Abstract
The growth arrest and DNA damage–inducible 45 (Gadd45) proteins act in many cellular processes. In the liver, Gadd45b (encoding Gadd45β) is the gene most strongly induced early during both compensatory regeneration and drug-induced hyperplasia. The latter response is associated with the dramatic and rapid hepatocyte growth that follows administration of the xenobiotic TCPOBOP (1,4-bis[2-(3,5)-dichoropyridyloxy] benzene), a ligand of the nuclear receptor constitutive androstane receptor (CAR). Here, we have shown that Gadd45b–/– mice have intact proliferative responses following administration of a single dose of TCPOBOP, but marked growth delays. Moreover, early transcriptional stimulation of CAR target genes was weaker in Gadd45b–/– mice than in wild-type animals, and more genes were downregulated. Gadd45β was then found to have a direct role in transcription by physically binding to CAR, and TCPOBOP treatment caused both proteins to localize to a regulatory element for the CAR target gene cytochrome P450 2b10 (Cyp2b10). Further analysis defined separate Gadd45β domains that mediated binding to CAR and transcriptional activation. Although baseline hepatic expression of Gadd45b was broadly comparable to that of other coactivators, its 140-fold stimulation by TCPOBOP was striking and unique. The induction of Gadd45β is therefore a response that facilitates increased transcription, allowing rapid expansion of liver mass for protection against xenobiotic insults.
Introduction
Because of its pivotal role in metabolism, adult liver has a remarkable capacity to adapt. Normally quiescent, 2 distinct classes of stimuli — injury and primary mitogens — push hepatocytes into active proliferation. The injury response is compensatory. After physical, chemical, nutritional, vascular, bacterial, or viral injury, or experimentally-induced partial hepatectomy (PH), the liver restores its original mass through a combination of proliferation and growth (1, 2). In contrast, TCPOBOP (1,4-bis[2-(3,5)-dichoropyridyloxy] benzene) is a powerful mitogen that induces hyperplasia and growth in the absence of injury, a response that is faster than liver regeneration (3–5).
At least 2 factors mediate the early signaling after PH, TNF, and IL-6, which on binding to their receptors activate NF-κB and Stat3 within the hepatocyte. TCPOBOP activates a completely different early transcriptional response, because it is a ligand of the so-called constitutive androstane receptor (CAR). This nuclear receptor transcription factor binds TCPOBOP, translocates into the nucleus, and directly activates target genes with functions that range from detoxification of drugs to cell cycle activation (6). The remarkable liver growth observed in this response reflects synthesis of detoxifying enzymes, their coregulators, and a supporting structure of smooth endoplasmic reticulum and membrane vesicles (7). DNA synthesis and cell division follow, presumably a further adaptation that makes the enlarged liver more efficient. CAR-mediated growth was first discovered as a response to phenobarbital, although this drug is not a direct ligand of CAR (7). Notably, both phenobarbital and TCPOBOP are powerful promoters of carcinogenesis (4), in contrast to the weak promotional effect of repeated partial hepatectomy (8).
Gene expression profiles originally identified Gadd45b (also known as MyD118) as the gene with the strongest early induction common to both PH and TCPOBOP treatment (9). Gadd45β is an 18-kDa protein closely related to Gadd45α and Gadd45γ/CR6—all 3 expressed by inducible genes and associated with diverse biological responses (10–13). Each growth arrest and DNA damage (Gadd45) protein has a different pattern of cell-specific and induced expression, presumably reflecting different signaling pathways (14, 15). Despite their similarity and small size, these proteins encompass a wide variety of activities. Gadd45β inhibits apoptosis by directly binding to the Jun kinase kinase MKK7/JNKK2 (16–19), but studies in other cells show increased apoptosis (20). Gadd45β binds MEK1/MEKK4 and activates p38 (21, 22). It also functions in terminal differentiation (23), cell cycle arrest (13), growth inhibition (24), and increased cell proliferation via binding to PCNA, Cdc2, and p21 (13). In addition, all 3 Gadd45 proteins can act as transcriptional coactivators of nuclear receptor transcription factors (25, 26).
In liver regeneration, 2 distinct pathways activate transcription of Gadd45b: NF-κB activates via a regulatory region upstream of the promoter (18, 19), and TGF-β activates via a downstream enhancer that responds to Smad3 and Smad4 (20, 27). NF-κB activation is an early response to TNF signaling, and TGF-β is released into the local circulation within 1 hour of PH (1). In contrast, TCPOBOP mediates direct stimulation of Gadd45β via CAR, independent of NF-κB and Smad3/4. This stimulation is absent from the CAR-null mouse and mediated by a specific CAR binding site (ref. 28 and our unpublished observations).
A recent study of liver regeneration in a Gadd45b–/– mouse demonstrated impaired proliferation, reduced growth, and increased cell death after PH (29). Here we report the investigation of TCPOBOP-induced hyperplasia in the Gadd45b–/– mouse (30, 31). In contrast to the findings after liver regeneration, loss of Gadd45β did not reduce hepatocyte proliferation. However, there was marked impairment of the rapid liver growth that characterizes this hyperplasia response, accompanied by reduced early transcription of most genes activated by TCPOBOP. CAR is a transcription factor, and the findings suggested that the absence of Gadd45β impaired its transcriptional stimulation. We therefore investigated direct interaction between Gadd45β and CAR, transcriptional effects of this interaction, and compensatory changes in the Gadd45b–/– mouse.
Results
Liver phenotype in untreated Gadd45b–/– mice.
We first analyzed the livers of untreated adult Gadd45b–/– mice and found no pathological changes or altered accumulation of fat or glycogen (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI38760DS1). However, Gadd45b–/– livers were slightly heavier and had smaller hepatocytes (P < 0.004), despite moderate increases in binucleate and tetraploid forms. The livers from Gadd45b–/– mice had a low basal level of proliferating hepatocytes expressing Ki67, essentially the same as those of wild-type mice (1.1%–1.2%; Supplemental Figure 2). Labeled stromal cells were more frequent than hepatocytes but also without obvious difference in Gadd45b–/–, although cells were not quantified. There was no significant apoptosis of hepatocytes — the TUNEL reaction showed less than 1 labeled hepatocyte per tissue section (data not shown).
Gene expression profiles of untreated mice showed moderate differences between Gadd45b–/– and wild-type mice (Supplemental Table 1). Gadd45b–/– mice had 30 upregulated and 23 downregulated genes with 2-fold differences of expression compared with wild-type. Among these genes, Ppara downregulation in Gadd45b–/– mice was accompanied by lower expression of 2 PPARα regulatory targets, Acox1 and Cd36. These changes could decrease fatty acid uptake and β-oxidation and thus reduce production of reactive oxygen. This effect might explain the observed downregulation of Mt1 and Mt2, which are related to lipid peroxidation, and Gsta1 and Gsta2, which respond to oxidative stress. In the group of up-regulated genes, the inclusion of Insig2, Apoa4, Apom, Fabp5, Mvk, Pmvk, Ugt2b37, Cyp2g1, Cyp2c39, and Hsd17b2 is suggestive of increased cholesterol synthesis and altered lipid metabolism and could represent compensatory responses to reduced PPARα signaling. In wild-type liver, the basal expression of Gadd45β was low but comparable to other transcriptional regulators (see below). The smaller hepatocytes and altered gene expression of Gadd45b–/– mice suggest that Gadd45β contributes to the normal liver phenotype either at its uninduced levels or after transient induction from various stimuli during normal liver homeostasis.
Proliferative response to TCPOBOP treatment.
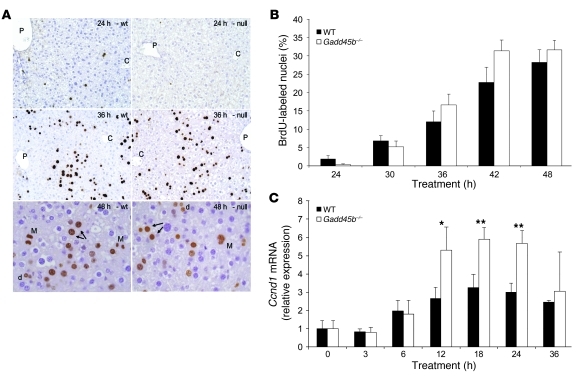
To investigate the proliferative response, mice received BrdU continuously in the drinking water starting 48 hours before TCPOBOP treatment (Figure 1). Analysis after treatment revealed only subtle differences between Gadd45b–/– and wild-type mice. Both showed midzonal proliferation with complete cell cycles in hepatocytes of different ploidy, including diploid cells. Wild-type animals showed more proliferating cells at 24 hours, whereas Gadd45b–/– mice had moderately increased proliferation at 42 and 48 hours. At 48 hours, mitoses were abundant, greater than 1 per high-power field in both wild-type and Gadd45b–/– mice (1.1 ± 0.7 and 1.5 ± 1.0, respectively; mean ± SEM); however, these differences were not statistically significant. Since cyclin D1 (encoded by Ccnd1) is a key regulator of the G1-S transition, we used real-time PCR and Western blot analysis to measure expression after TCPOBOP treatment (Figure 1C and Supplemental Figure 3). Induction was apparent in both wild-type and Gadd45b–/– mice at 6 hours with peak levels at 12–18 hours, but was twice as strong in the Gadd45b–/– mouse. Apoptosis was not prominent in the response induced by TCPOBOP, and no increase was apparent in Gadd45b–/– mice (Supplemental Figure 4). The inhibition of cell proliferation and increased cell death previously found in Gadd45b–/– mice after PH (29) were not observed in TCPOBOP-induced hyperplasia.
Figure 1. Proliferative response to TCPOBOP in wild-type and Gadd45b–/– mice.
(A) Histological patterns of proliferation. Early labeling of a few hepatocytes was apparent at 24 hours (top) only in the wild-type mouse, but labeling was similar at 36 hours (middle). Both early and later labeling was predominantly midzonal (P, portal; C, central). At 48 hours (bottom), labeled mitoses (M) and daughter cell pairs (arrows) were apparent in both wild-type and Gadd45b–/– animals. These could be identified in hepatocytes of varying ploidy, including diploid cells (d). Original magnification, ×200 (top and middle); ×400 (bottom). (B) Cell counts. S-phase (BrdU-labeled) nuclei are shown as a percentage of total hepatocytes, counted from 20 random high-power fields from 3–5 livers per group per time point. (C) Ccnd1 mRNA expression. Real-time RT-PCR analysis was carried out on RNA from 3–5 livers per time point. Data are mean ± SD. *P < 0.05, **P < 0.01, Gadd45b–/– vs. wild-type, t test.
Growth and global transcription.
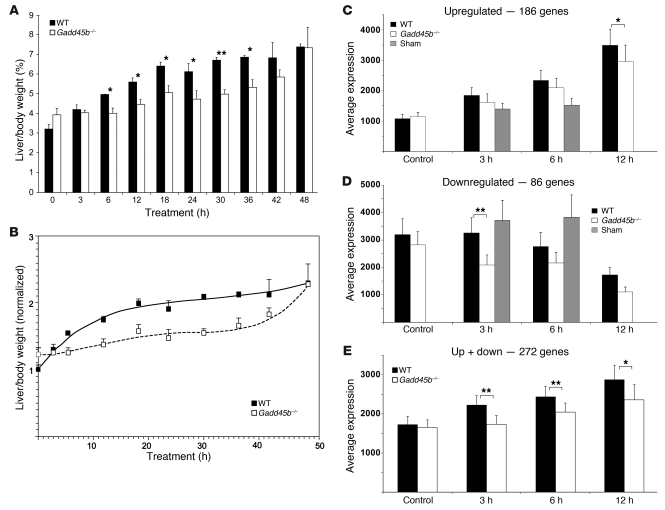
In addition to effects on proliferation, TCPOBOP also induced rapid liver growth. Treatment caused a 30% increase in wild-type liver mass after only 3 hours (P < 0.04), which doubled by 18 hours (P < 0.002; Figure 2, A and B). Growth temporarily paused as the cells underwent S phase, but then resumed after cell division. The massive early growth reflects high-level synthesis of detoxification proteins, their coregulators, and supporting organelles. Although livers from untreated Gadd45b–/– mice were 22% larger than those of wild-type mice, their induced growth prior to S phase was greatly impaired. The wild-type mouse took 6 hours to increase liver mass by 50%, whereas Gadd45b–/– mice required 18 hours for the same increase. Nevertheless, growth caught up by 48 hours.
Figure 2. Growth and transcriptional effects of TCPOBOP treatment.
(A) Average liver mass as a percentage of body weight. 3–5 livers were studied per time point. Data are mean ± SEM. *P < 0.05, **P < 0.01, Gadd45b–/– vs. wild-type, unpaired t test. (B) Growth curves were calculated from the data in A. (C) Upregulation of a set of 186 genes (Supplemental Table 2). (D) Downregulation of a set of 86 genes (Supplemental Table 3), analyzed as in C. (E) Average total transcription. Averaging of expression from both sets of regulated changes demonstrated net upregulation that was significantly greater in the wild-type mouse at all 3 time points. (C–E) All arrays were studied using 2-color hybridization, with experimental RNA as the red probe and pooled RNA from normal wild-type livers as the green probe. For this analysis, the intensities detected on individual arrays were normalized to the average green intensity of the entire set of arrays. Hybridization intensities ranged from a low intensity threshold of 100 to a maximum of 60,000. The values of all genes in the set were averaged at each time point. Data are mean ± SEM. *P < 0.05; **P < 0.01, Gadd45b–/– vs. wild-type, paired t test.
In PH-induced liver regeneration, TNF signaling activates Jnk phosphorylation. Because Gadd45β inhibits this phosphorylation by binding Mkk7, the Gadd45b–/– mouse shows sustained phosphorylation of Jnk1/2 after PH (29). However, TNF activation does not occur in TCPOBOP-induced hyperplasia (3, 28). Consistent with these prior observations, we were unable to demonstrate Jnk phosphorylation at any time up to 48 hours after treatment in either Gadd45b–/– or wild-type livers (Supplemental Figure 5). This negative result highlighted an important difference between hyperplasia and liver regeneration and ruled out increased Jnk activation as the mechanism of altered growth.
Since TCPOBOP is a ligand of CAR, the early synthetic response consists predominantly of direct transcriptional targets. We used comprehensive genome-wide expression profiles to characterize the transcriptional responses of Gadd45b–/– and wild-type mice at 3, 6, and 12 hours (Supplemental Figure 6 and Supplemental Tables 2 and 3) and compared these with untreated controls and with sham treatments of 3 and 6 hours. We chose a 2-fold change in gene expression (i.e., >2× or <0.5× normal liver) as the cutoff, because this threshold usually discriminates statistically significant changes of gene expression detected on microarrays (32). Genes with 2-fold expression changes — present in any treatment interval of either wild-type or Gadd45b–/– liver — comprised sets of 186 upregulated and 86 downregulated genes. The down responses were presumably indirect, since CAR is a transcriptional activator; indeed, the early (3 hours) downregulation response in the wild-type was very limited. This large set of expressions allowed comparison of the general transcriptional responses to TCPOBOP (Figure 2, C–E, and Supplemental Figure 6). Both the number of upregulated genes and their average expression were greater in wild-type mice at all time points (Figure 2C). In contrast, more genes were downregulated in Gadd45b–/– mice, and their average expression was lower (Figure 2D). When the 2 sets were averaged together, the effect was an increase in total transcription consistent with liver growth (Figure 2E), but the net average was significantly less in Gadd45b–/– animals at all time points.
Specific transcriptional responses.
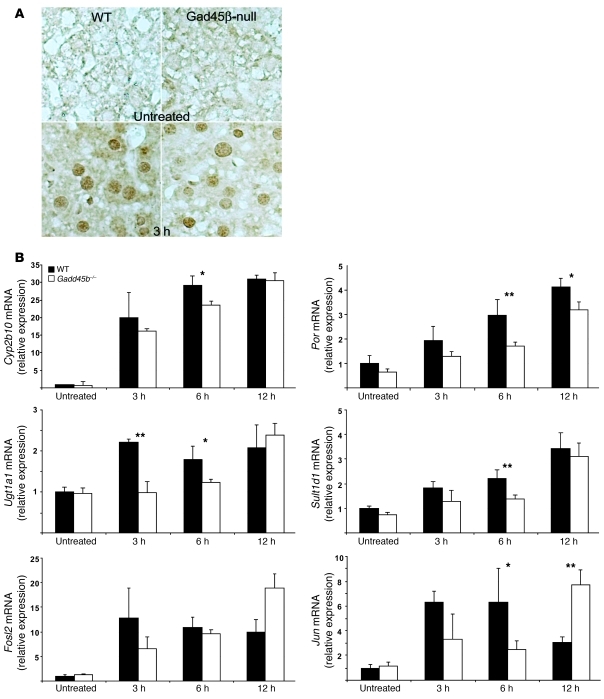
Although the extensive changes in transcription suggested that Gadd45β deficiency affects CAR function, we found that TCPOBOP treatment induced equivalent translocation to the nucleus in both Gadd45b–/– and wild-type mice (Figure 3A and Supplemental Figure 7). Analysis by quantitative RT-PCR then confirmed significantly reduced expression of 4 classical CAR target genes, selected from the microarray profiles to represent strong and weak transcriptional induction or strong and weak effects of the Gadd45b mutation (Figure 3B). Phenobarbital-responsive cytochrome p450 2b10 (Cyp2b10), the prototype gene for direct regulation by CAR (33), had the third-strongest induction in the dataset, while P450 oxidoreductase (Por) and Sulfotransferase 1d (Sult1d1) had intermediate expression levels. UDP glucuronyl transferase 1a1 (Ugt1a1) had low basal expression and weak induction. All 4 genes showed reduced stimulation in Gadd45b–/– mice at 3 and 6 hours that was moderated by 12 hours. Further analysis of the array datasets failed to show correlation of the degree of inhibition in the Gadd45b–/– mouse with the strength of gene expression or with the magnitude of induction (data not shown). The exact pattern of change therefore differed for each gene.
Figure 3. Transcriptional regulation by CAR.
(A) CAR translocation. Immunoperoxidase detected a weak diffuse reaction to CAR in the cytoplasm of untreated hepatocytes. 3 hours after treatment with TCPOBOP, there was clear demonstration of nuclear CAR. Gadd45b–/– and wild-type genotypes were indistinguishable. Original magnification, ×400. (B) Quantification of 6 representative mRNAs. Real time RT-PCR analysis was carried out as in Figure 1C. Data are mean ± SD. *P < 0.05; **P < 0.01, Gadd45b–/– vs. wild-type, t test.
Jun and Fos-like 2 (Fosl2) are early transcription factors, direct transcriptional targets of CAR that contributed to subsequent cell cycle activation (our unpublished observations). These both showed attenuated stimulation in Gadd45b–/– mice at 3 hours, but overstimulation at 12 hours.
Coactivation.
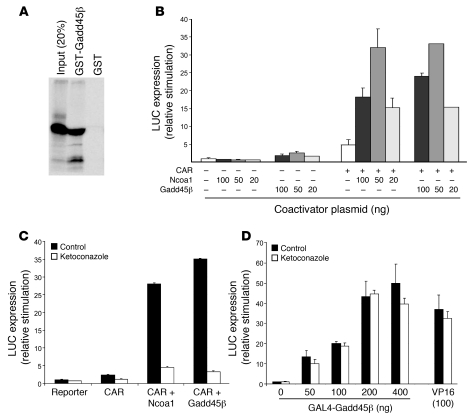
Gadd45β is a multifunctional protein with a variety of cellular binding partners. Reduced early transcription in Gadd45b–/– mice suggested that Gadd45β has a role in transcriptional stimulation of CAR-regulated genes. Indeed, the protein has LXXLL motifs that are found in many transcriptional coactivators, and Yi et al. showed direct binding of Gadd45 proteins to nuclear receptor transcription factors, with transcriptional coactivation (25). Moreover, a recent study showed that Gadd45β coactivates CAR-mediated transcription in reporter assays (26). To explore coactivation, we first used a GST fusion protein to demonstrate strong direct binding of CAR and Gadd45β (Figure 4A). Cotransfection assays combined Gadd45β and CAR expression plasmids with a reporter construct derived from Cyp2b10, the best-characterized CAR target gene. Gadd45β synergistically coactivated this reporter with activity comparable to that of Ncoa1 (also known as Src1), a primary coactivator of nuclear receptors (Figure 4B and Supplemental Figure 8). Ketoconazole, which binds to a region of the CAR ligand-binding domain and blocks binding of Ncoa1 (34, 35), also blocked Gadd45β-mediated coactivation (Figure 4C). To determine whether Gadd45β has intrinsic transcriptional activity, a Gal4 DNA-binding domain (Gal4-DBD) was fused to full-length Gadd45β. The fusion protein activated a Gal4-specific reporter plasmid (Figure 4D), comparable to the strong generic activator Gal4-VP16. Ketoconazole did not block this activation, which indicates that the drug had no direct inhibitory effect on Gadd45β and that the activation function of Gadd45β was independent of CAR. Gadd45β therefore coactivates via an intrinsic activation function that it brings to the transcription factor.
Figure 4. Transcriptional coactivation by Gadd45β.
(A) Direct binding of CAR to Gadd45β. A plasmid expressing a GST-binding domain fused to full-length Gadd45β was expressed in E. coli. Fusion protein or a control GST-binding domain protein was bound to glutathione agarose beads and then incubated with 35S-CAR prepared by cell free translation. After wash, the bound protein was eluted and resolved by acrylamide gel electrophoresis. The control lane contained a 20% input fraction. (B) Comparison of coactivation by Gadd45β and Ncoa1. Cotransfection experiments were set up with a limiting amount of CAR expression plasmid (10 ng) to display maximal coactivation. (C) Inhibition of coactivation by ketoconazole (25 μM). Transfection assays used 10 ng CAR, 100 ng Ncoa1, or Gadd45β expression plasmids. (D) Intrinsic activation by DNA-bound Gadd45β. A plasmid expressing a Gal4-DBD fused to the N terminus of full-length Gadd45β was cotransfected with a Gal4 site LUC reporter, with and without ketoconazole. The strong generic activator Gal4-VP16 (100 ng) is also shown for comparison. (B–D) Data are mean ± SD of duplicate assays in HepG2 cells.
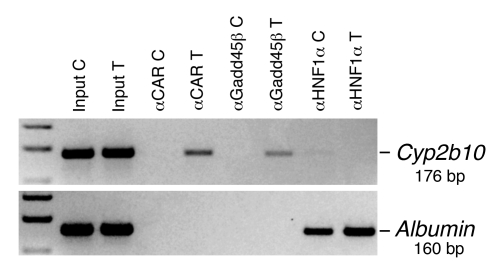
ChIP demonstrated that Gadd45β binds CAR in vivo on the phenobarbital response element of Cyp2b10 (36). Since Gadd45β and CAR Ab have been problematic, we first screened a series to find reagents with adequate affinity (Supplemental Figure 9), after which ChIP was performed on nuclei isolated from untreated and TCPOBOP-treated livers (Figure 5). Both CAR and Gadd45β Abs precipitated the Cyp2b10 regulatory region, but only after TCPOBOP treatment. Control immunoprecipitation verified the specificity of these assays. Under both conditions, Cyp2b10 was negative for HNF1α and the Albumin promoter was positive, confirming that the chromatin preparations were of equal quality. Thus, Gadd45β has all essential properties of a coactivator and collaborates with CAR in vivo to regulate transcription after TCPOBOP treatment.
Figure 5. Recruitment of CAR and Gadd45β to an activated gene in vivo.
Control untreated (C) and TCPOBOP-treated (T) liver segments were rapidly fixed and then subjected to ChIP analysis using Abs to CAR, Gadd45β, and HNF1α. The phenobarbital response element of Cyp2b10, an upstream enhancer, was detected as a 176-bp PCR amplimer of the gene region from –2,103 to –2,128 bp. The HNF1α-binding site of the Albumin gene promoter was detected as a 160-bp amplimer of the gene region from –210 to –51.
Transcriptional regulatory domains of Gadd45β.
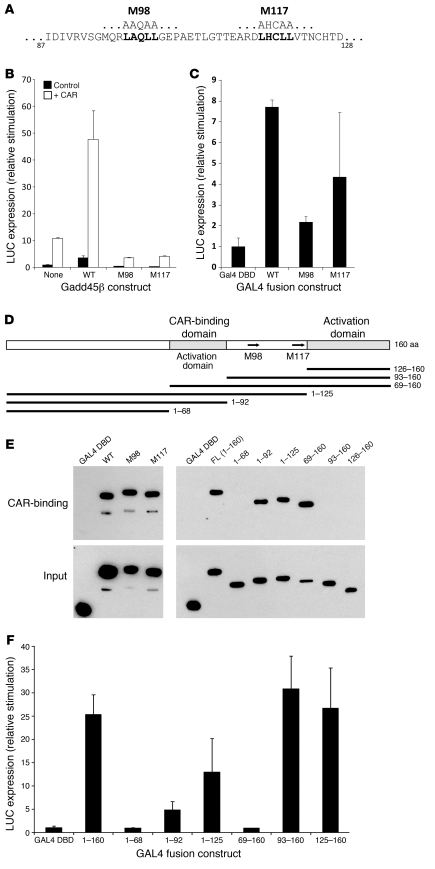
To substantiate the mechanism of coactivation, we first mutated the 2 LXXLL motifs (Figure 6, A–D). Each mutation converted Gadd45β into a dominant negative that inhibited CAR-mediated activation (Figure 6B). Intrinsic activation function did not require CAR binding (Figure 4D), and both mutations weakened — but did not abolish — CAR-independent activation by Gal4 fusion proteins (Figure 6C). To study direct binding to CAR, we initially used cell-free translation of GST-domain fusion proteins, reversing the strategy of Figure 4A, (i.e., binding multiple forms of Gadd45β to GST-CAR immobilized on glutathione agarose). However, there was a high level of nonspecific binding that might have obscured small differences among the constructs (data not shown). An alternate approach used proteins synthesized in vivo in 293T cells, 6His-CAR and Gadd45β constructs fused to the Gal4-DBD (Figure 6, D and E). The binding assays confirmed that mutation of the LXXLL motifs did not affect binding to CAR, consistent with their dominant-negative effect. A series of truncations then enabled mapping of CAR-binding and activation domains. The aa 69–92 region bound CAR (Figure 6, E and F). This region also mediated weak activation, but the main activation domain was the C-terminal region, aa 125–160. However, it should be noted that the aa 69–160 construct failed to show activation, which suggests that its altered structure interfered with coactivation but not with binding to CAR. The detailed analysis confirmed that Gadd45β is a true coactivator by showing that distinct functions required for coactivation reside in different parts of the peptide.
Figure 6. Localization of transcriptional functions to specific regions of Gadd45β.
(A) LXXLL motifs in Gadd45β. Motifs at aa 98 or 117 were mutated (M) by substituting Ala for each Leu in the motif. (B) Reporter assays of coactivation by wild-type and mutant Gadd45β. Expression plasmids were cotransfected with CAR or empty vector, in combination with the Cyp2b10 LUC reporter construct. (C) Direct activation assays. Full-length wild-type or mutant Gadd45β was fused with a Gal4-DBD, and assayed for transcriptional activation using a Gal4-binding site LUC reporter. (D) Map of deletion constructs, with deduced functional domains. Arrows show the position of LXXLL domains within the 160-aa Gadd45β peptide. (E) Mapping of the CAR-binding domain. 293T cells were transfected with expression plasmids for fusion proteins, the GAL4-DBD combined with full-length wild-type or mutated Gadd45β protein, or segments retained in deletion constructs (FL, full length). A plasmid expressing 6His-CAR was transfected into separate cells. Cell extracts were mixed, and 6His-tagged proteins were captured by passing over a Ni-affinity matrix. Eluted proteins were resolved on SDS-acrylamide gels, and products were detected by Western blot using an Ab for the Gal4-DBD. (F) Mapping of the activation domain. Transcriptional activation by Gal4-DBD fusion constructs was assayed as above. (B, C, and F) Data are mean ± SD calculated from duplicate assays in HepG2 cells.
Compensatory changes in coactivation?
Because growth of Gadd45b–/– liver catches up to the wild-type liver by 48 hours, we investigated whether altered expression of other coactivators might compensate for the lack of Gadd45β (Figures 7 and 8 and Supplemental Figure 10), starting with a comprehensive list of 18 coactivators that contain the LXXLL motif. The list included the 3 homologous Gadd45b factors as well as coactivators found by microarray studies to be induced by TCPOBOP treatment, notably Gadd45a and Nrbf2. Carm1 and Kat2b (also known as Pcaf), 2 additional coactivators that lack LXXLL, were also included because they interact with nuclear receptors (37).
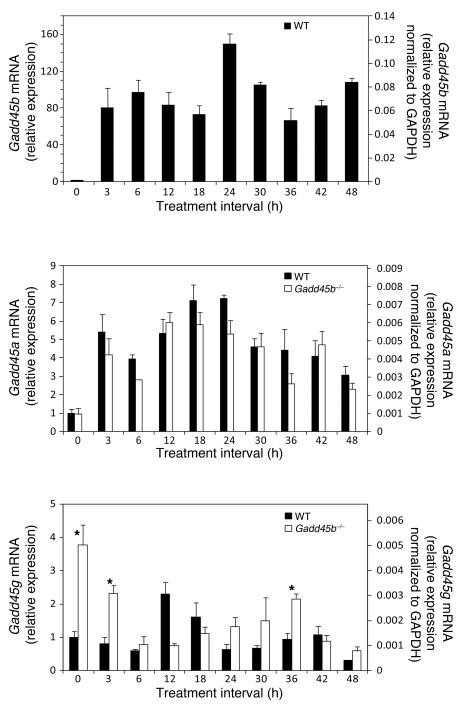
Figure 7. Expression of Gadd45b, Gadd45a, and Gadd45g mRNA in TCPOBOP-induced hyperplasia.
Quantitative RT-PCR measurements were normalized to both untreated wild-type control liver and GAPDH mRNA (left and right y axes, respectively). Data are mean ± SD, calculated from assays of 3–5 specimens per group. *P < 0.05, Gadd45b–/– vs. wild-type, t test.
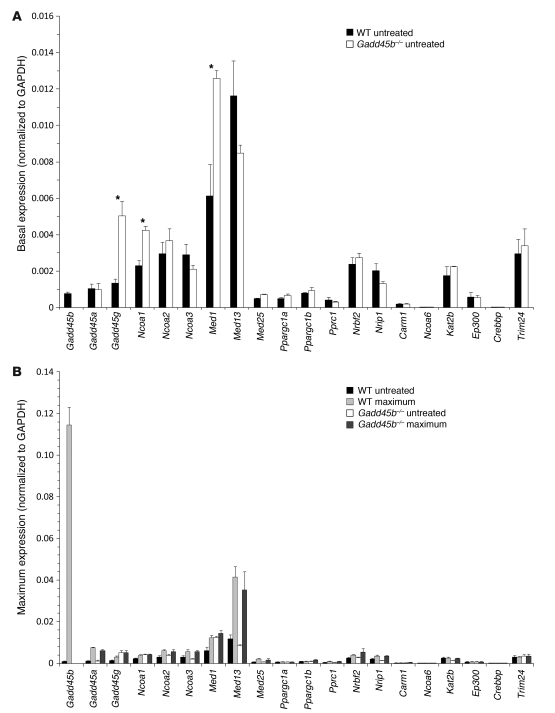
Figure 8. Coactivator mRNA responses in TCPOBOP-induced hyperplasia.
Comparison of basal (A) and maximal (B) mRNA levels of 20 coactivators after TCPOBOP treatment in untreated wild-type and Gadd45b–/– livers. The full time course for each mRNA is shown in Supplemental Figure 10. Data are mean ± SD, calculated from assays of 3–5 specimens per group. *P < 0.05, Gadd45b–/– vs. wild-type, t test.
In untreated normal liver, Gadd45b was expressed at the level of many other coactivators (Figure 7, Figure 8A, and Supplemental Figure 10), which may explain the phenotypic changes in the Gadd45b–/– liver. Gadd45b and Gadd45a were both induced by TCPOBOP, but induction of Gadd45b was extraordinary (140-fold vs. 7-fold). Gadd45g — which was increased in untreated Gadd45b–/– mice — showed moderate induction in both genotypes, although with different time courses. Gadd45b–/– mice also showed significant baseline increases in Ncoa1 and Med1, suggestive of compensation for the absence of Gadd45β. However, the maximal induced levels of all coactivators were comparable in Gadd45b–/– and wild-type animals. The stimulated levels of Gadd45b mRNA far exceeded that of any other coactivator; indeed, this induced level exceeded the sum of all other coactivators in the cell.
Discussion
Biological significance of CAR-mediated growth.
Broad interest in liver regeneration has overshadowed drug-induced hyperplasia, except for tumor promotion. Hyperplasia and regeneration are mediated by different pathways so that each has distinctive kinetics of growth and cell proliferation (3, 5, 9), exemplified by the rapid growth after TCPOBOP treatment.
TCPOBOP selectively activates CAR, and much of the growth is caused by induction of enzymes that mediate detoxification. The induced cell division is most likely a mechanism to stabilize and further increase the new liver mass. CAR-induced hyperplasia is thus adaptive, a rapid response to toxic xenobiotic agents. The pure response, as mediated by TCPOBOP, occurs without measurable liver injury or induction of inflammatory mediators. Nevertheless, some of the natural agents that selected this adaptation might simultaneously induce hyperplasia and injure hepatocytes. CAR indirectly mediates the hepatic toxicity of acetominophen — not itself a ligand or inducer — because p450 enzymes stimulated by CAR metabolize acetominophen to N-acetyl-p-benzoquinone imine (NAPQI) (38). This coincidental toxicity may be clinically important, but it is maladaptive and presumably had little effect on the natural selection that facilitated CAR-induced hyperplasia.
In addition to TCPOBOP, numerous CAR activators are known ligands and indirect activators, and some are species specific. Natural CAR activators include quercetin, kaempferol, and diallyl sulfide (39, 40). Manmade inducers include phenobarbital, phenytoin, chlorinated pesticides, polychlorinated biphenyls, meclizine, chlorpromazine, nonylphenol, metamizole, pthalate, di(2-ethylhexyl) phthalate, and 6-(4-chlorophenyl:imidazo[2,1-b]thiazole-5carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO) (41–49). None is an obvious toxin. Thus, no agent seems to fit the scenario that combines direct liver injury and CAR-induced hyperplasia. The survival value of this hepatic response therefore goes beyond its local effect on liver cells because many organs are subject to drug-induced intoxication or injury. The rapid CAR-induced synthesis that deactivated incapacitating or injurious agents was a striking adaptive response to a single exposure. The CAR-induced transcriptional response was also striking, and Gadd45β was at the top of the list of induced genes. Our results indicate that this induction is a feed-forward mechanism, immediate synthesis of a coactivator that facilitates CAR-mediated transcription.
Gadd45β expression in hepatocyte proliferation and survival.
The nearly normal DNA labeling in Gadd45b–/– mice during hyperplasia was particularly important, because it showed that Gadd45β was not a fundamental regulator of proliferation (Figure 1). Moreover, the eventual proliferation and growth were the same in Gadd45b–/– and wild-type mice, which indicates that homeostatic regulation overcomes differences in the rates of transcription and synthesis. In a recent study of liver regeneration, a Gadd45b–/– mutant had selective overactivation of the MKK7/JNK pathway, but combination with a Jnk2-null mutation blocked this activation and restored normal proliferation without Gadd45β (29). The reduced proliferation observed after PH is therefore likely to be an indirect effect. Moreover, this study characterized proliferation and injury markers only at a single time point, 48 hours after treatment. Thus, an effect on early growth and transcription may still be present after PH.
Gadd45b–/– mice showed increased proliferation, apparent in both labeling index and mitotic count, although the differences compared with wild-type in our small experimental groups were not statistically significant. However, the significant overexpression of cyclin D1 in the Gadd45b–/– liver confirmed a stronger proliferative signal. Since AP1 factors are known regulators of Ccnd1, the increased stimulation could result from increased expression of Jun and Fosl2. These latter genes are CAR targets (our unpublished observations), but the delayed onset of their increased expression suggested that Gadd45β normally stimulates partial repression of these genes, or alternatively inhibits a second activation mechanism that potentiates CAR-mediated stimulation. In either case, the findings suggest coordination that allows growth via synthesis of CAR regulatory targets to precede cell proliferation, since the latter process will reprioritize cellular resources. Earlier or increased proliferation might therefore attenuate the rapid adaptive response to xenobiotics.
CAR signaling also protects hepatocytes from cell death, presumably with the collaboration of Gadd45β (50). Nevertheless, the finding that TCPOBOP did not induce apoptosis highlights a fundamental difference in the 2 models of proliferation. Liver regeneration occurs in a setting of impaired liver function and injury — the liver must maintain function and compensate for injury before it can proliferate. In contrast, hyperplasia is a simpler response by a healthy liver.
Coactivation.
Although recent papers have emphasized p38/MAPK signaling and antiapoptotic functions, the widespread transcriptional changes in Gadd45b–/– mice led us to consider the role of Gadd45β as a coactivator, an effect demonstrated in an early paper by Yi et al. (25). Our studies demonstrated relationships with CAR, and a recent paper by Kodama and Negishi showed that yet another nuclear receptor, PXR, stimulates Gadd45β expression and then interacts with the protein after it is expressed (51).
The 3 closely related 18-kDa Gadd45 proteins all bind nuclear receptors and coactivate them, effects suggested by the presence of LXXLL peptide motifs. Yet they are atypical, much smaller than 160-kDa Ncoa1, which mediates comparable coactivation of CAR. Gadd45β has 2 such motifs, at aa 98 and aa 117, but only the former is conserved in Gadd45γ and the latter in Gadd45α. In Ncoa1, the LXXLL motifs are part of the domain that binds to CAR. However, mutation of the LXXLL motifs of Gadd45β did not block its robust binding to CAR. Mutation instead converted Gadd45β to a dominant negative, an effect also found for the coactivator Nrbf2 (52, 53). Further analysis resolved separate domains that mediated binding to CAR and transcriptional activation and showed that the LXXLL-containing region between them was essential for integration of these functions. A prior study of Mkk7 binding showed that a nearly identical Gadd45β mutation at aa 98 did not block interaction (16), so this mutant may be invaluable for distinguishing the effects of Gadd45β on different regulatory pathways.
Compared with other coactivators of nuclear receptors, Gadd45β did not have a chromatin-modifying domain, like the histone acetyl transferase domain of Ncoa1 or the histone arginine methyltransferase of CARM1. The intrinsic activation function of the Gadd45β C-terminal domain therefore suggests that this region acts by recruiting another coactivating partner. Defining this second partner will be critical for explaining how such a small protein mediates strong coactivation.
Liver growth, compensatory effects, and pathway integration.
As a transcriptional activator, CAR stimulated expression of numerous genes in the liver, and most showed reduced stimulation in the absence of Gadd45β. The effect varied from gene to gene, but without Gadd45β, fewer were stimulated and average stimulation was reduced. TCPOBOP also induced CAR-dependent downregulation of some genes, but discerning the mechanism of downregulation is problematic — presumably indirect — since CAR is a transcriptional activator. More genes were downregulated in the Gadd45b–/– mouse, even at 3 hours (Figure 2, Supplemental Figure 4, and Supplemental Table 3), and it is difficult to relate the functions of the diverse genes that showed mutant-specific effects. The relationship may be only that these genes competed poorly for coactivators with the newly stimulated genes, a competition that was accentuated in the Gadd45b–/– mutant.
A single dose of TCPOBOP had a powerful and persistent effect on liver mass. By 48 hours, after completion of the first cell cycle, Gadd45b–/– liver mass was 2.3 times that of control, although the growth temporarily leveled from about 18–42 hours, presumably an accommodation for S phase and cell division. A previous study showed a 3-fold increase in liver mass at 5 days after treatment and demonstrated that phenobarbital, more widely studied than TCPOBOP, mediates a similar but somewhat weaker response (54). Studies from the Michalopoulos laboratory have introduced the hepatostat concept to objectify the homeostatic processes that normalize liver weight in response to body mass and physiological processes (54–56). CAR signaling might seem to reset the hepatostat, but studies with phenobarbital show the effect is temporary. Prolonged treatment with phenobarbital causes the liver to reach a larger steady-state size, but it shrinks to its original mass after treatment is stopped. The hepatostat apparently regulates size and synthesis, but not cell death, because the effect occurs by reduction of cell size, but not cell number (57).
Liver mass in the Gadd45b–/– mouse caught up to that of wild-type by 48 hours, which suggests that the level of growth is determined by a combination of intrinsic signal from CAR and extrinsic signals from the hepatostat. We therefore conclude that the hepatostat responds to liver function, not growth rate. In Gadd45b–/– mice, the growth delay occurred prior to S phase, and although several coactivators responded to CAR stimulation, there was little compensatory increase of any coactivator to replace Gadd45β. Its dramatic induction in the wild-type mouse more than doubles the total of all coactivators in the cell, altering the capacity for activating CAR target genes. Since Gadd45β definitely functions in pathways other than transcriptional coactivation (13, 16–23, 51), this unusually high expression might provide sufficient protein to impact transcription and also contribute to these other processes.
Methods
Reagents.
TCPOBOP, BrdU, and ketoconazole were obtained from Sigma-Aldrich. Abs to CAR (sc-13065), Gadd45β (sc-8776, lot C3007), HNF1α (sc-6547), cyclin D1 (sc-450), and the Gal4-DBD (sc-510) were from Santa Cruz Biotechnology; Ab to Ki-67 was from Novacastra; Abs to Jnk (catalog no. 9528) and pJnk (catalog no. 4668) were from Cell Signaling Technology; and Ab to BrdU was from Roche. Protein A and protein G agarose were obtained from Upstate.
Animals and histology.
Gadd45b–/– mice (30, 31) in a C57BL/6 genetic background and control wild-type mice were maintained in the barrier facility of the Institute for Animal Studies of Albert Einstein College of Medicine. 5- to 7-month-old female mice received a single dose of TCPOBOP dissolved in dimethylsulfoxide-corn oil, 3 mg/kg by gavage. Sham controls received an equivalent amount of vehicle. Starting 48 hours before treatment, the mice continuously received BrdU (500 μg/ml) in their drinking water. At 3, 6, 12, 18, 24, 36, 42, and 48 hours after treatment, groups of 3–4 mice were euthanized by exsanguination during ether anesthesia. Control mice were untreated, and sham-treated mice were studied 3 and 6 hours after gavage. Liver segments were frozen for molecular studies or fixed in 10% buffered neutral formalin and embedded in paraffin for histological analysis. Immunohistochemistry used standard immunoperoxidase methods. Morphometric analysis of photomicrographs was carried out using NIS-Elements D software (Nikon). To quantify mitoses, 20 random high-power fields were counted from each 48-hour liver.
RNA studies.
Total mRNA was isolated from livers using the guanidium isothiocyanate method (TRIzol; Invitrogen), followed by precipitation with LiCl (9). For microarray studies, 20 μg total RNA was labeled with the Superscript Plus Direct cDNA Labeling System and Alexa Fluor aha-dUTPs (Invitrogen). Labeled product was purified and concentrated with a Microcon YM-50 column (Millipore). 2-color hybridization was always carried out with red-labeled experimental sample and green-labeled pooled RNA from normal C57BL/6 livers. The labeled cDNAs were hybridized overnight at 50°C on aminosilane-coated Corning glass slides spotted with 33,000 selected mouse oligonucleotide sequences (70-mer Operon mouse 3.0 series), which were printed by the Microarray Core Facility of AECOM. Microarrays were scanned with an Axon GenePix 4000A scanner, and data were acquired through GenePix Pro 6.0 software (Molecular Devices). Hybridization data were further normalized without background subtraction, using locally weighted linear regression (LOWESS) analysis via an in-house program. For each experimental condition, separate arrays from 3–5 livers were analyzed, and the results were averaged for each array spot. The datasets were compiled in Microsoft Access for further analysis. Hybridization intensities ranged from 0 to 60,000 before a background cutoff of 100. Expression levels were normalized to a 0–100 scale for some data presentations (see Supplemental Tables 1–3). Individual spots were included in a compilation if they met the following criteria in any single experimental condition: average expression >2 or <0.5 times control and SD <0.68 times the averaged value. The array data were deposited in the Gene Expression Omnibus (GEO) database ( http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE31087).
For real-time PCR analysis, cDNA was synthesized from 1 μg RNA using a High Capacity cDNA Archive Kit (Applied Biosystems). PCR reactions contained cDNA synthesized from 0.05 μg, and assays were done in triplicate. Real-time PCR analysis used SYBR Green PCR Master Mix (Applied Biosystems) with the primer pairs listed in Supplemental Table 4. Gene expression was normalized to the level of GAPDH mRNA in the same RNA specimen.
Plasmids and transfection.
All reporter assays were carried out in HepG2 cells, using standard calcium phosphate transfection and luciferase (LUC) methods. For assays carried out on 6-well plates, each transfection used 1.25 μg total DNA and 1 μg reporter plasmid (or 850 ng Gal4 reporters) plus variable amounts of expression plasmid and carrier (see Results). Plasmids were generated to express CAR and Gadd45β; GST, Gal4, and 6His fusion constructs; and a CAR LUC reporter containing the 53-bp phenobarbital response element from the mouse Cyp2b10 gene (36). Detailed cloning is described in Supplemental Methods.
Protein analysis.
Bacterial GST-Gadd45β and control GST proteins were induced and purified according to standard procedures. Mouse CAR protein was translated from plasmid pCI-CAR with TNT T7 Quick Coupled Transcription/Translation System (Promega) in the presence of 35S-methionine according to the manufacturer’s protocols. 2 μg GST fusion protein bound to glutathione-sepharose beads (GE Healthcare) was incubated with 2 μl in vitro translated protein in 250 μl binding buffer (50 mM Tris, pH 8.0; 100 mM NaCl; 0.1% NP-40; 0.3 mM DTT; 10 mM MgCl2; and 10% Glycerol) plus Inhibitor Cocktails for General Use (Sigma-Aldrich) for 4 hours at 4°C. The beads were then washed several times with binding buffer. Proteins were eluted with 2× SDS loading buffer and resolved on SDS-PAGE gels.
Plasmids expression 6His-mCAR and various Gal4-Gadd45β fusions were transfected into 293T cells with Lipofectamine 2000 (Invitrogen). After 48 hours, cells were lysed in RIPA (Santa Cruz Biotechnologies) with protease inhibitor cocktails (Sigma-Aldrich). 100 μl lysate from His-mCAR–transfected cells and 100 μl lysate from Gal4-Gadd45β fusion plasmid–transfected cells were mixed with 600 μl TBSN (10 mM Tris, pH 7.4; 150 mM NaCl; and 0.1% NP-40) and 25 μl Ni-NTA agarose (Qiagen) and rotated at 4°C for 2 hours. The complexes were then washed 4 times with TBSN. Ni-NTA bound proteins were eluted by boiling in SDS loading buffer and resolved on 12% SDS-PAGE. Western blots were carried out with a Gal4-DBD Ab using standard methods and signal detection with Western Lightning Ultra (Perkin Elmer).
ChIP.
For study of intact liver, the method of Boyd and Farnham (58, 59) was used as modified by Nguyen et al. (60). To fragment chromatin, liver homogenates were sonicated with Fisher Sonic Dismembranator, Model 500, set at 50% amplitude, with a microtip. Sonication on ice was carried out in 30-second bursts for a total of 8 minutes. Chromatin was then immediately treated or aliquoted and stored in liquid nitrogen. Chromatin from 10 mg liver was used for each immunoprecipitation. To reduce average fragment size to 400–500 bp, chromatin was treated with micrococcal nuclease (90 U/ml; Worthington) for 15 minutes at 25°C in 80 mM KCl, 20 mM Hepes (pH 7.5), 2.8 mM ATP, 6 mM CaCl2, 500 mM sucrose. EGTA and EDTA were then added to final concentrations of 6 mM. Subsequent immunoprecipitation, wash, DNA purification, and PCR analysis were described previously (61). For PCR detection, the region around the Cyp2b10 phenobarbital response element was amplified with primers 5′-CTCCAGTGACTTAGGAGGAAGGTCAGAA-3′ and 5′-CTGGGAATCCTGGGCTGGAATACT-3′, and the Albumin gene promoter region was amplified with 5′-CAGATGGCAAACATACGCAAGGG-3′ and 5′-GTGATCTGTGTGCAGAAAGACTCGCTC-3′.
Statistics.
Sets of data were compared using unpaired (Figure 1, Figure 2A, Figure 3, Figure 7, Figure 8, and Supplemental Figure 10) or paired (Figure 2, C–E) 2-tailed Student’s t tests. A P value less than 0.05 was considered significant.
Study approval.
Animal experiments were performed with approval of the Animal Care and Use Committees of Albert Einstein College of Medicine.
Supplementary Material
Acknowledgments
We are grateful to Aldo Massimi for assistance with the microarray studies and to Radma Mahmood for immunohistochemistry. The work was supported by NIH grants CA104292 to J. Locker and CA122376 to D.A. Liebermann and by Associazione Italiana Ricerca sul Cancro (AIRC) and Ministero Università e Ricerca Scientifica (PRIN ex 40%, and 60%) grants to A. Columbano.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(11):4491–4502. doi:10.1172/JCI38760.
References
- 1.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213(2):286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fausto N, Riehle KJ. Mechanisms of liver regeneration and their clinical implications. J Hepatobiliary Pancreat Surg. 2005;12(3):181–189. doi: 10.1007/s00534-005-0979-y. [DOI] [PubMed] [Google Scholar]
- 3.Ledda-Columbano GM, et al. In vivo hepatocyte proliferation is inducible through a TNF and IL-6- independent pathway. Oncogene. 1998;17(8):1039–1044. doi: 10.1038/sj.onc.1202018. [DOI] [PubMed] [Google Scholar]
- 4.Dragani TA, Manenti G, Galliani G, Della Porta G. Promoting effects of 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene in mouse hepatocarcinogenesis. Carcinogenesis. 1985;6(2):225–228. doi: 10.1093/carcin/6.2.225. [DOI] [PubMed] [Google Scholar]
- 5.Ledda-Columbano GM, Pibiri M, Loi R, Perra A, Shinozuka H, Columbano A. Early increase in cyclin-D1 expression and accelerated entry of mouse hepatocytes into S phase after administration of the mitogen 1, 4-Bis[2- (3,5-Dichloropyridyloxy)] benzene. Am J Pathol. 2000;156(1):91–97. doi: 10.1016/S0002-9440(10)64709-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20(9):2951–2958. doi: 10.1128/MCB.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kodama S, Negishi M. Phenobarbital confers its diverse effects by activating the orphan nuclear receptor CAR. Drug Metab Rev. 2006;38(1–2):75–87. doi: 10.1080/03602530600569851. [DOI] [PubMed] [Google Scholar]
- 8.Shirai T, Lee MS, Wang CY, King CM. Effects of partial hepatectomy and dietary phenobarbital on liver and mammary tumorigenesis by two N-hydroxy-N-acylaminobiphenyls in female CD rats. Cancer Research. 1981;41(6):2450–2456. [PubMed] [Google Scholar]
- 9.Locker J, et al. A common set of immediate–early response genes in liver regeneration and hyperplasia. Hepatology. 2003;38(2):314–325. doi: 10.1053/jhep.2003.50299. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Up-regulation of estrogen responsive genes in hypospadias: microarray analysis. J Urol. 2007;177(5):1939–1946. doi: 10.1016/j.juro.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Qiu W, Zhou B, Chu PG, Luh F, Yen Y. The induction of growth arrest DNA damage-inducible gene 45 beta in human hepatoma cell lines by S-adenosylmethionine. Am J Pathol. 2007;171(1):287–296. doi: 10.2353/ajpath.2007.070121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Q, Shen N, Li XM, Chen SL. Genomic view of IFN-alpha response in pre-autoimmune NZB/W and MRL/lpr mice. Genes Immun. 2007;8(7):590–603. doi: 10.1038/sj.gene.6364421. [DOI] [PubMed] [Google Scholar]
- 13.Liebermann DA, Hoffman B. Gadd45 in the response of hematopoietic cells to genotoxic stress. Blood Cells Mol Dis. 2007;39(3):329–335. doi: 10.1016/j.bcmd.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu B, Ferrandino AF, Flavell RA. Gadd45beta is important for perpetuating cognate and inflammatory signals in T cells. Nat Immunol. 2004;5(1):38–44. doi: 10.1038/ni1020. [DOI] [PubMed] [Google Scholar]
- 15.Liebermann DA, Hoffman B. Myeloid differentiation (MyD) primary response genes in hematopoiesis. Blood Cells Mol Dis. 2003;31(2):213–228. doi: 10.1016/S1079-9796(03)00160-8. [DOI] [PubMed] [Google Scholar]
- 16.Papa S, et al. Insights into the structural basis of the GADD45beta-mediated inactivation of the JNK kinase, MKK7/JNKK2. J Biol Chem. 2007;282(26):19029–19041. doi: 10.1074/jbc.M703112200. [DOI] [PubMed] [Google Scholar]
- 17.Papa S, et al. Gadd45 beta mediates the NF-kappa B suppression of JNK signalling by targeting MKK7/JNKK2. Nat Cell Biol. 2004;6(2):146–153. doi: 10.1038/ncb1093. [DOI] [PubMed] [Google Scholar]
- 18.Jin R, et al. Regulation of the gadd45β promoter by NF-κB. DNA Cell Biol. 2002;21(7):491–503. doi: 10.1089/104454902320219059. [DOI] [PubMed] [Google Scholar]
- 19.De Smaele E, et al. Induction of gadd45beta by NF-kappaB downregulates pro-apoptotic JNK signalling. Nature. 2001;414(6861):308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- 20.Yoo J, et al. Transforming growth factor-beta-induced apoptosis is mediated by Smad-dependent expression of GADD45b through p38 activation. . J Biol Chem. 2003;278(44):43001–43007. doi: 10.1074/jbc.M307869200. [DOI] [PubMed] [Google Scholar]
- 21.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95(4):521–530. doi: 10.1016/S0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 22.Gupta M, Gupta SK, Hoffman B, Liebermann DA. Gadd45a and Gadd45b protect hematopoietic cells from UV-induced apoptosis via distinct signaling pathways, including p38 activation and JNK inhibition. J Biol Chem. 2006;281(26):17552–17558. doi: 10.1074/jbc.M600950200. [DOI] [PubMed] [Google Scholar]
- 23.Abdollahi A, Lord KA, Hoffman-Liebermann B, Liebermann DA. Sequence and expression of a cDNA encoding MyD118: a novel myeloid differentiation primary response gene induced by multiple cytokines. Oncogene. 1991;6(1):165–167. [PubMed] [Google Scholar]
- 24.Wang J, et al. GADD45B inhibits MKK7-induced cardiac hypertrophy and the polymorphisms of GADD45B is associated with inter-ventricular septum hypertrophy. Biochem Biophys Res Comm. 2008;372(4):623–628. doi: 10.1016/j.bbrc.2008.05.122. [DOI] [PubMed] [Google Scholar]
- 25.Yi YW, Kim D, Jung N, Hong SS, Lee HS, Bae I. Gadd45 family proteins are coactivators of nuclear hormone receptors. Biochem Biophys Res Commun. 2000;272(1):193–198. doi: 10.1006/bbrc.2000.2760. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto Y, Negishi M. The antiapoptotic factor growth arrest and DNA-damage-inducible 45 beta regulates the nuclear receptor constitutive active/androstane receptor-mediated transcription. Drug Metab Dispos. 2008;36(7):1189–1193. doi: 10.1124/dmd.108.020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Major MB, Jones DA. Identification of a gadd45beta 3’ enhancer that mediates SMAD3- and SMAD4-dependent transcriptional induction by transforming growth factor beta. J Biol Chem. 2004;279(7):5278–5287. doi: 10.1074/jbc.M311517200. [DOI] [PubMed] [Google Scholar]
- 28.Columbano A, et al. Gadd45beta is induced through a CAR-dependent, TNF-independent pathway in murine liver hyperplasia. Hepatology. 2005;42(5):1118–1126. doi: 10.1002/hep.20883. [DOI] [PubMed] [Google Scholar]
- 29.Papa S, et al. Gadd45beta promotes hepatocyte survival during liver regeneration in mice by modulating JNK signaling. J Clin Invest. 2008;118(5):1911–1923. doi: 10.1172/JCI33913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amanullah A, et al. Cell signalling: cell survival and a Gadd45-factor deficiency. Nature. 2003;424(6950):741. doi: 10.1038/424741b. [DOI] [PubMed] [Google Scholar]
- 31.Gupta M, et al. Hematopoietic cells from Gadd45a- and Gadd45b-deficient mice are sensitized to genotoxic-stress-induced apoptosis. Oncogene. 2005;24(48):7170–7179. doi: 10.1038/sj.onc.1208847. [DOI] [PubMed] [Google Scholar]
- 32.Shi L, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics. 2008;9(suppl 9):S10. doi: 10.1186/1471-2105-9-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol. 1998;18(10):5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H, et al. Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene. 2007;26(2):258–268. doi: 10.1038/sj.onc.1209788. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, et al. Activated pregnenolone X-receptor is a target for ketoconazole and its analogs. Clin Cancer Res. 2007;13(8):2488–2495. doi: 10.1158/1078-0432.CCR-06-1592. [DOI] [PubMed] [Google Scholar]
- 36.Sueyoshi T, Negishi M. Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol. 2001;41:123–143. doi: 10.1146/annurev.pharmtox.41.1.123. [DOI] [PubMed] [Google Scholar]
- 37.Mckenna NJ, O’Malley BW. SnapShot: NR coregulators. Cell. 2010;143(1):172. doi: 10.1016/j.cell.2010.09.032. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Huang W, Chua SS, Wei P, Moore DD. Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science. 2002;298(5592):422–424. doi: 10.1126/science.1073502. [DOI] [PubMed] [Google Scholar]
- 39.Fisher CD, et al. Induction of drug-metabolizing enzymes by garlic and allyl sulfide compounds via activation of constitutive androstane receptor and nuclear factor E2-related factor 2. Drug Metab Dispos. 2007;35(6):995–1000. doi: 10.1124/dmd.106.014340. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Stanton JD, Tolson AH, Luo Y, Wang H. Bioactive terpenoids and flavonoids from Ginkgo biloba extract induce the expression of hepatic drug-metabolizing enzymes through pregnane X receptor, constitutive androstane receptor, and aryl hydrocarbon receptor-mediated pathways. Pharm Res. 2009;26(4):872–882. doi: 10.1007/s11095-008-9788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274(10):6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 42.Honkakoski P, Jaaskelainen I, Kortelahti M, Urtti A. A novel drug-regulated gene expression system based on the nuclear receptor constitutive androstane receptor (CAR). Pharm Res. 2001;18(2):146–150. doi: 10.1023/A:1011068015301. [DOI] [PubMed] [Google Scholar]
- 43.Huang W, Zhang J, Wei P, Schrader WT, Moore DD. Meclizine is an agonist ligand for mouse constitutive androstane receptor (CAR) and an inverse agonist for human CAR. Mol Endocrinol. 2004;18(10):2402–2408. doi: 10.1210/me.2004-0046. [DOI] [PubMed] [Google Scholar]
- 44.Jackson JP, Ferguson SS, Moore R, Negishi M, Goldstein JA. The constitutive active/androstane receptor regulates phenytoin induction of Cyp2c29. Mol Pharmacol. 2004;65(6):1397–1404. doi: 10.1124/mol.65.6.1397. [DOI] [PubMed] [Google Scholar]
- 45.Poso A, Honkakoski P. Ligand recognition by drug-activated nuclear receptors PXR and CAR: structural, site-directed mutagenesis and molecular modeling studies. Mini Rev Med Chem. 2006;6(8):937–947. doi: 10.2174/138955706777935008. [DOI] [PubMed] [Google Scholar]
- 46.Hernandez JP, Huang W, Chapman LM, Chua S, Moore DD, Baldwin WS. The environmental estrogen, nonylphenol, activates the constitutive androstane receptor. Toxicol Sci. 2007;98(2):416–426. doi: 10.1093/toxsci/kfm107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saussele T, et al. Selective induction of human hepatic cytochromes P450 2B6 and 3A4 by metamizole. Clin Pharmacol Ther. 2007;82(3):265–274. doi: 10.1038/sj.clpt.6100138. [DOI] [PubMed] [Google Scholar]
- 48.DeKeyser JG, Stagliano MC, Auerbach SS, Prabhu KS, Jones AD, Omiecinski CJ. Di(2-ethylhexyl) phthalate is a highly potent agonist for the human constitutive androstane receptor splice variant CAR2. Mol Pharmacol. 2009;75(5):1005–1013. doi: 10.1124/mol.108.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeKeyser JG, Laurenzana EM, Peterson EC, Chen T, Omiecinski CJ. Selective phthalate activation of naturally occurring human constitutive androstane receptor splice variants and the pregnane X receptor. Toxicol Sci. 2011;120(2):381–391. doi: 10.1093/toxsci/kfq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baskin-Bey ES, et al. Constitutive androstane receptor (CAR) ligand, TCPOBOP, attenuates Fas-induced murine liver injury by altering Bcl-2 proteins. Hepatology. 2006;44(1):252–262. doi: 10.1002/hep.21236. [DOI] [PubMed] [Google Scholar]
- 51.Kodama S, Negishi M. Pregnane X receptor PXR activates the GADD45beta gene, eliciting the p38 MAPK signal and cell migration. J Biol Chem. 2011;286(5):3570–3578. doi: 10.1074/jbc.M110.179812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yasumo H, Masuda N, Furusawa T, Tsukamoto T, Sadano H, Osumi T. Nuclear receptor binding factor–2 (NRBF–2), a possible gene activator protein interacting with nuclear hormone receptors. Biochimica et Biophysica Acta. 2000;1490(1–2):189–197. doi: 10.1016/s0167-4781(99)00244-4. [DOI] [PubMed] [Google Scholar]
- 53.Flores AM, Li L, Aneskievich BJ. Isolation and functional analysis of a keratinocyte-derived, ligand-regulated nuclear receptor comodulator. J Invest Dermatol. 2004;123(6):1092–1101. doi: 10.1111/j.0022-202X.2004.23424.x. [DOI] [PubMed] [Google Scholar]
- 54.Lin CW, et al. Hepatocyte proliferation and hepatomegaly induced by phenobarbital and TCPOBOP is suppressed in hepatocyte–targeted glypica 3 transgenic mice. Hepatology. 2011;54(2):620–630. doi: 10.1002/hep.24417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Apte U, et al. Enhanced liver regeneration following changes induced by hepatocyte-specific genetic ablation of integrin-linked kinase. Hepatology. 2009;50(3):844–851. doi: 10.1002/hep.23059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu B, et al. Suppression of liver regeneration and hepatocyte proliferation in hepatocyte-targeted glypican 3 transgenic mice. Hepatology. 2010;52(3):1060–1067. doi: 10.1002/hep.23794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bursch W, et al. Role of apoptosis for mouse liver growth regulation and tumor promotion: comparative analysis of mice with high (C3H/He) and low (C57Bl/6J) cancer susceptibility. Toxicol Lett. 2004;149(1–3):25–35. doi: 10.1016/j.toxlet.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 58.Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci U S A. 1998;95(23):13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyd KE, Farnham PJ. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol Cell Biol. 1999;19(12):8393–8399. doi: 10.1128/mcb.19.12.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen TT, Cho K, Stratton SA, Barton MC. Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene. Mol Cell Biol. 2005;25(6):2147–2157. doi: 10.1128/MCB.25.6.2147-2157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kajiyama Y, Tian J, Locker J. Characterization of distant enhancers and promoters in the albumin-alpha-fetoprotein locus during active and silenced expression. J Biol Chem. 2006;281(40):30122–30131. doi: 10.1074/jbc.M603491200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.