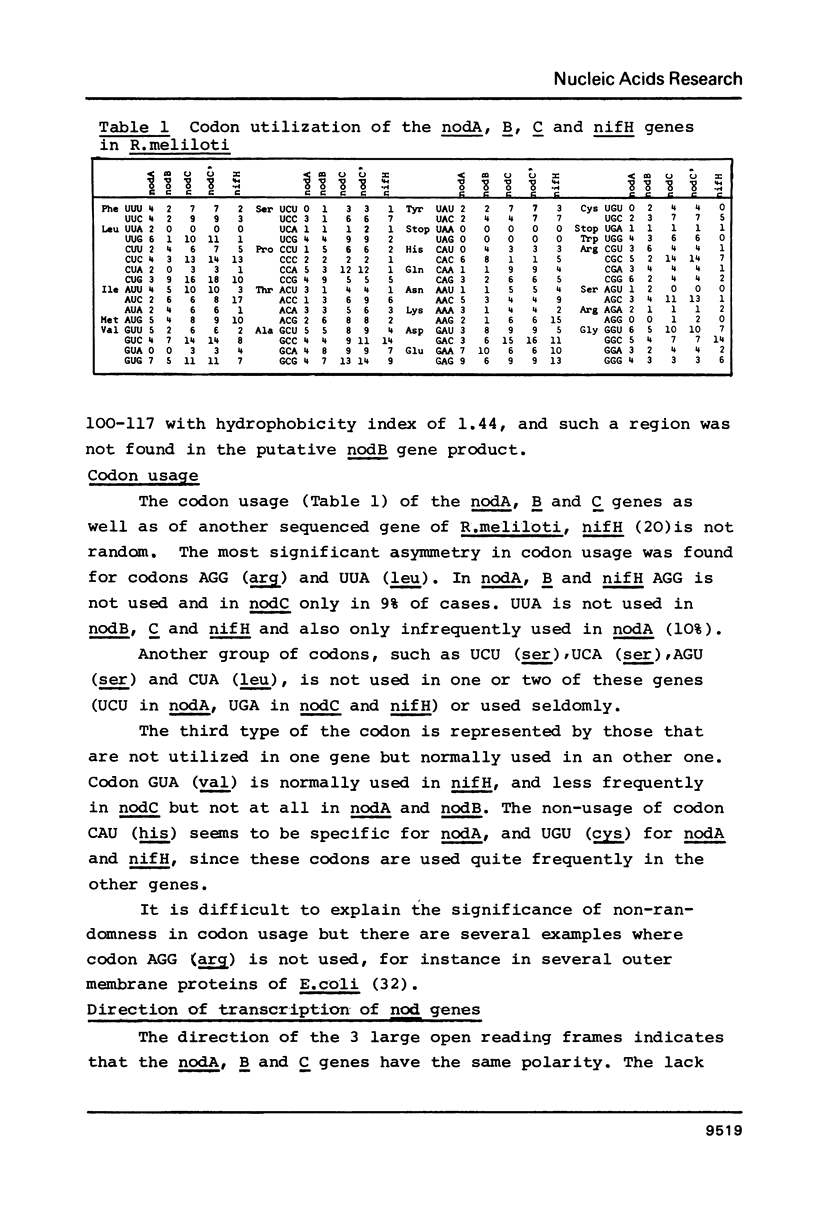
Abstract
A Rhizobium meliloti DNA region, determining nodulation functions common in different Rhizobium species, has been delimited by directed Tn5 mutagenesis and its nucleotide sequence has been determined. The sequence data indicates three large open reading frames with the same polarity coding for three proteins of 196, 217 and 402 (or 426) amino acid residues, respectively. We suggest the existence of three nod genes on this region, which were designated as nodA, B and C, respectively. Comparison of the R. meliloti nodA, B, C nucleotide and amino acid sequences with those from R. leguminosarum, as reported in the accompanying paper, shows 69-72% homology, clearly demonstrating the high degree of conservation of common nod genes in these Rhizobium species.
Full text
PDF













Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg D. E., Davies J., Allet B., Rochaix J. D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema W. J., Long S. R., Brown S. E., van den Bos R. C., Earl C., Ausubel F. M. Physical and genetic characterization of Rhizobium meliloti symbiotic mutants. J Mol Appl Genet. 1983;2(3):249–260. [PubMed] [Google Scholar]
- Bánfalvi Z., Sakanyan V., Koncz C., Kiss A., Dusha I., Kondorosi A. Location of nodulation and nitrogen fixation genes on a high molecular weight plasmid of R. meliloti. Mol Gen Genet. 1981;184(2):318–325. doi: 10.1007/BF00272925. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T., Grundström T., Jaurin B., Robinson J. J., Weiner J. H. Location and nucleotide sequence of frdB, the gene coding for the iron-sulphur protein subunit of the fumarate reductase of Escherichia coli. Eur J Biochem. 1982 Aug;126(1):211–216. doi: 10.1111/j.1432-1033.1982.tb06768.x. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J., Reeves P. Primary structure of the tolC gene that codes for an outer membrane protein of Escherichia coli K12. Nucleic Acids Res. 1983 Sep 24;11(18):6487–6495. doi: 10.1093/nar/11.18.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. V., Dixon B. Metastasis of a transplantable mammary tumour in rats treated with cyclophosphamide and/or irradiation. Br J Cancer. 1977 Aug;36(2):221–226. doi: 10.1038/bjc.1977.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S., Bergström S., Edlund T., Grundström T., Jaurin B., Lindberg F. P., Olsson O. Overlapping genes. Annu Rev Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- Overbeeke N., Bergmans H., van Mansfeld F., Lugtenberg B. Complete nucleotide sequence of phoE, the structural gene for the phosphate limitation inducible outer membrane pore protein of Escherichia coli K12. J Mol Biol. 1983 Feb 5;163(4):513–532. doi: 10.1016/0022-2836(83)90110-9. [DOI] [PubMed] [Google Scholar]
- Rosenberg C., Boistard P., Dénarié J., Casse-Delbart F. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet. 1981;184(2):326–333. doi: 10.1007/BF00272926. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Schmidt J., John M., Kondorosi E., Kondorosi A., Wieneke U., Schröder G., Schröder J., Schell J. Mapping of the protein-coding regions of Rhizobium meliloti common nodulation genes. EMBO J. 1984 Aug;3(8):1705–1711. doi: 10.1002/j.1460-2075.1984.tb02035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrest J. P., Feldmann R. J. Membrane proteins: amino acid sequence and membrane penetration. J Mol Biol. 1974 Aug 25;87(4):853–858. doi: 10.1016/0022-2836(74)90090-4. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Taylor W. R., Thornton J. M. Recognition of super-secondary structure in proteins. J Mol Biol. 1984 Mar 15;173(4):487–512. [PubMed] [Google Scholar]
- Török I., Kondorosi A. Nucleotide sequence of the R.meliloti nitrogenase reductase (nifH) gene. Nucleic Acids Res. 1981 Nov 11;9(21):5711–5723. doi: 10.1093/nar/9.21.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. H., Pankhurst C. E., Kondorosi A., Broughton W. J. Morphology of root nodules and nodule-like structures formed by Rhizobium and Agrobacterium strains containing a Rhizobium meliloti megaplasmid. J Cell Biol. 1983 Sep;97(3):787–794. doi: 10.1083/jcb.97.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Tamura T., Yokota T., Takano T. Overlapping genes in the heat-labile enterotoxin operon originating from Escherichia coli human strain. Mol Gen Genet. 1982;188(2):356–359. doi: 10.1007/BF00332701. [DOI] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]


