SUMMARY
The COP10-DET1-DDB1 (CDD) complex is an evolutionarily conserved protein complex discovered for its role in the repression of photomorphogenesis in Arabidopsis, and is important in many cellular and developmental processes in both plants and animals. However, its molecular mode of action remains poorly understood. Here, we show that the CDD component, DET1, possesses transcriptional repression activity and physically interacts with two closely related MYB transcription factors, CCA1 and LHY, which are core components of the plant circadian clock. DET1 associates with the promoter of CCA1/LHY target genes, such as TOC1, in a CCA1/LHY-dependent manner and is required for their repression, suggesting a recruitment of DET1 by the central oscillator components to regulate the clock. Our results reveal DET1 as a core transcriptional repression factor important for clock progression. Overall, the CDD complex may function as a transcriptional co-repressor in diverse processes through direct interaction with distinct transcription factors.
INTRODUCTION
The nine CONSTITUTIVE PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA (COP/DET/FUS) proteins are evolutionarily conserved regulators that are important in diverse cellular and developmental processes (Schwechheimer and Deng, 2000; Sullivan et al., 2003; Wei et al., 2008; Yi and Deng, 2005). These proteins were originally identified as repressors in seedling photomorphogenesis by genetic screens for Arabidopsis mutants displaying light-grown phenotypes in the dark, and were later found in many eukaryotes including humans. In the light signaling pathway of plants, this group of proteins integrates light signals perceived by various photoreceptors and is important for the proteolysis of photomorphogenesis-promoting transcription factors, such as ELONGATED HYPOCOTYL 5 (HY5) (Hoecker, 2005; Osterlund et al., 2000). Biochemical and genetic evidence have defined the COP/DET/FUS proteins as components of three distinct biochemical entities, the COP1 complex, the COP9 signalsome (CSN) and the COP10-DET1-DDB1 (CDD) complex, and revealed their close tie to the ubiquitin (Ub)-proteasome system (Wei et al., 2008; Yanagawa et al., 2004; Yi and Deng, 2005). While COP1 acts as an E3 Ub ligase and the CSN is a general regulator for all cullin-RING E3 ligases (CRLs), the molecular role of the CDD complex has remained unclear.
The Arabidopsis CDD complex consists of COP10, DET1 and DDB1, and a similar complex is also present in humans (Pick et al., 2007; Schroeder et al., 2002; Yanagawa et al., 2004). In plants, besides its role in seedling photomorphogenesis, the CDD complex is likely involved in other processes as mutations in DET1 or COP10 result in pleiotropic phenotypes (Chory et al., 1989; Wei et al., 1994). Further, various cellular and developmental defects in social amoeba, fruit flies, and mammals were observed when orthologs of the genes were mutated (Berloco et al., 2001; Dubin et al., 2011; Plafker et al., 2008). These observations suggest that the CDD complex may play a fundamental role in signal transduction and/or gene regulation. Two of the CDD components, COP10 and DDB1, are established members of the ubiquitin system. COP10 is a Ub E2 variant (UEV), which has no E2 activity, but can enhance the activity of other E2s (Lau and Deng, 2009; Suzuki et al., 2002; Yanagawa et al., 2004). DDB1 is the core adaptor component of the CULLIN4 (CUL4)-based E3 Ub ligase, a member of the CRL family (Jackson and Xiong, 2009). It acts as a linker that connects diverse WDXR motif-containing DWD/DCAF/CDW substrate receptors to the CUL4 scaffold. DET1, on the other hand, is a nuclear protein with no recognizable domains (Pepper et al., 1994). Through DDB1, CDD can associate with CUL4 and RBX1 to form a CUL4-based E3 ligase (Chen et al., 2006; Pick et al., 2007). However, it is unclear whether COP10 and DET1, which are not conventional DDB1-interacting DWD proteins, can serve as bona fide substrate receptors. In the context of a CUL4-based E3 ligase, human DET1 has been shown to act as another linker protein that connects the substrate adaptor hCOP1 to the CUL4A-DDB1 core (Wertz et al., 2004). However, in Arabidopsis, DET1 does not appear to serve this function (Chen et al., 2010). Further, DET1 may be involved in the regulation and/or function of some CUL4-based ligases (Castells et al., 2011; Pick et al., 2007). Besides the connection to the ubiquitin system, DET1 has been implicated in chromatin regulation. DET1 associates with the histone gene cluster in Drosophila embryos and has been shown to interact with the non-acetylated tail of histone H2B in plants (Benvenuto et al., 2002; Berloco et al., 2001). However, the significance of DET1’s role in transcriptional regulation still remains to be explored.
In this study, we demonstrate that DET1 can function as a transcriptional co-repressor and is involved in transcriptional repression in the central circadian clock of Arabidopsis. The circadian clock is an endogenous, approximately 24-hr timekeeper in living systems that provides adaptive advantage, and in most organisms, is based on interlocked negative feedback loops of transcriptional activators and repressors (Zhang and Kay, 2010). In plants, the central loop operates through the reciprocal regulation between two morning-phased partially redundant MYB transcription factors, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), and the evening-phased pseudoresponse regulator TIMING OF CAB EXPRESSION 1 (TOC1) (Harmer, 2009; McClung and Gutierrez, 2010). CCA1 and LHY are the transcriptional repressors in the loop, and they bind to the promoter of TOC1 and negatively regulate its transcription (Alabadi et al., 2001; Schaffer et al., 1998; Wang and Tobin, 1998). On the other hand, TOC1 indirectly promotes the expression of CCA1 and LHY, partly through a CCA1-binding transcription factor, CCA1 HIKING EXPEDITION (CHE) (Alabadi et al., 2001; Pruneda-Paz et al., 2009). Although it is conceivable that in mediating the transcriptional repression, CCA1 and LHY would recruit additional cellular factors, little is known about their identity. Here, we show that DET1 can direct transcriptional repression, and through a hypothesis-driven approach, identify a physical interaction between DET1 and both CCA1 and LHY. Our data support a model in which DET1 is recruited by CCA1 and LHY to their target genes, including TOC1, and mediates their repression for proper clock progression. These results not only establish DET1’s role in transcriptional control, but also discover a direct involvement of DET1 in the central circadian clock and offer insights into the mechanism of clock progression.
RESULTS
DET1 possesses transcriptional repression activity
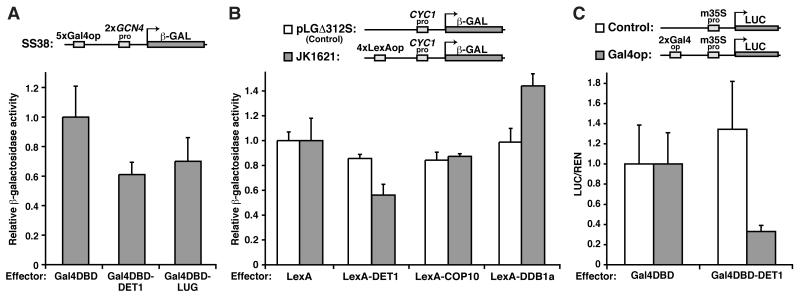
To test if DET1 has a direct effect on transcription, we first performed a transcriptional assay in yeast (Figure 1A). In this system, the transcriptional reporter, SS38, contains a β-galactosidase-encoding lacZ gene, which is driven by two active GCN4 promoters and can be modulated through the Gal4 operators (Gal4op) (Figure 1A). We fused DET1 to the Gal4 DNA binding domain (Gal4DBD) and expressed it in a SS38-containing yeast strain. As shown in Figure 1A, the presence of Gal4DBD-DET1 resulted in a 40% reduction in reporter activity compared with the Gal4DBD control, suggesting that DET1 can repress transcription. A similar reduction was observed when we used LUG, a well-characterized transcriptional co-repressor in plants (Sridhar et al., 2004), as a positive control. To further confirm the repression activity and test the other components of the CDD complex, another yeast transcriptional assay system was employed. Similar to the results above, expression of LexA-DET1 caused a 40% decrease in the activity of a LexA operator (LexAop)-dependent reporter, JK16121, compared with the LexA alone control (Figure 1B, grey bars). The effect of DET1 is also specific as the repression activity of LexA-DET1 in the control reporter, pLGΔ312S, which lacks LexAops, was similar to its LexA counterpart (Figure 1B, white bars). COP10 and DDB1a, however, do not appear to repress transcription. The enhancement observed in DDB1a may be due to its interaction with a transcriptional activator(s) in yeasts. Besides its likely recruitment of an array of CUL4 substrate receptors, many of which may play a role in chromatin regulation, DDB1 has been shown to associate with the STAGA coactivator complex in human cells (Jackson and Xiong, 2009; Martinez et al., 2001). Finally, the in planta transcriptional repression activity of DET1 was tested in a transient dual luciferase assay in Nicotiana benthamiana (Figure 1C). In this assay, the firefly luciferase reporter was driven by the minimal 35S promoter (m35S) with or without two copies of Gal4op. As shown in Figure 1C, Gal4DBD-DET1 caused a 70% reduction in the Gal4op reporter activity compared with the Gal4DBD control. Also, the control reporter without Gal4ops indicates that the repression is specific. Taken together, the results suggest that DET1 can suppress transcription when recruited to promoters.
Figure 1. DET1, a component of the CDD complex, directs transcriptional repression.
(A and B) Transcriptional repression assays in yeast with a lacZ reporter driven by basal promoters with either Gal4 operators (Gal4op) [SS38 in (A)], LexA operators (LexAop) [JK1621 in (B)], or no operator [pLGΔ312S in (B)] upstream. In (A), the reporter was integrated into the yeast genome and plasmids encoding Gal4-DNA binding domain (Gal4DBD) alone (negative control) and Gal4DBD fused with DET1 or LUG (positive control) were independently transformed into yeast. β-galactosidase activities with the different effectors are shown (mean±s.d.; n≥6). In (B), the two reporters were co-transformed with different LexA-fusion proteins. pLGΔ312S is a control reporter for detecting potential effector’s non-specific activity, while LexA alone acts as a negative control. Reporter activities were measured as in (A). (C) Transient transcriptional repression assays in plants with a luciferase (LUC) reporter driven by a minimal CaMV 35S promoter (m35Spro) with or without Gal4ops upstream. The reporter vectors also contain a 35S-driven Renilla luciferase (REN) for signal normalization. The reporters were co-transformed independently with a plasmid containing either the GAL4DBD alone or GAL4DBD-DET1 into the leaves of Nicotiana benthamiana. Activities of firefly luciferase (LUC) normalized to Renilla luciferase (REN) are shown (mean±s.d.; n≥4). Activities with the control effectors: Gal4DBD alone (A and C) or LexA alone (B), are set to 1.
DET1 interacts with two closely related MYB transcription factors, CCA1 and LHY, which are central clock components
Since neither DET1 nor the other components of the CDD complex contain any DNA-binding domain, we further hypothesized that, to exert its effect on transcription, the CDD complex must be recruited to target promoters through interaction of its components with specific transcription factors. In search of interacting partners, we looked for clues from a classic light-activated gene, CHLOROPHYLL A/B-BINDING PROTEIN 2 (CAB2 or Lhcb1*1). CAB2 is overexpressed in dark-grown det1 mutants and it has been suggested that DET1 mediates its regulation on CAB2 via the transcription factors, HY5 and CCA1 (Maxwell et al., 2003; Pepper et al., 1994). Thus, we speculated that DET1 might directly regulate CAB2 through interaction with HY5 and/or CCA1.
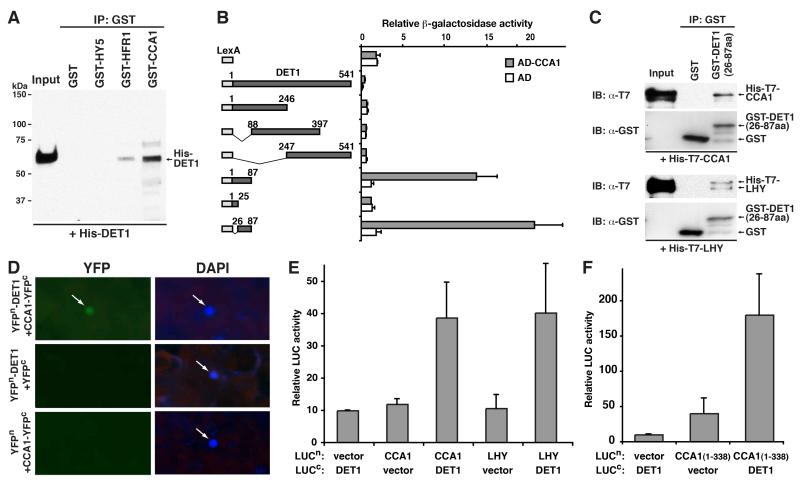
To test for interaction between DET1 and the two transcription factors, we performed an in vitro-pull down assay between recombinant His-DET1 and GST-tagged HY5, CCA1 and another light-regulated transcription factor, HFR1, which works downstream of the COP/DET/FUS proteins and may be a potential interactor of DET1 (Jiao et al., 2007) (Figure 2A). We found that DET1 can directly interact with CCA1 and, to a lesser extent, HFR1, but not HY5. To further confirm the DET1-CCA1 interaction and identify the interacting domains on the two proteins, yeast two-hybrid assays were carried out (Figure 2B and Figure S1A). Using a series of deletion constructs of DET1, the CCA1-interacting domain on DET1 was mapped to its N-terminal end, encompassing its 26th to 87th amino acid (a.a.) residues (Figure 2B). Interestingly, neither full-length DET1 nor its three larger fragments, 1-246, 88-397 and 247-541 a.a., showed positive reporter activities. Rather, their activities were even lower than the LexA alone control. Since yeast two-hybrid is a transcription-based assay, we attribute this to the transcriptional repression activity in the full-length and the three fragments of DET1. Using the mapped N-terminal end of DET1 (26-87a.a.), we performed a reverse in vitro-pull down assay using a GST-tagged version of the DET1 fragment against either His-T7-tagged CCA1 or LHY (Figure 2C). This short 61 a.a.-long peptide of DET1 was able to pull down both CCA1 and LHY, supporting the yeast two-hybrid results. Further in vitro pull-down assays with COP10 also suggest that COP10 can interact with CCA1 and LHY (Figure S1B-S1D). Next, we tested in vivo interaction between DET1 and the two transcription factors. Bimolecular fluorescence complementation (BiFC) assays in N. benthamiana leaves demonstrate a direct interaction between DET1 and CCA1 in the nucleus of plant cells, where co-expression of YFPn-DET1 and CCA1-YFPc reconstituted a functional YFP but controls did not (Figure 2D). Similarly, firefly luciferase complementation imaging (LCI) assays gave further evidence for an interaction between DET1 and both CCA1 and LHY in vivo, where in this case, the activity of luciferase (LUC) activity was reconstituted (Figure 2E). Also, the DET1-interaction domain on CCA1 is likely within its N-terminal half (Figure 2F and Figure S1A). These in vitro and in vivo results identify the two transcription factors, CCA1 and LHY, as direct interacting partners of DET1.
Figure 2. DET1 interacts with two MYB transcription factors, CCA1 and LHY, which are core components of the plant circadian clock.
(A) In vitro GST pull-down assay between recombinant His-DET1 and GST-fused HY5, HFR1 and CCA1. His-DET1 was incubated with immobilized GST or the GST-fused proteins, and immunoprecipitated fractions were probed with an anti-DET1 antibody. (B) Liquid yeast two-hybrid assays for interaction between CCA1 and deletion series of DET1. Left panel shows the constructs encoding LexA alone (control) and LexA-fused DET1 and its fragments. Right panel shows the corresponding β-galactosidase activities with either Activation Domain (AD) alone or AD-CCA1 (mean±s.d.; n≥4). (C) In vitro GST pull-down assay between His-T7-tagged CCA1 or LHY and a GST-fused N-terminal fragment of DET1 (26-87 a.a.). The His-T7-tagged proteins were incubated with immobilized GST or the GST-fused DET1 fragment, and immunoprecipitated fractions were probed with an anti-T7 and anti-GST antibody. IB: Immunoblot. (D) Bimolecular fluorescence complementation (BiFC) analysis for in vivo interaction between DET1 and CCA1. N- and C-terminal fragments of yellow fluorescence protein (YFP; YFPN and YFPC) were fused to DET1 and CCA1, respectively. Combinations of their encoding plasmids and controls (indicated on the left panel) were transiently transformed into leaves of Nicotiana benthamiana. Presence of YFP signal (YFP) indicates reconstitution of YFP through protein interaction of the tested pairs. Positions of the nuclei were stained with DAPI and marked by arrows. (E and F) Firefly luciferase complementation imaging (LCI) assays for in vivo interaction between DET1, CCA1 and LHY. CCA1, LHY and an N-terminal fragment of CCA1 (1-338 a.a.) were fused with the N-terminus of luciferase (LUCn), and DET1 was fused with the C-terminal part (LUCc). Plasmids were transformed into plants as in (D) and LUC activities above background indicate LUC reconstitution and interaction. Relative LUC activities (luminescence intensity/leaf area) of the indicated transformation (lower panel) are shown (mean±s.d.; n≥4).
DET1 binds to CCA1 and LHY-controlled genes
The interaction between the DET1 subunit of the CDD complex and the two core oscillator components, CCA1 and LHY, suggests that DET1 may be one of the factors that CCA1 and LHY recruit for transcriptional repression in the circadian clock. As mentioned earlier, in the central loop, CCA1 and LHY bind to the promoter of TOC1 and negatively regulate its expression (Alabadi et al., 2001). Since DET1 has transcriptional repression activity and interacts with both CCA1 and LHY, we suspected that DET1 might contribute to the repression function of CCA1 and LHY on TOC1 and other CCA1/LHY-regulated genes and play an active role in the plant circadian clock.
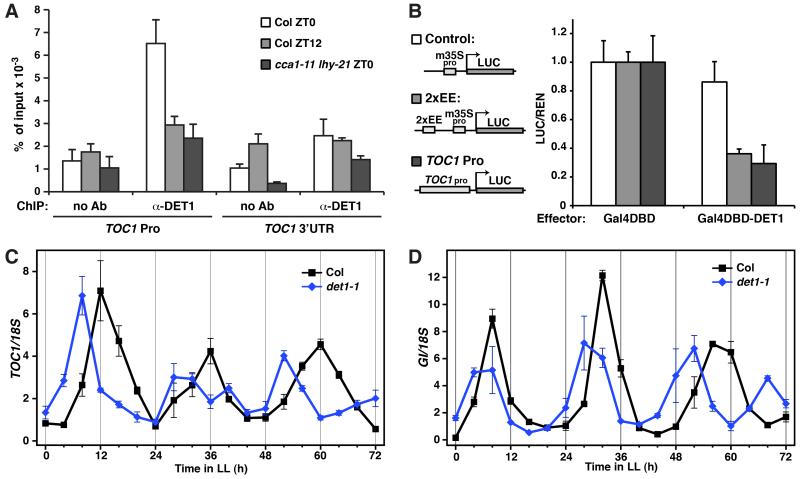
We first tested whether DET1 can associate with the promoter of TOC1 and GIGANTEA (GI), another important clock gene, and likely an evening-loop component, that CCA1 and LHY may bind and repress (McClung and Gutierrez, 2010; Park et al., 1999). The promoters of TOC1 and GI, along with many evening-phased genes, contains a sequence motif, the evening element (EE), that CCA1 and LHY can recognize and bind (Alabadi et al., 2001). Through chromatin immunoprecipitation (ChIP)-PCR, a specific binding of DET1 to the EE-containing region of TOC1 and GI promoter was detected (Figure S2A). As a positive control, LHY also binds to the same regions with stronger detectable signal, as expected for a direct interaction. Because expression of CCA1 and LHY oscillates, highest at dawn and lowest at dusk (Schaffer et al., 1998; Wang and Tobin, 1998), we examined if DET1 binding on the TOC1 promoter exhibits a similar oscillation through ChIP followed by quantitative real-time PCR (ChIP-qPCR), as the qualitative PCRs in Figure S2A likely did not provide enough resolution. From the quantitative analysis, binding of DET1 to the TOC1 promoter was most evident at zeitgeber time 0 (ZT0) (subjective dawn; white bars), while at ZT12 (subjective dusk; grey bars), the ChIP signal was reduced to a level slightly above background (Figure 3A). This pattern correlates with the rhythmic expression of CCA1 and LHY. The reduction in binding is also promoter-specific as the signals at the 3′ untranslated region (UTR) of TOC1 at ZT0 and ZT12 did not exhibit the change and were at a low level. To test the dependency of DET1 binding to the TOC1 promoter on CCA1 and LHY, an anti-DET1 ChIP with a cca1 lhy double mutant was also performed. Consistent with our model, the signal on the TOC1 promoter in the double mutant was the lowest and was at a level comparable to that of Col ZT12, indicating a lack of binding (Figure 3A, black bars). These ChIP results suggest that DET1 can bind to direct targets of CCA1 and LHY, and the binding, at least for TOC1, is CCA1- and LHY-dependent.
Figure 3. DET1 binds to target genes of CCA1 and LHY and regulates their circadian expressions.
(A) Chromatin Immunoprecipitation-quantitative PCR (ChIP-qPCR) analyses of DET1 binding on TOC1 promoter. ChIP assays were performed on seedlings of wild-type (Col) collected at zeitgeber time (ZT) 0 and 12 and the cca1-11 lhy-21 double mutant collected at ZT 0, with and without an anti-DET1 antibody. ChIP DNA was quantified by quantitative real-time PCR with primers and probes specific to the evening element (EE)-containing region of TOC1 promoter (TOC1 Pro) and the 3′ untranslated region of TOC1 (TOC1 3′UTR) (control). Signals were normalized to the input DNA (mean±s.d.; n≥3). Ab: Antibody (B) Transient transcriptional repression assays in plants for DET1 effect on an EE-containing reporter (2xEE) and a TOC1 promoter-driven reporter (TOC1 Pro). Assays were carried out as in Figure 1C. Relative reporter activities (LUC/REN) with the indicated effectors are shown (mean±s.d.; n≥4). Activities of reporters with the control effector, Gal4DBD, are set to 1. (C and D) Circadian rhythms of TOC1 (C) and GI (D) expression in wild-type (Col) and det1-1 under continuous light (LL). Plants were grown with long day cycles (16L8D) for 8 days and then released to LL. RNA levels were quantified by TaqMan real-time RT-PCR and signals were normalized to 18S (mean±s.d.; two biological and two technical replicates).
DET1 represses targets genes of CCA1 and LHY for their normal progression
We next examined the effect of DET1 on the transcription of CCA1/LHY-regulated genes. By using the in planta transcriptional assay described earlier, we found that overexpression of DET1 resulted in a 70% reduction in the activity of an EE-regulated reporter, while having a modest effect on the control (Figure 3B). DET1 also had a similar negative effect on a reporter driven by the TOC1 promoter. These data suggest that DET1 can mediate its transcriptional repression activity on the target genes of CCA1 and LHY. Subsequently, we studied how DET1 affects the transcription of endogenous CCA1/LHY-controlled genes, TOC1 and GI (Figure 3C and 3D). The two genes exhibit circadian cycles of transcription and peak in the evening. Loss-of-function mutations in CCA1 and LHY cause an early shift in the peak and a shorter period in both TOC1 and GI expression (Mizoguchi et al., 2002). To test if a similar pattern happens when DET1 is lost, we analyzed the circadian profile of these two clock genes in a moderate loss-of-function mutant of DET1, det1-1, which produces ~2% of wild-type (WT) transcript (Pepper et al., 1994). As shown in Figure 3C and 3D, in WT seedlings, expression of TOC1 and GI peaked at ZT 12 and 8, respectively, which is consistent with the previous report (Mizoguchi et al., 2002). In det1-1, however, the expression peak of the two genes shifted earlier and their period was shortened. These changes in the circadian profile of TOC1 and GI in det1-1 closely resemble that in the cca1 and lhy mutants (Mizoguchi et al., 2002). We also examined if this may be due to altered stability of CCA1 and LHY proteins in det1-1, which was suggested by another study (Song and Carré, 2005). However, our in vivo examination of LHY and CCA1 protein level in the det1-1 background did not reveal any noticeable changes, arguing against a role of DET1 in their turnover (Figure S2B and S3) (comparison of CCA1 was between CCA1-OX and CCA1-OX;det1-1 due to difficulties in its detection at WT level). Rather, a forward shift in the circadian profile of LHY protein in det1-1 was detected and is likely due to a similar shift in its mRNA, which was also observed in CCA1 (Figure S2C and S2D). We speculate that the shift in CCA1 and LHY expression may be due to the corresponding shift in the circadian profiles of TOC1 and GI or the proposed negative auto-regulatory feedback loop of CCA1 and LHY, which might also require DET1 function (Schaffer et al., 1998; Wang and Tobin, 1998). These results, along with the above data, indicate that DET1, CCA1 and LHY work cooperatively in regulating the expression of CCA1/LHY target genes and it is likely the activity, rather than any stabilizing effect, of DET1 that is important for the functional consequences of the interaction.
DET1 is important for CCA1 function in the circadian clock
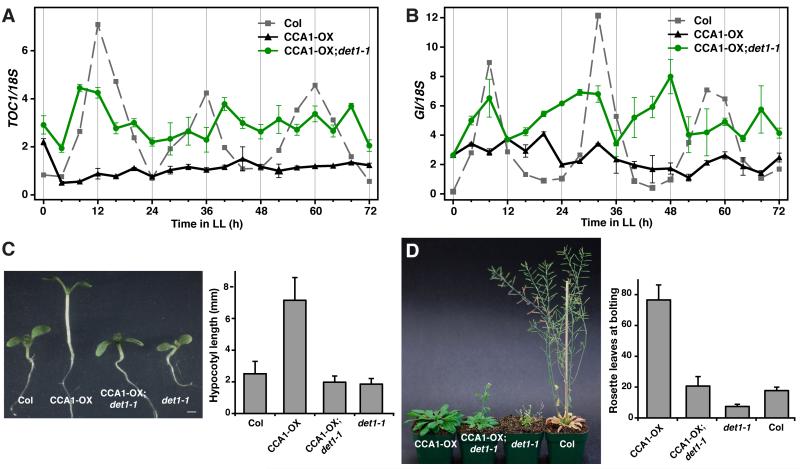
To address how important DET1 is to the function of CCA1, we further analyzed if the dominant gain-of-function phenotypes in CCA1-overexpression lines (CCA1-OX) can be suppressed in the det1-1 background (Maxwell et al., 2003; Wang and Tobin, 1998). Overexpression of CCA1 by the constitutive CaMV 35S promoter disrupts the daily oscillation of TOC1 expression and results in a constant low level of TOC1 transcripts (Figure 4A, compare grey and black plots) (Alabadi et al., 2001). In the det1-1 background (CCA1-OX;det1-1), however, we found that the repressive effect of CCA1 observed in CCA1-OX is considerably alleviated, indicating that CCA1 is not fully functional without DET1 (Figure 4A, compare green and black plots). A similar de-repression effect of CCA1-OX in det1-1 background was also observed in GI expression (Figure 4B). These data support that the transcriptional repression activity of CCA1 depends on the level of DET1. Importantly, the protein level of CCA1 in CCA1-OX and CCA1-OX;det1-1 remains similar, again indicating that DET1 does not alter the protein level of CCA1 (Figure S3). As a result of a disruption in the circadian rhythm, light-grown CCA1-OX seedlings develop long hypocotyls and the adult plants show a substantial delay in flowering (Wang and Tobin, 1998). Both of these gain-of-function phenotypes were largely suppressed in the det1-1 background. While the delay in flowering was substantially less severe but still evident, the hypocotyl length defect was rescued fully (Figure 4C and 4D). From these molecular and phenotypic analyses, we conclude that DET1 is important for the function of CCA1 and the proper progression of the plant circadian clock.
Figure 4. DET1 is important for CCA1 function.
(A and B) Circadian rhythms of TOC1 (A) and GI (B) expression in CCA1-OX and CCA1-OX;det1-1 under continuous light (LL). Experiments were performed as in Figure 3C and 3D. (C) 9-day old seedlings of wild-type (Col), CCA1-OX, CCA1-OX;det1-1 and det1-1 grown under long day (LD; 16L8D). Hypocotyl length measurements of the seedlings are shown (mean±s.d.; n≥20). (D) 5-week old plants of CCA1-OX, CCA1-OX;det1-1, det1-1 and Col grown under LD. The number of rosette leaves at the time of flowering is shown (mean±s.d.; n≥12).
DISCUSSION
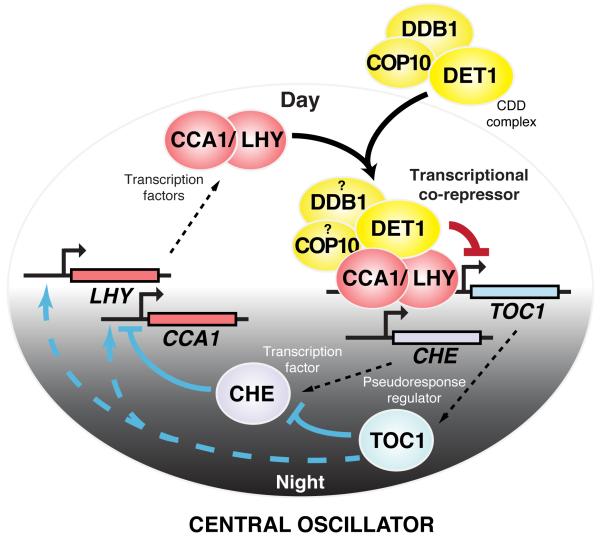
We demonstrate here that DET1 can repress transcription, physically interacts with the core circadian clock components, CCA1 and LHY, for binding and repressing CCA1/LHY target genes, and is required for the gain-of-function phenotypes in CCA1-overexpression lines, suggesting a direct involvement of DET1 in mediating the transcriptional repression in the circadian clock. Based on the molecular and genetic data, we propose a model where DET1, possibly in the form of a CDD complex, is recruited by the morning-phased CCA1 and LHY through direct interaction at dawn (Figure 5). The recruitment brings the transcriptional repression activity of DET1 to the evening-phased CCA1/LHY target genes, such as TOC1 and GI, and represses their expression. In the evening, the levels of CCA1 and LHY drop, which leads to a decrease in the binding of the CDD complex on TOC1 and the evening-phased genes and an increase in their transcription. High levels of TOC1 in turn activate CCA1 and LHY and the cycle repeats. This functional interaction between DET1 and the central clock components identifies DET1 as an important factor in the circadian clock and illustrates DET1’s role as a transcriptional co-repressor.
Figure 5. A model for the functional interaction between the CDD complex with CCA1 and LHY in regulating the central loop of the Arabidopsis circadian clock.
At dawn, the morning-expressed CCA1 and LHY physically interact with DET1, likely in the form of the CDD complex, and recruit it to their evening-phased gene targets, such as TOC1. The CDD complex then exerts its repression activity on these CCA1/LHY targets and suppresses their expression. Gradual reduction in the level of CCA1 and LHY throughout the day reduces the association of the CDD complex on the genes and allows accumulation of TOC1 transcripts, which peak at dusk. TOC1 in turn induces expression of LHY and CCA1, where their proteins reach their peak at dawn, and the cycle continues. CHE, a CCA1-binding transcription factor, forms a reciprocal negative loop with CCA1. TOC1 can directly interact with CHE and antagonize its repression on CCA1.
Although our results show that DET1 plays an important role in the circadian rhythm, the use of the intermediate det1-1 mutant likely did not reveal the full extent of DET1’s effect in this process. The shortened period of circadian rhythms of TOC1 and GI in det1-1 is more similar to the single mutants of cca1 and lhy than to the cca1 lhy double mutant, which displays a more dramatic shortening of period and eventual dampening of rhythms (Figure 3C and 3D) (Mizoguchi et al., 2002). The milder circadian defect in det1-1 could be due to the presence of a low level of DET1, which may still partially support the function of CCA1 and LHY. Alternatively, other unknown co-repressors may contribute to the transcriptional repression. Although technically challenging, analyzing the circadian rhythm in seedling-lethal null mutants of DET1 would help clarify the situation.
The de-repression of TOC1 and GI expression observed in CCA1-OX;det1-1 clearly shows a requirement of DET1 in mediating the repression function of CCA1 (Figure 4A and 4B). However, the circadian profiles of the evening genes do not resemble those in WT or det1-1, and they fluctuated around the mid-level between the peak and trough of WT expression with reduced amplitudes. The inability of det1-1 to restore circadian oscillation of TOC1 and GI indicates that the overexpression of CCA1 was not fully suppressed, resulting in a reduced gain-of-function phenotype. Again, this may be explained by a residual amount of DET1 protein present in det1-1 or other co-repressors that support CCA1 function. Another possible explanation is the likely continual occupancy of TOC1 and GI promoters by CCA1, which may hinder binding and/or function of other transcriptional regulators necessary for normal expression.
Since the CDD complex plays a central role in the light signaling pathway, it is appealing to hypothesize that the DET1-CCA1/LHY interaction might act as an integration point that connects light signals to the clock. However, unlike COP1, the CDD complex has not been shown to be light-regulated in its localization or activity, and transcript and protein levels of DET1 are not regulated by light (Pepper et al., 1994) (data not shown). Thus, it remains to be explored whether light signals can modulate the circadian clock through DET1 or the CDD complex. We also tested whether expression of DET1 might oscillate, potentially adding a layer of control of DET1 function in the clock, but no circadian rhythm of transcript or protein levels was detected (Figure S4). This suggests that transcriptional repression of CCA1/LHY targets by DET1 is regulated through the abundance of CCA1 and LHY, as proposed in our model. Another possibility is a time-dependent change in the affinity in the DET1-CCA1/LHY interaction, but it was not tested in this study. Interestingly, it was recently noted that transcripts of the ortholog of DET1 in Ostreococcus tauri, a marine phytoplankton, showed a weak oscillating pattern with peaks during the day (Monnier et al., 2010; van Ooijen et al., 2011). Although further studies will be needed to establish a role of Ostreococcus DET1 in the circadian clock, control of DET1 level may indeed be employed by the primitive plants in regulating the clock.
Our findings that DET1 can act as a transcriptional co-repressor offer insights into its role and importance in both plants and animals. In the light signaling pathway, DET1, possibly in the context of a CUL4-CDD E3 ligase, may repress light-activated genes through a recruitment by the light-regulating transcription factors, such as CCA1 and LHY. One such example may be CAB2, where DET1, via its interaction with CCA1, may directly repress the light-activated, CCA1 target gene. This additional mode of regulation by the COP/DET/FUS proteins at the transcriptional level, together with proteolysis of photomorphogenesis-promoting transcription factors at the post-translational level, could ensure tight control of photomorphogenic development in darkness (Jiao et al., 2007; Sullivan et al., 2003).
Besides the involvement of the CDD complex in light signaling, mutants of DET1 and COP10 also display severe and pleiotropic phenotypes, such as adult lethality in strong mutants, and early flowering in short day (SD), dwarfism and loss of apical dominance in det1-1 (Pepper et al., 1994; Pepper and Chory, 1997; Wei et al., 1994). Since the circadian clock regulates the transition to flowering, and mutants of CCA1 and LHY also flower early in SD (de Montaigu et al., 2010; Mizoguchi et al., 2002), the disruption of the functional interaction between DET1 and CCA1/LHY in the circadian clock in det1-1 may underlie its early flowering phenotype. However, with the many mutant phenotypes beyond light and circadian defects, it is plausible that the CDD complex is recruited broadly by additional transcription factors, and act as a general transcriptional co-repression complex in other cellular and developmental processes. Furthermore, because DET1 is highly conserved in eukaryotes, we speculate that orthologs of Arabidopsis DET1 might function in a similar manner. Indeed, in Drosophila, DmDET1/ABO was found to bind to the promoters of histone genes and its mutants accumulate higher level of the transcripts (Berloco et al., 2001). Future efforts to identify DET1-interacting transcription factors in plants and different organisms will likely reveal more life processes that are controlled by DET1 and the CDD complex.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
The wild-type Arabidopsis ecotype used in this study is Columbia-0 (Col). The mutant lines: det1-1 (Pepper et al., 1994), cca1-11 lhy-21 double mutant (ecotype Ws) (Hall et al., 2003), CCA1-OX (Wang and Tobin, 1998) and CCA1-OX;det1-1 (Maxwell et al., 2003) were previously described. For ChIP assays and hypocotyl measurements, seedlings were grown on MS plates under long day conditions (LD; 16 h light/ 8 h dark) in a growth chamber at 21°C. For circadian rhythm experiments under continuous light (LL), seedlings were first grown in LD for 8 days and then transferred to LL at dawn (Mizoguchi et al., 2002). For flowering time measurements, plants were grown on soil under LD.
Yeast transcriptional repression assays
For the Gal4-based assays, the DET1 effector was constructed by cloning its full-length cDNA into the pMAN vector. pMAN-LUG was kindly provided by Z. Liu. These constructs were transformed into the yeast strain 122 carrying the reporter SS38 (Saha et al., 1993). For the LexA-based assays, LexA fusion constructs were constructed with the pLexA vector. The two reporters, pLGΔ312S and JK1621, are previously described (Guarente and Hoar, 1984; Keleher et al., 1992). The repression assays were carried out according to previous reports (Mosher et al., 2006; Sridhar et al., 2004). Quantitative β-galactosidase activity was assayed with the yeast β-galactosidase assay kit (Thermo Scientific).
Transcriptional repression assays in plants
The transient transcriptional repression assays in the leaves of Nicotiana benthamiana were performed based on a previous report (Hellens et al., 2005). The various reporters were constructed by cloning regulatory sequences: m35Spro (−67 to +8), Gal4op-m35Spro [Gal4op: 2x17MX (Webster et al., 1988)], 2xEE-m35Spro (2xEE: 2xCTGTGATTCTGCAGTTTTTTTGTGT) and TOC1pro (1009 bp fragment upstream of ATG), into the multiple cloning site of the reporter vector, pGreenII 0800-LUC, which drive transcription of a firefly luciferase (LUC) (Hellens et al., 2005). The reporter vector also contains a 35S-driven Renilla luciferase (REN), allowing signal normalizations. Effector constructs were constructed with the pPZPY122 plasmid. The reporter activities (LUC/REN) were measured with the Dual-Luciferase® Reporter (DLR™) assay system (Promega) on a GloMax® 20/20 luminometer (Promega).
In vitro pull-down assays
The bacterial expression vectors for GST-HY5, CCA1 and HFR1 are described previously (Hardtke et al., 2000; Wang et al., 1997; Yang et al., 2005). Full-length DET1, CCA1 and LHY cDNAs were cloned into pET-28a, generating N-terminal His-T7-tagged fusions. Expression vector for GST-DET1(26-87aa) was generated by cloning a corresponding DET1 cDNA fragment into pGEX-4T-1. Recombinant proteins were purified as described, except that His-DET1 was purified from ArcticExpress RIL cells (Stratagene) (Lau and Deng, 2009). For pull-down assays, purified His-fusion proteins were incubated with either GST or GST fusion proteins in GST binding buffer (20 mM Tris/HCl, pH 7.5, 150mM NaCl and 0.1% Nonidet P40) for 2 h at 4°C. Glutathione–Sepharose 4B beads (GE Healthcare) were then added and the mixture were incubated for 1 h. After washing the beads three times with the GST binding buffer, bound proteins were eluted in 2×SDS loading buffer and analyzed by Western blotting with anti-DET1 (Chen et al., 2010), anti-T7 antibody (Novagen) or anti-GST antibody (GE Healthcare).
Yeast two-hybrid assays
Full-length and various fragments of DET1 cDNA were cloned into pEG202 as bait constructs, while full-length CCA1 cDNA was cloned into pJG4-5 as a prey construct. The two-hybrid interaction assays were performed as described previously (Chen et al., 2006). Quantitative β-galactosidase activity was assayed with the yeast β-galactosidase assay kit (Thermo Scientific).
Bimolecular fluorescence complementation (BiFC) analysis
Full-length DET1 and CCA1 were cloned into the binary pBiFC vectors containing either N- or C-terminal fragments of yellow fluorescence protein (YFP; YFPN and YFPC) (de Lucas et al., 2008). Transient BiFC assays in Nicotiana benthamiana were performed according to our previous report (Feng et al., 2008).
Firefly luciferase complementation imaging (LCI) assays
Full-length CCA1, LHY and fragments of CCA1 were cloned into 35S::NLuc, while full-length DET1 was cloned into 35S::CLuc (Chen et al., 2008). Transient LCI assays in Nicotiana benthamiana were performed as described previously (Chen et al., 2008). Luciferase signals were viewed in an IVIS® 200 optical imaging system (Caliper LifeSciences) and quantified with the Living Image® 4.0 software.
Chromatin immunoprecipitations (ChIP) and quantitative/qualitative PCR amplifications
Chromatin isolation was performed with 5 g of whole Arabidopsis seedling tissues according to a previous report (Bowler et al., 2004). The resuspended chromatin pellet was sonicated with a Bioruptor™ UCD 200 (Diagenode) at high intensity for 10 min (cycles of 30 sec on and 30 sec off) at 4°C. The sheared DNA was approximately 0.3-0.8 kb in length. Immunoprecipitation was carried out with either anti-DET1 (Chen et al., 2010), anti-LHY(Daniel et al., 2004) and anti-H3 (Millipore 07-690), followed by incubation with DYNAL® magnetic beads (Dynabeads® Protein G; Invitrogen). ChIPed DNAs were washed and reverse crosslinked according to the Affymetrix’ Chromatin Immunoprecipitation Assay Protocol Rev.3. An aliquot of untreated sonicated chromatin was also reverse cross-linked and used as total input DNA control. For qPCR, reactions were performed using Applied Biosystems Taqman® PCR reagents and custom Taqman® primers and probes targeted to promoter and 3′ untranslated region of TOC1 (Table S1). A CFX96™ Real-Time PCR detection system (Bio-Rad) and its interface software were used to analyze the data. For qualitative PCR analysis, amplification was performed with 32 cycles with annealing temperature at 53°C. PCR primers used were listed in Table S2. PCR products were resolved on a 2% agarose gel and stained with SYBR green.
RNA extraction and quantitative RT-PCR
Total RNA from 8-day-old Arabidopsis seedlings harvested at the indicated times in LL was extracted with the RNeasy plant mini kit (Qiagen). Reverse transcription using random hexamer primers was performed using Taqman® reverse transcription reagents (Applied Biosystems) according to the manufacturer’s protocol. Quantitative PCR reactions were performed using Applied Biosystems Taqman® PCR reagents and Taqman® gene expression probes for TOC1 (At02270094_g1), GI (At02305660_g1), LHY (At02174363_m1), CCA1 (At02173356_g1), DET1 (At02298846_g1) and COP10 (At02246513_g1). RT products from RNA samples without adding reverse transcriptase were used as negative controls. A 7900HT Fast Real-Time PCR system (Applied Biosystems) and its interface software were used to analyze the samples and data. Transcript expression levels were normalized to 18S.
Hypocotyl length and flowering time measurements
For hypocotyl measurements, at least 20 seedlings of the same genotype were scanned on a flatbed scanner and the hypocotyl lengths were measured using the ImageJ software (NIH). Flowering time measurements, as determined by the number of rosette leaves at bolting, were carried out as described previously (Wang and Tobin, 1998).
Supplementary Material
HIGHLIGHTS.
DET1, a CDD complex component, can direct transcriptional repression
DET1 interacts with CCA1 and LHY, which are core circadian components in plants
DET1 binds to target genes of CCA1/LHY and regulates their oscillations
DET1 is required for the repressive function of CCA1 in the circadian clock
ACKNOWLEDGMENTS
We thank E. Tobin for the GST-CCA1 plasmid, CCA1-OX seeds and antibodies to CCA1 and LHY; J. Chory for the CCA1-OX;det1-1 seeds; I. Carré for the anti-LHY antibody; X. Dong for yeast strain 122; Z. Liu for the LUG vector; M. Johnston for the LexA repression reporters; R.P. Hellens for the repression reporter in plants; X. Mo for the LUC constructs; X. Tian for the use of the luciferase imager; P.C.Y. Tang for advice on quantitative RT-PCR; N.-C. Paek and J. Zhou for discussion of unpublished data; and E. Abrash, A. Elling and N. Wei, for comments on the manuscript. O.S.L. was supported by the Croucher Foundation, the National Science Foundation (NSF) and the National Institutes of Health (NIH). J.-B.C. was in part supported by the Natural Sciences and Engineering Research Council of Canada. This research was supported by a NIH grant (GM-47850) and a NSF 2010 grant (MCB- 0929100) to X.W.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alabadi D, Oyama T, Yanovsky M, Harmon F, Mas P, Kay S. Reciprocal Regulation Between TOC1 and LHY/CCA1 Within the Arabidopsis Circadian Clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C. The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr Biol. 2002;12:1529–1534. doi: 10.1016/s0960-9822(02)01105-3. [DOI] [PubMed] [Google Scholar]
- Berloco M, Fanti L, Breiling A, Orlando V, Pimpinelli S. The maternal effect gene, abnormal oocyte (abo), of Drosophila melanogaster encodes a specific negative regulator of histones. Proc Natl Acad Sci USA. 2001;98:12126–12131. doi: 10.1073/pnas.211428798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. Chromatin techniques for plant cells. Plant J. 2004;39:776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
- Castells E, Molinier J, Benvenuto G, Bourbousse C, Zabulon G, Zalc A, Cazzaniga S, Genschik P, Barneche F, Bowler C. The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2 to maintain genome integrity upon UV stress. EMBO J. 2011;30:1162–1172. doi: 10.1038/emboj.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, Deng XW. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22:108–123. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, et al. Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development. Plant Cell. 2006;18:1991–2004. doi: 10.1105/tpc.106.043224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:3292–3297. doi: 10.1073/pnas.0400163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- de Montaigu A, Toth R, Coupland G. Plant development goes like clockwork. Trends Genet. 2010;26:296–306. doi: 10.1016/j.tig.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Dubin MJ, Kasten S, Nellen W. Characterization of the Dictyostelium homolog of chromatin binding protein DET1 suggests a conserved pathway regulating cell type specification and developmental plasticity. Eukaryot Cell. 2011;10:352–362. doi: 10.1128/EC.00196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the “TATA box”. Proc Natl Acad Sci U S A. 1984;81:7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Bastow RM, Davis SJ, Hanano S, McWatters HG, Hibberd V, Doyle MR, Sung S, Halliday KJ, Amasino RM, Millar AJ. The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell. 2003;15:2719–2729. doi: 10.1105/tpc.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW. HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 2000;19:4997–5006. doi: 10.1093/emboj/19.18.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. The circadian system in higher plants. Annu Rev Plant Bio. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U. Regulated proteolysis in light signaling. Curr Opin Plant Biol. 2005;8:469–476. doi: 10.1016/j.pbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW. Effect of Arabidopsis COP10 ubiquitin E2 enhancement activity across E2 families and functional conservation among its canonical homologues. Biochem J. 2009;418:683–690. doi: 10.1042/BJ20081943. [DOI] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell BB, Andersson CR, Poole DS, Kay SA, Chory J. HY5, Circadian Clock-Associated 1, and a cis-element, DET1 dark response element, mediate DET1 regulation of chlorophyll a/b-binding protein 2 expression. Plant Physiol. 2003;133:1565–1577. doi: 10.1104/pp.103.025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR, Gutierrez RA. Network news: prime time for systems biology of the plant circadian clock. Curr Opin Genet Dev. 2010;20:588–598. doi: 10.1016/j.gde.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- Monnier A, Liverani S, Bouvet R, Jesson B, Smith JQ, Mosser J, Corellou F, Bouget FY. Orchestrated transcription of biological processes in the marine picoeukaryote Ostreococcus exposed to light/dark cycles. BMC Genomics. 2010;11:192. doi: 10.1186/1471-2164-11-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher RA, Durrant WE, Wang D, Song J, Dong X. A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell. 2006;18:1750–1765. doi: 10.1105/tpc.105.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J. DET1, a negative regulator of light-mediated development and gene expression in arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- Pepper AE, Chory J. Extragenic suppressors of the Arabidopsis det1 mutant identify elements of flowering-time and light-response regulatory pathways. Genetics. 1997;145:1125–1137. doi: 10.1093/genetics/145.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick E, Lau OS, Tsuge T, Menon S, Tong Y, Dohmae N, Plafker SM, Deng XW, Wei N. Mammalian DET1 regulates Cul4A activity and forms stable complexes with E2 ubiquitin conjugating enzymes. Mol Cell Biol. 2007;27:4708–4719. doi: 10.1128/MCB.02432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plafker KS, Farjo KM, Wiechmann AF, Plafker SM. The human ubiquitin conjugating enzyme, UBE2E3, is required for proliferation of retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2008;49:5611–5618. doi: 10.1167/iovs.08-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Brickman JM, Lehming N, Ptashne M. New eukaryotic transcriptional repressors. Nature. 1993;363:648–652. doi: 10.1038/363648a0. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol. 2002;12:1462–1472. doi: 10.1016/s0960-9822(02)01106-5. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Deng XW. The COP/DET/FUS proteins-regulators of eukaryotic growth and development. Semin Cell Dev Biol. 2000;11:495–503. doi: 10.1006/scdb.2000.0203. [DOI] [PubMed] [Google Scholar]
- Song H-R, Carré IA. DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol Biol. 2005;57:761–771. doi: 10.1007/s11103-005-3096-z. [DOI] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z. Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA. 2004;101:11494–11499. doi: 10.1073/pnas.0403055101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Shirasu K, Deng XW. The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat Rev Genet. 2003;4:948–958. doi: 10.1038/nrg1228. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Yanagawa Y, Kwok SF, Matsui M, Deng XW. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 2002;16:554–559. doi: 10.1101/gad.964602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G, Dixon LE, Troein C, Millar AJ. Proteasome Function Is Required for Biological Timing throughout the Twenty-Four Hour Cycle. Curr Biol. 2011;21:869–875. doi: 10.1016/j.cub.2011.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Webster N, Jin JR, Green S, Hollis M, Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988;52:169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- Wei N, Kwok SF, von Arnim AG, Lee A, McNellis TW, Piekos B, Deng XW. Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell. 1994;6:629–643. doi: 10.1105/tpc.6.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O’Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303:1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- Yanagawa Y, Sullivan JA, Komatsu S, Gusmaroli G, Suzuki G, Yin J, Ishibashi T, Saijo Y, Rubio V, Kimura S, et al. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 2004;18:2172–2181. doi: 10.1101/gad.1229504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan JA, Hoecker U, Liu B, Xu L, Deng XW, Wang H. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell. 2005;17:804–821. doi: 10.1105/tpc.104.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Deng XW. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 2005;15:618–625. doi: 10.1016/j.tcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.