer pathway. We next performed biochemical and microscopy experiments to address whether these proteins interact. Analysis of cytosolic fractions from wild-type cells by fast-performance liquid chromatography size-exclusion chromatography suggested that Ere1 and Ere2 do not form a stable complex in the cytoplasm (unpublished data). However, analysis of solubilized membrane fractions from wild-type cells on glycerol velocity sizing gradients indicated that Ere1 and Ere2 are both present in two distinct protein complexes with sizes of ∼200 and 500 kDa, respectively (Figure 7A). To further investigate whether Ere1 and Ere2 interact in vivo, we performed coimmunoprecipitation and microscopy experiments using cells expressing differentially tagged forms of Ere1 and Ere2. First, Ere2-3xHA was specifically coimmunoprecipitated with Ere1-FLAG (Figure 7B). Second, Ere1-RFPmars and Ere2-GFP colocalized to endosomal compartments in ESCRT-mutant cells (Figure 7C). Because these results indicated that Ere1 and Ere2 associate, we then tested a potential role for Ere2 in regulating Ere1–cargo interactions. For this, we monitored Ere1 cross-linking to internalized Can1 in ESCRT-mutant cells either expressing or lacking Ere2 (Figure 7D). Surprisingly, Ere2 was not required for Ere1-13xmyc to interact with the cargo Can1-3xHA; instead, Ere1 cargo binding was increased (approximately twofold) in cells lacking Ere2 (Figure 7D). Thus Ere2 appears to regulate Ere1 function in cargo-specific recycling in the retromer-mediated sorting pathway.
FIGURE 7:
Ere1 and Ere2 physically interact and colocalize on endosomal compartments. (A) Glycerol velocity gradient sizing analysis of Ere1-FLAG and Ere2-FLAG in membrane fractions (P13 and P100) from wild-type cells. Major peak fractions for known molecular weight standards are indicated. The predicted molecular weight of monomeric Ere1 is ∼46 kDa and that of monomeric Ere2 is ∼114 kDa. (B) Ere2-3xHA specifically coimmunoprecipitates with Ere1-FLAG. Wild-type cells expressing an integrated Ere2-3xHA fusion or coexpressing integrated Ere1-FLAG and Ere2-3xHA fusions were grown to mid-log. Ere1-FLAG was immunoprecipitated from solubilized cell lysates, and the resulting immunopurified material was analyzed by Western blotting using antisera against the FLAG and HA epitopes to detect Ere1 and Ere2, respectively. (C) Ere1 and Ere2 colocalize on endosomal compartments. ESCRT-mutant (vps23Δ) cells coexpressing Ere1-RFPmars and Vps5-GFP were examined by confocal microscopy. (D) Interaction between Ere1 and Can1 is increased in cells lacking Ere2. Cross-linkers were added to vps23Δ mutant and vps23Δ ere2Δ double-mutant cells expressing Ere1-13xmyc alone or coexpressing both Ere1-13xmyc and Can1-3xHA following treatment with cycloheximide for 1.5 h. Can1-3xHA was immunoprecipitated from solubilized cell lysates and analyzed by Western blotting to detect Ere1 and Can1 in the input and immunoprecipitate fractions.
Discussion
The recycling of internalized PM proteins is essential for proper cell growth and development. We identified new components in the pathways that drive cell-surface protein recycling. We used the yeast arginine transporter Can1 as a model cell-surface protein and found that two pathways independently facilitate its recycling. One pathway requires the Snx4/41/42 complex, and the other pathway requires the retromer complex. Thus our study highlights the need for multiple pathways that mediate recycling from endosomes. Moreover, we found two novel proteins, Ere1 and Ere2, that function in the retromer-mediated recycling pathway and provide cargo specificity within this pathway. Thus, even while major pathways for recycling from endosomes (e.g., the Snx4/41/42 and retromer complexes) have been described, our findings suggest that additional proteins or complexes regulate cargo specificity for these pathways.
Multiple pathways guide the efficient recycling of PM proteins
Previous reports demonstrated that the Snx4/41/42 and retromer complexes mediate distinct recycling pathways from different classes of endosomes to the TGN (Bonifacino and Rojas, 2006). The Snx4/41/42 complex sorts proteins out of early endosomes, whereas the retromer functions on late endosomes (Hettema et al., 2003). Our genetic results further support these previous studies, as cells impaired in both the Snx4/41/42 and retromer pathways displayed additive phenotypes in the canavanine resistance assay and increased rates of Can1 degradation (Figure 3 and Table 1). In addition, our data suggest that both recycling pathways can recognize common cargo proteins (e.g., Can1). This redundancy in pathways may ensure the efficient recycling of certain internalized cell-surface proteins. Although our results suggest that Ere1 may facilitate Can1 recognition by the retromer, it remains unclear how the Snx4/41/42 complex recognizes Can1. Our findings indicate that Ere1 does not function in the Snx4/41/42 pathway, as 1) double deletion of ERE1 and SNX41 results in an additive canavanine-resistant phenotype, and 2) the recycling of the Snc1 SNARE occurs normally in cells lacking Ere1 (Supplemental Figure S1). Possibly, additional unknown factors related to Ere1 provide this function.
Ere1 and Ere2 function in cargo-specific recycling from endosomes
In addition to known recycling factors, we identified two novel WD40 repeat–containing proteins, Ere1 and Ere2, in our screen. Multiple lines of evidence indicate that both Ere1 and Ere2 function in recycling from endosomes. First, we observed impaired recycling of Can1 in ESCRT-mutant cells lacking Ere1 or Ere2 (Figure 2). Second, cells lacking Ere1 showed increased turnover rates of Can1 (Figure 3). Third, an additional PM cargo protein, the methionine transporter Mup1, showed impaired recycling in ESCRT-mutant cells lacking Ere1 (Supplemental Figure S1). Finally, both Ere1 and Ere2 associate with membranes and are stabilized on endosomal compartments in ESCRT mutant cells (Figure 5 and Supplemental Figure S2). Both the accumulation of PM cargo proteins at abnormal endosomes and increased recycling of internalized PM proteins from endosomal compartments in ESCRT-mutant cells may result in the stabilization of Ere1 and Ere2 at endosomes in these mutant cells. Consistent with this idea, we found Ere1 in a complex with internalized Can1 in ESCRT-mutant cells (Figure 6).
Our study further demonstrated that Ere1 provides cargo-specific functions in the retromer pathway. First, as mentioned, Ere1 physically interacted with Can1 under conditions that stimulate Can1 endocytosis (Figure 6). Future studies are needed to address whether Ere1 directly binds Can1 or requires the retromer complex for binding. Of interest, although Ere1 was also necessary for the recycling of an additional PM protein, Mup1, it was not required for the recycling of another PM cargo protein, Ftr1 (Supplemental Figures S1 and S4). Consistent with a cargo-specific role, Ere1 also was not required for the transport of another retromer cargo, the sorting receptor Vps10, that traffics between the Golgi and endosomes (Figure 4).
In addition to the Snx4/41/42 complex and retromer, clathrin and the adaptor complex AP-1 have been implicated in the sorting of proteins between the Golgi and endosomes (Bonifacino and Rojas, 2006). Previous work suggested that clathrin and the retromer might act sequentially to mediate endosome-to-TGN trafficking of the Shiga toxin receptor in mammalian cells (Johannes and Romer, 2010). Future studies will be needed to investigate the potential role of clathrin and its adaptor proteins (such as AP-1) in Can1 recycling and in the function of the Ere1 and Ere2 proteins. However, we found that cells lacking AP-1 function did not display a canavanine-resistant phenotype (apl2Δ mutant cells; Table 1).
We found that two independent pathways mediate the recycling of the cell-surface protein Can1. One pathway requires the Snx4/41/42 complex, which is well conserved from yeast to human cells. Human homologues of Snx4/41/42 are known: SNX4, SNX7, and SNX30, respectively (Leprince et al., 2003; Skanland et al., 2007; Traer et al., 2007). Likewise, the Snx3 and retromer-mediated pathway also is well conserved, as human orthologues of Snx3 and the retromer complex have been identified. For example, previous work implicated human SNX3 and SNX1 (the human orthologue of yeast Vps5) in the proper trafficking of internalized cell-surface proteins (Xu et al., 2001; Worby and Dixon, 2002). It is thus not surprising that the yeast Ere1 and Ere2 proteins are also conserved in mammalian cells (Figure 2). However, functions for human WDR85 (the Ere1 orthologue) and WDR6 (the Ere2 orthologue) are largely unknown (see later discussion), and future work is needed to test whether these proteins are involved in cell-surface protein recycling.
A previous study suggested that the human Ere1 orthologue, WDR85, functions as an adaptor molecule for a protein modification complex (the Dph1–4 proteins) necessary for the conversion of a histidine residue on EF2 into diphthamide for ADP-ribosylation by diphtheria toxin (Carette et al., 2009). This study also indicated that Ere1 is required for ADP-ribosylation of EF2 in yeast lysates, suggesting that Ere1 might also be involved in diphthamide biosynthesis (Carette et al., 2009). However, we found that diphthamide biosynthesis is not necessary for proper recycling of Can1, as cells lacking the protein Dph2 were not resistant to canavanine (unpublished data). However, Ere1 may still be involved in other uncharacterized protein modifications that influence recycling from endosomes. In addition, ERE1 was identified in a previous genetic screen for factors that modulate ribosomal DNA transcription (and named RRT2 in this study; Hontz et al., 2009). Likewise, ERE2 was identified in a genetic screen for factors implicated in retrotransposon transposition (and named RTT10 in this study; Nyswaner et al., 2008). However, both of these genetic screens identified known factors involved in membrane trafficking pathways, suggesting roles for vesicle transport in regulating these processes. Thus, although Ere1 and Ere2 might have additional targets or roles, we propose that Ere1 and Ere2 function as regulators of recycling from endosomes. Our results showed that Ere1 localizes to endosomes, colocalizes with the retromer subunit Vps5, and interacts with the cargo protein Can1. In addition, ERE1 was identified in a previous genome-wide screen for factors involved in endosomal and vacuolar sorting (Bonangelino et al., 2002).
Mechanisms for Ere1-regulated recycling
Our study suggests that the WD40-repeat proteins Ere1 and Ere2 are regulators of the retromer complex. Several previous studies showed that additional coat complexes, such as COP-I, COP-II, clathrin (Lee and Goldberg, 2010), and the recently discovered BBsome complex (Jin et al., 2010), also consist of WD40-repeat or β-propeller domain–containing proteins. These β-propeller–containing proteins share similar functions in cargo selection and sorting. It is thus interesting that Ere1 cross-linked to the cargo protein Can1. Ere1 is a protein with four predicted WD40 repeats, whereas Ere2 is predicted to contain nine WD40 repeats. WD40 repeats usually assemble to form a β-propeller structure with seven or eight blades. Proteins with fewer than seven WD40 repeats, such as Ere1, may need to homo-oligomerize or assemble with other WD40-repeat proteins to form a complete β-propeller (Xu and Min, 2011). Our study demonstrated that Ere1 and Ere2 exist in protein complexes on endosomal membranes by velocity sizing gradients and coimmunoprecipitation experiments (Figure 7). We suggest that Ere1 and Ere2 are recruited to endosomal membranes, where they form a complex required for cargo-specific recycling in the retromer pathway. Future experiments are needed to determine the stoichiometry of Ere1 and Ere2 complexes, as well as to address whether additional factors associate with the Ere1/Ere2 complexes on endosomes.
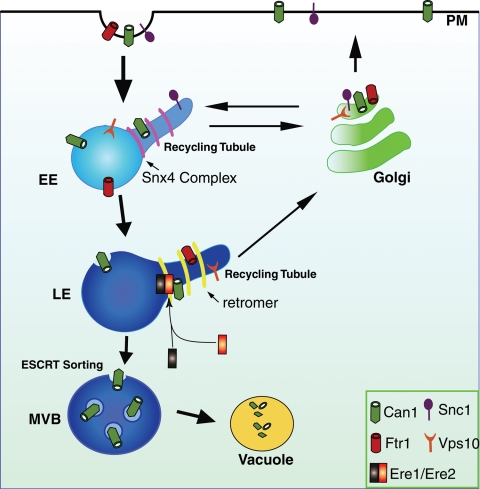
We propose that the Ere1 and Ere2 proteins function in the retromer pathway for the selection of specific cargoes for sorting into recycling tubules at endosomes (Figure 8). Recent studies indicated that separate recycling tubules enriched in distinct cargo proteins (the transferrin receptor and β2-adrenergic receptor) can be generated from a common endosomal compartment (Puthenveedu et al., 2010) and that the SNX27 protein is required for sorting β2-adrenergic receptors into retromer-mediated recycling tubules (Temkin et al., 2011). However, it is not fully known how cargo proteins are initially selected and sorted into distinct tubules that emanate from endosomes. Cargo-specific factors, such as the Ere1 and Ere2 proteins, may act at an early step in cargo recognition and thereby direct selectivity that drives further sorting of cargoes into distinct recycling tubules (Figure 8). Alternatively, Ere1 and Ere2 may act at a later step to stabilize cargo proteins in specific domains following initial selection and sorting by the retromer.
FIGURE 8:
Model for Ere1 and Ere2 cargo-specific function in retromer-mediated recycling from endosomes. After stimulation, Can1 is internalized from the plasma membrane (PM) and delivered to early endosomes (EE). On early endosomes, a pool of Can1 is selected and sorted by the Snx4/41/42 complex for recycling to the Golgi and subsequent delivery back to the cell surface. Additional cargoes, such as the SNARE protein Snc1, are recycled from early endosomes in a retromer- and Ere1/Ere2–independent process. The remaining fraction of Can1 that is not recycled by the Snx4/41/42 complex is transported to late endosomes (LE). At late endosomes, Can1 is subject to different fates. First, Can1 is recognized by the Ere1/Ere2 complex for sorting into the retromer-mediated recycling pathway (see Discussion). The Ere1/Ere2 complex is not required for the sorting of additional retromer cargoes, such as Ftr1 and Vps10, into recycling tubules generated at late endosomes. Internalized Can1 that is not recycled by the Snx4/41/42 or retromer pathways is sorted into the ESCRT-mediated MVB pathway and eventually delivered to the vacuole for degradation.
Previous studies found that the retromer subunit Vps35 binds a conserved motif, (F/W)L(M/V), that is found in the cation-independent mannose 6-phosphate receptor and sortilin sorting receptor in mammalian cells (McGough and Cullen, 2010). However, it is unclear how the retromer recognizes and sorts cargo proteins that lack this motif. In mammalian cells, internalized GPCRs that lack the (F/W)L(M/V) motif are believed to be recognized by the retromer subunit Vps26, which contains an arrestin-like domain (Shi et al., 2006; McGough and Cullen, 2010). In yeast, both the arginine transporter Can1 and the iron transporter Ftr1 lack the (F/W)L(M/V) sorting motif but are recycled by the retromer. In the case of Ftr1, the retromer subunit Snx3 recognizes a sorting signal in the cytoplasmic tail of Ftr1, and thus Snx3 acts as an Ftr1 adaptor for the retromer. Our study showed that Ere1 is not required for Ftr1 sorting (Figure 4), but Ere1 function was necessary for the recycling of additional cargoes (e.g., Can1 and Mup1) by Snx3 and the retromer, and thus Ere1 could modulate Snx3 function as a cargo adaptor. Future studies will likely uncover mechanisms for the cargo-specific functions of Ere1 and Ere2 in the retromer pathway. In addition, it will be interesting to examine whether the mammalian WDR85 and WDR6 proteins also are involved in PM protein recycling from endosomes.
MATERIALS AND METHODS
Yeast strains and plasmid construction
Strains used for the canavanine resistance screen were obtained from the single-gene-deletion libraries constructed by the International Deletion Consortium (Winzeler et al., 1999) constructed in BY4741 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0; Brachmann et al., 1998). Lists of additional Saccharomyces cerevisiae strains and plasmids used in this study are shown in Tables 2 and 3. Gene deletions and tagging in yeast were performed by homologous recombination (Longtine et al., 1998). Standard techniques were used for yeast growth.
TABLE 2:
Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| SEY6210 | MATα leu2-3112 ura3-52 his3Δ200 trp1-Δ901 lys2-801 suc2Δ9 | Robinson et al. (1988) |
| SEY6210.1 | MATa leu2-3112 ura3-52 his3Δ200 trp1-Δ901 lys2-801 suc2Δ9 | Robinson et al. (1988) |
| YSY15 | SEY6210 snx3Δ::TRP1 | This study |
| BHY152 | SEY6210 vps5Δ::HIS3 | Seaman et al. (1998) |
| SRY149 | SEY6210.1 snx41Δ::TRP | This study |
| SRY34 | SEY6210 snx4Δ::TRP1 | This study |
| YSY12 | SEY6210 ere1Δ::TRP1 | This study |
| MBY22 | SEY6210.1 vps23Δ::HIS3 | Katzmann et al. (2003) |
| YSY21 | SEY6210 vps23Δ::HIS3 ere1Δ::TRP1 | This study |
| YSY21 | SEY6210 vps23Δ::HIS3 snx41Δ::TRP1 | This study |
| YSY83 | SEY6210 vps23Δ::HIS3 snx3Δ::TRP1 | This study |
| EEY9 | SEY6210 snf7Δ::HIS3 | Teis et al. (2008) |
| YSY163 | SEY6210 snf7Δ::HIS3 ere1Δ::TRP1 | This study |
| YSY105 | SEY6210 snf7Δ::HIS3 snx41Δ::TRP1 | This study |
| YSY138 | SEY6210 vps5Δ::HIS3 snx3Δ::TRP1 | This study |
| YSY76 | SEY6210 snx41Δ::HIS3 snx3Δ::TRP1 | This study |
| YSY128 | SEY6210 ere1Δ::HIS3 snx3Δ::TRP1 | This study |
| YSY19 | SEY6210 ere1Δ::HIS3 snx41Δ::TRP1 | This study |
| PBY34 | SEY6210 VPS10-GFP::TRP1 | Burda et al. (2002) |
| PBY126 | SEY6210 VPS10-GFP::TRP1 vps5Δ::HIS3 | Burda et al. (2002) |
| YSY211 | SEY6210.1 VPS10-GFP::TRP1 ere1Δ::TRP1 | This study |
| YSY90 | SEY6210 ERE1-GFP::TRP1 | This study |
| YSY74 | SEY6210 ERE1-GFP::TRP1 vps27Δ::HIS3 | This study |
| YSY120 | SEY6210.1 ERE1-Flag::HIS3MX | This study |
| YSY227 | SEY6210 ERE1-Flag::HIS3MX vps23Δ::TRP1 | This study |
| YSY273 | SEY6210 ERE1-13xmyc::TRP1 vps23Δ::HIS3 | This study |
| YSY293 | SEY6210 ERE1-RFPmars::TRP1 vps23Δ::HIS3 VPS5-GFP::TRP1 | This study |
| YSY284 | SEY6210 ERE1-13xmyc::TRP1 vps23Δ::HIS3MX VPS5-HA::TRP1 | This study |
| YSY101 | SEY6210 ERE2-Flag::HIS3MX | This study |
| YSY294 | SEY6210 ERE1-Flag::HIS3MX ere2Δ::TRP1 | This study |
| YSY156 | SEY6210 ERE1-Flag::HIS3MX ERE2-HA::TRP1 | This study |
| YSY285 | SEY6210 ERE1-RFPmars::TRP1 vps23Δ::HIS3 ERE2-GFP::TRP1 | This study |
| YSY282 | SEY6210 ERE1-13xyc::TRP1 vps23Δ::HIS3 ere2Δ::TRP1 | This study |
| YSY87 | SEY6210 vps23Δ::HIS3 ere2Δ::TRP1 | This study |
| YSY271 | SEY6210 snf7Δ::HIS3 ere2Δ::HIS3 | This study |
| SRY442 | SEY6210.1 snx42Δ::TRP1 | This study |
| YSY14 | vps26Δ::LEU2 | This study |
| PSY1-29 | SEY6210 vps29Δ::HIS3 | Seaman et al. (1998) |
| MSY0435 | SEY6210 vps35Δ::HIS3 | Horazdovsky et al. (1997) |
| vps17Δ | SEY6210 vps17Δ::HIS3 | Emr lab collection |
| YSY32 | SEY6210.1 ere2Δ::TRP1 | This study |
| SRY213 | SEY6210 snx4Δ::HIS3 snx41Δ::HIS3 snx42Δ::HIS3 | This study |
| YSY99 | SEY6210 vps5Δ::HIS3 ere1Δ::TRP1 | This study |
| YSY16 | SEY6210 snx3Δ::TRP1 ere2Δ::TRP1 | This study |
| YSY45 | SEY6210 ere1Δ::HIS3 ere2Δ::TRP1 | This study |
| BPY182 | SEY6210 snx3Δ::TRP1 snx41Δ::HIS3 | Emr lab collection |
| BPY152 | SEY6210 snx3Δ::TRP1 snx4Δ::HIS3 | Emr lab collection |
| BPY133 | SEY6210 snx4Δ::HIS3 vps35Δ::HIS3 | Emr lab collection |
| YSY6 | SEY6210 snx41Δ::TRP1 ere2Δ::TRP1 | This study |
| YSYS19 | SEY6210 snx41Δ::TRP1 ere1Δ::HIS3 | This study |
| YSY91 | SEY6210 snx4Δ::TRP1 ere1Δ::HIS3MX | This study |
| YSY163 | SEY6210 snf7Δ::HIS3 ere1Δ::TRP1 | This study |
| YSY105 | SEY6210 snf7Δ::HIS3 snx41Δ::TRP1 | This study |
| YSY295 | SEY6210 GFP-SNC1::URA3 | This study |
| YSY296 | SEY6210 snx41Δ::HIS3 GFP-SNC1::URA3 | This study |
| YSY297 | SEY6210 ere1Δ::TRP1 GFP-SNC1::URA3 | This study |
| YSY298 | SEY6210 ERE1-RFPmars::TRP1 vps23Δ::HIS3 SNX41GFP::TRP1 | This study |
| vps34tsf | SEY6210 vps34Δ::TRP1 pRS416.vps34tsf | Stack et al. (1995) |
TABLE 3:
Plasmids used in this study.
| Plasmid | Description | Source |
|---|---|---|
| pCHL571 | pRS416-Can1-GFP | Lin et al. (2008) |
| pCHL642 | pRS416-Mup1-GFP | Lin et al. (2008) |
| pCHL654 | pRS416-Ftr1-GFP | Lin et al. (2008) |
| pGO45 | pRS416-GFP-CPS | Katzmann et al. (2003) |
| pRS416-DsRed-FYVE | Katzmann et al. (2003) | |
| pRS416-Can1-3xHA | Lin et al. (2008) | |
| pCHL582 | pRS416-Can1-Flag | Lin et al. (2008) |
| pYS001 | pRS415-Ere1-HA | This study |
Genetic screen for canavanine-resistant mutants
We first determined the concentration of canavanine (5 μg/ml) that impaired the growth of wild-type cells (BY4741), using replication techniques for the screens. Following these control conditions, the collection of the ∼4700 viable yeast deletion mutants was replicated onto media plates containing no canavanine or plates containing 5 μg/ml canavanine. Plates were scored for relative growth of the deletion-mutant strains compared with wild-type cells after 2 and 4 d of incubation. The screens were performed three times, and we required that a deletion-mutant strain scored as resistant at least twice to be included in the final list of mutants identified in the screen. Using this approach, we identified 173 cvr mutant strains that displayed a canavanine-resistant phenotype (Figure 1B; see Supplemental Table for the entire list).
Fluorescence microscopy
Living cells expressing fluorescent fusion proteins were grown in minimal media to an A600 of 0.4–0.6. Fluorescence microscopy was performed at room temperature in synthetic media. Images in Figures 2, 4, and 5 and Supplemental Figures S1 and S2 were captured by a microscopy system (DeltaVision RT; Applied Precision, Issaquah, WA) equipped with a microscope (IX7; Olympus, Center Valley, PA). Data were deconvolved using Softworx (DeltaVision RT). Images for Figures 6 and 7 were collected with a spinning disk confocal microscopy system (3I, Yokogawa, Tokyo, Japan), using a microscope (DMI3000B) equipped with a digital camera (QuantEM; Photometrics, Tucson, AZ). For Figure 2C and Supplemental Figure S1A, fluorescence intensity (IF) was measured for 25 cells, according to a previous study (Teis et al., 2008). Briefly, IF for five different regions on the PM and endosomes (E compartments) in each cell were measured. Quantification of IF on the E compartment and at the PM allowed the calculation of a florescence ratio (FR; IF at the E compartment divided by IF at the PM) as a measure of Can1-GFP subcellular distribution. A high FR represents predominant localization of Can1-GFP to endosomes and E compartments. For the FM4-64 labeling assays, yeast cells were labeled with FM4-64 for 10 min, washed twice, and incubated for 45 min before imaging (Vida and Emr, 1995).
Subcellular fractionation
Twenty A600 equivalents of yeast cells were spheroplasted and lysed in 4.3 ml of ice-cold lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.2, 50 mM potassium acetate, 1 mM EDTA, and 200 mM sorbitol) containing 0.1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 1 μM pepstatin A, and protease inhibitor cocktail (cOmplete EDTA-free; Roche, Indianapolis, IN). One-milliliter aliquots of lysates were precleared for 5 min at 500 × g and were then centrifuged at 13,000 × g for 10 min at 4°C to generate pellets (P13) and supernatant fractions (S13). The 1-ml S13 fraction was centrifuged at 100,000 × g for 1 h at 4°C in an ultracentrifuge to generate the P100 and S100 fractions. Proteins were trichloroacetic acid (TCA) precipitated and analyzed by SDS–PAGE and immunoblotting.
Cross-linking of Ere1-13xmyc to Can1-3xHA
Forty A600 equivalents of yeast cells were spheroplasted, treated with 50 mg/ml cycloheximide, and incubated with 25 μM dithiobis(succinimidylpropinoate) (DSP) and dimethyl 3,3′-dithiobispropiomnimidate (DTBP) for 30 min. Complexes were immunoisolated, reduced to cleave cross-linkers, and analyzed by SDS–PAGE and immunoblotting.
Velocity sedimentation on 10–40% glycerol gradients
Membrane fractions of 100 A600 equivalents of the yeast cells were generated as described earlier and solubilized in lysis buffer (20 mM HEPES, pH 7.2, 50 mM potassium acetate, 1 mM EDTA, 200 mM sorbitol) containing 0.1 mM AEBSF, 1 μM pepstatin A, protease inhibitor cocktail, and 0.5% Triton. Linear glycerol gradients (10–40%) with 0.5% Triton were prepared, and the solubilized protein samples were loaded and sedimented for 4 h at 100,000 × g. One-milliliter fractions were collected from the top of the gradient and TCA precipitated. Calibration of the gradient was performed using aldolase, catalase, ferritin, and thyroglobulin.
Immunodetection
Antibodies used in this study include the following: α-Myc (9E10, Santa Cruz Biotechnology, Santa Cruz, CA), α-FLAG (M2, Sigma-Aldrich, St. Louis, MO), α-HA (12CA5, Roche), G6PDH (Sigma-Aldrich), and α-Pep12 (Invitrogen, Carlsbad, CA). For Can1-FLAG degradation analysis, cells expressing FLAG-tagged Can1 were grown to mid-log phase and treated with cycloheximide to induce Can1 endocytosis. At the indicated times, cells were collected, precipitated, resolubilized, resolved by SDS–PAGE, and analyzed by blotting with the indicated antibodies. The quantification of half-life times (t1/2) was determined by analyzing band intensities (using ImageJ [National Institutes of Health, Bethesda, MD]) on Western blots (developed by enhanced chemiluminescence). The t1/2 value of Can1-FLAG in each strain was determined from three independent experiments, using multiple exposures from each experiment to measure half-life times.
Supplementary Material
Acknowledgments
We thank members of the Emr laboratory, Y. Mao, C. Fromme, and M. Smolka, for helpful discussions. We are grateful to Bill Parrish for construction of yeast strains and to Steven Padilla for technical assistance in performing the canavanine screen. This work was supported by funds from the Weill Institute for Cell and Molecular Biology and a Cornell University Research Grant (to S.D.E.).
Abbreviations used:
- ESCRT
endosomal complexes required for transport
- GPCR
G protein–coupled receptor
- TGN
trans-Golgi network
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0440) on August 31, 2011.
REFERENCES
- Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, Sharma YV, Yan D, Selva EM, Lin X. The retromer complex influences Wnt secretion by recycling wntless from endosomes to the trans-Golgi network. Dev Cell. 2008;14:120–131. doi: 10.1016/j.devcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brown MS, Anderson RG, Goldstein JL. Recycling receptors: the round-trip itinerary of migrant membrane proteins. Cell. 1983;32:663–667. doi: 10.1016/0092-8674(83)90052-1. [DOI] [PubMed] [Google Scholar]
- Burda P, Padilla SM, Sarkar S, Emr SD. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J Cell Sci. 2002;115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- Chen KY, Tsai PC, Hsu JW, Hsu HC, Fang CY, Chang LC, Tsai YT, Yu CJ, Lee FJ. Syt1p promotes activation of Arl1p at the late Golgi to recruit Imh1p. J Cell Sci. 2010;123:3478–3489. doi: 10.1242/jcs.074237. [DOI] [PubMed] [Google Scholar]
- Dittman J, Ryan TA. Molecular circuitry of endocytosis at nerve terminals. Annu Rev Cell Dev Biol. 2009;25:133–160. doi: 10.1146/annurev.cellbio.042308.113302. [DOI] [PubMed] [Google Scholar]
- Franch-Marro X, Wendler F, Guidato S, Griffith J, Baena-Lopez A, Itasaki N, Maurice MM, Vincent JP. Wingless secretion requires endosome-to-Golgi retrieval of Wntless/Evi/Sprinter by the retromer complex. Nat Cell Biol. 2008;10:170–177. doi: 10.1038/ncb1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XD, Albert S, Tcheperegine SE, Burd CG, Gallwitz D, Bi E. The GAP activity of Msb3p and Msb4p for the Rab GTPase Sec4p is required for efficient exocytosis and actin organization. J Cell Biol. 2003;162:635–646. doi: 10.1083/jcb.200302038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G, Malinsky J, Stahlschmidt W, Loibl M, Weig-Meckl I, Frommer WB, Opekarova M, Tanner W. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J Cell Biol. 2008;183:1075–1088. doi: 10.1083/jcb.200806035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyaloglu AC, von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu Rev Pharmacol Toxicol. 2008;48:537–568. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Lewis MJ, Black MW, Pelham HR. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003;22:548–557. doi: 10.1093/emboj/cdg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman NJ, Elmendorf JS. Signaling, cytoskeletal and membrane mechanisms regulating GLUT4 exocytosis. Trends Endocrinol Metab. 2011;22:110–116. doi: 10.1016/j.tem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontz RD, Niederer RO, Johnson JM, Smith JS. Genetic identification of factors that modulate ribosomal DNA transcription in Saccharomyces cerevisiae. Genetics. 2009;182:105–119. doi: 10.1534/genetics.108.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, Davies BA, Seaman MN, McLaughlin SA, Yoon S, Emr SD. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol Biol Cell. 1997;8:1529–1541. doi: 10.1091/mbc.8.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, Popoff V. Tracing the retrograde route in protein trafficking. Cell. 2008;135:1175–1187. doi: 10.1016/j.cell.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Johannes L, Romer W. Shiga toxins—from cell biology to biomedical applications. Nat Rev Microbiol. 2010;8:105–116. doi: 10.1038/nrmicro2279. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Goldberg J. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 2010;142:123–132. doi: 10.1016/j.cell.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince C, Le Scolan E, Meunier B, Fraisier V, Brandon N, De Gunzburg J, Camonis J. Sorting nexin 4 and amphiphysin 2, a new partnership between endocytosis and intracellular trafficking. J Cell Sci. 2003;116:1937–1948. doi: 10.1242/jcs.00403. [DOI] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Longtine M, Mckenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Marzioch M, Henthorn DC, Herrmann JM, Wilson R, Thomas DY, Bergeron JJ, Solari RC, Rowley A. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol Biol Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough IJ, Cullen PJ. Recent advances in retromer biology. Traffic. 2010;12:963–971. doi: 10.1111/j.1600-0854.2011.01201.x. [DOI] [PubMed] [Google Scholar]
- Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008;451:919–928. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
- Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics. 2008;178:197–214. doi: 10.1534/genetics.107.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan CL, Baum PD, Gu M, Jorgensen EM, Clark SG, Garriga G. C. elegans AP-2 and retromer control Wnt signaling by regulating mig-14/Wntless. Dev Cell. 2008;14:132–139. doi: 10.1016/j.devcel.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Kuster M, Herr P, Furger E, Banziger C, Hausmann G, Basler K. Wingless secretion promotes and requires retromer-dependent cycling of Wntless. Nat Cell Biol. 2008;10:178–185. doi: 10.1038/ncb1687. [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Lauffer B, Temkin P, Vistein R, Carlton P, Thorn K, Taunton J, Weiner OD, Parton RG, von Zastrow M. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell. 2010;143:761–773. doi: 10.1016/j.cell.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Barrowman J, Schieltz D, Yates JR, 3rd, Ferro-Novick S. Identification and characterization of five new subunits of TRAPP. Eur J Cell Biol. 2000;79:71–80. doi: 10.1078/S0171-9335(04)70009-6. [DOI] [PubMed] [Google Scholar]
- Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Sato K, Nakano A. Emp47p and its close homolog Emp46p have a tyrosine-containing endoplasmic reticulum exit signal and function in glycoprotein secretion in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2518–2532. doi: 10.1091/mbc.E02-01-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN. Recycle your receptors with retromer. Trends Cell Biol. 2005;15:68–75. doi: 10.1016/j.tcb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne GS. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J Cell Biol. 1992;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Strochlic TI, Tong AH, Boone C, Burd CG. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat Cell Biol. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- Sherwood PW, Carlson M. Efficient export of the glucose transporter Hxt1p from the endoplasmic reticulum requires Gsf2p. Proc Natl Acad Sci USA. 1999;96:7415–7420. doi: 10.1073/pnas.96.13.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Rojas R, Bonifacino JS, Hurley JH. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nat Struct Mol Biol. 2006;13:540–548. doi: 10.1038/nsmb1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skanland SS, Walchli S, Utskarpen A, Wandinger-Ness A, Sandvig K. Phosphoinositide-regulated retrograde transport of ricin: crosstalk between hVps34 and sorting nexins. Traffic. 2007;8:297–309. doi: 10.1111/j.1600-0854.2006.00527.x. [DOI] [PubMed] [Google Scholar]
- Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177:115–125. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale RD, Loci D, Houghton F, Karlsson L, Gleeson PA. A large family of endosome-localized proteins related to sorting nexin 1. Biochem J. 2001;358:7–16. doi: 10.1042/0264-6021:3580007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin P, Lauffer B, Jager S, Cimermancic P, Krogan NJ, von Zastrow M. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol. 2011;13:717–723. doi: 10.1038/ncb2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traer CJ, Rutherford AC, Palmer KJ, Wassmer T, Oakley J, Attar N, Carlton JG, Kremerskothen J, Stephens DJ, Cullen PJ. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat Cell Biol. 2007;9:1370–1380. doi: 10.1038/ncb1656. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol. 2002;3:919–931. doi: 10.1038/nrm974. [DOI] [PubMed] [Google Scholar]
- Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Hortsman H, Seet L, Wong SH, Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- Yang PT, Lorenowicz MJ, Silhankova M, Coudreuse DY, Betist MC, Korswagen HC. Wnt signaling requires retromer-dependent recycling of MIG-14/Wntless in Wnt-producing cells. Dev Cell. 2008;14:140–147. doi: 10.1016/j.devcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.