Abstract
The elucidation of extra-nuclear lysine acetylation has been of growing interest, as the co-substrate for acetylation, acetyl CoA, is at a key metabolic intersection. Our hypothesis was that mitochondrial and cytoplasmic protein acetylation may be part of a fasted/re-fed feedback control system for the regulation of the metabolic network in fuel switching, where acetyl CoA would be provided by fatty acid oxidation, or glycolysis, respectively. To test this we characterized the mitochondrial and cytoplasmic acetylome in various organs that have a high metabolic rate relative to their mass, and/or switch fuels, under fasted and re-fed conditions (brain, kidney, liver, skeletal muscle, heart muscle, white and brown adipose tissues). Using immunoprecipitation, coupled with LC-MSMS label free quantification, we show there is a dramatic variation in global quantitative profiles of acetylated proteins from different organs. In total, 733 acetylated peptides from 337 proteins were identified and quantified, out of which 31 acetylated peptides from the metabolic proteins that may play organ-specific roles were analyzed in detail. Results suggest that fasted/re-fed acetylation changes coordinated by organ-specific (de-)acetylases in insulin-sensitive versus insensitive organs may underlie fuel use and switching. Characterization of the tissue-specific acetylome should increase understanding of metabolic conditions wherein normal fuel switching is disrupted, such as in Type II diabetes.
Introduction
The function and physical properties of proteins can be regulated by protein post-translational modifications (PTMs). Lysine acetylation is a reversible PTM that affects a variety of biological processes, such as protein-DNA interactions, enzyme activation/inactivation, subcellular localization and protein stability, etc 1, 2. In the past four decades, histones have been the primary focus of acetylation studies because of their high abundance and high frequency of lysine acetylation. Only studies of specific acetylation in individual proteins were possible in the past due to the lack of robust detection technologies3–5. Improvements of modern mass spectrometers, specifically improved accurate mass detection, sensitivity and dynamic range, higher resolution, and faster scan rates, all serve to greatly facilitate global acetylation studies6 as has been reported recently7–9. In 2006, Kim et al. published the first proteome-wide report by using immunoprecipitation enrichment of acetylated peptides with an anti-acetylated lysine antibody and HPLC-MSMS detection8 to identify 195 acetylated proteins from mouse liver. Choudhary et al. and Zhao et al. advanced this approach and demonstrated the largest data set of acetylated peptides identified from human cells (1750 acetylated proteins) and liver tissue (1047 acetylated proteins)7, 9. These reports show large numbers of acetylated non-histone proteins, many of which are mitochondrial or cytoplasmic species. Thus, lysine acetylation is involved in a greater diversity of functional roles and subcellular localizations than previously recognized.
As lysine acetylation is dynamic and regulates protein function in ways not fully understood, global quantitative analyses of lysine acetylation are critical to better understand the functional impact and use of this modification in biological systems. For example, Kendrick et al. showed fatty liver was associated with reduced sirtuin activity and increased acetylation levels of mitochondrial proteins10. They isolated acetylated proteins from total liver proteome samples from mice on a high fat diet (HFD, 4 months) and control animals. Acetylated proteins were immunoprecipitated with immobilized anti-acetylated lysine antibodies, purified proteins were separated on 1-D gels, scanned, and relative quantification between HFD-fed and control animal samples was performed. Bands that exhibited significant differential staining between HFD and control samples were subjected to in-gel digestion and mass spectrometry analysis. In total, 193 proteins were differentially acetylated in HFD samples, including proteins involved in gluconeogenesis, mitochondrial protein oxidation, methionine metabolism, liver injury and endoplasmic reticulum stress response. Importantly, the work of Kendrick et al. shows that differences in levels of acetylated liver proteins relevant to altered feeding status in mice can be observed by proteomic methods. Previously, Choudhary et al. used SILAC (Stable isotope labeling with amino acids in cell culture)7 and Zhao et al. used iTRAQ labeling (isobaric tag for relative and absolute quantification)9 coupled with LC-MSMS techniques to quantify peptide-level, or site-specific acetylation in human cell lines and liver tissue respectively. Schwer and co-workers11 reported label free quantification (LFQ) of the acetylated peptides from mouse liver tissue. LFQ technology is beneficial for studies of live animal tissues because incorporation of heavy isotopes is not required. In this way, Schwer et al. studied how calorie restriction (CR) can alter the mitochondrial protein acetylation levels in mouse liver. Approximately 300 proteins were quantified, and 72 were determined to have at least a 2.5 fold change in acetylation during CR. SILAM (Stable Isotope Labeling in Mammals)12 can in principle allow isotope incorporation in these studies with live animal tissues, although the additional expense significantly limits the applicability. In this work, a further advance of these approaches includes measurement of differences of acetylation on specific peptides by quantitative LC-MSMS methods, and comparison of level changes observed among several tissues or organs.
Acetylation is dynamic and cellular acetylation status is dependent on the activities of acetylases and deacetylases. Intracellular acetyl CoA, used by acetylases, sits on the metabolic crossroads of glycolysis, fatty acid oxidation, ketogenesis, amino acid metabolism, and TCA cycle utilization for ATP synthesis, making acetyl CoA an ideal quantity for sensing (via the non-nuclear acetylome) metabolic network function. Intracellular NAD+/NADH ratio, a central determinant of nutritional status also critically regulates the activity of NAD+ deacetylases. Metabolic inflexibility, or the inability of an organism to adapt and modify fuel oxidation in response to changes in nutrient availability, is characteristic of dysfunction seen in metabolic syndrome and Type II Diabetes13–18. The hypothesis for this study is that fasted/re-fed characterization of the organ specific mitochondrial and cytoplasmic acetylome may yield insights into both normal functional roles of tissue specific fuel switching, as well as abnormalities seen in situations, like diabetes mellitus, where metabolic inflexibility is evident. Therefore, analyzing the acetylation pattern of major mammalian tissues that greatly differ metabolically and play a diverse role in energy homeostasis under different nutritional status would be highly informative. Previous studies have reported murine fasted and re-fed hepatic acetylome only8. Our efforts in this study focus on the characterization of the fasted/re-fed acetylome patterns of tissues that are known to switch fuels between the fasted/fed states (liver, skeletal muscle, heart muscle, white adipose and brown adipose) or have a high metabolic rate relative to their mass (brain and kidney)19. No one organ is responsible for the metabolic rate, but some organs (brain, kidney, heart, and gastrointestinal tract) contribute a much larger fraction of their metabolic rate than their fractional mass or volume of the body, whereas others (bone, white adipose tissue, skin, and skeletal muscle) contribute much less19. Brown adipose tissue contributes to the metabolic rate in neonatal mammals, and small mammals adapted to cold environments21. Metabolic consequences of the presence or absence of the thermogenic capacity of brown adipose tissue in mice has been reported21, 22, so brown adipose was also used for this study; white adipose tissue was included for comparison. The metabolic rate may fall 10% during sleep, a normal fasted condition, and may fall by 40% during long-term starvation in humans20, supporting the relevance of acetylome changes on energy metabolism found for metabolic network enzymes during dietary shifts.
In this study, the acetylation levels of peptides from mitochondrial proteins in each tissue under differential feeding status were of primary interest. In total, 733 acetylated peptides from 337 proteins were identified, out of which the levels of 58 acetylated peptides changed 3-fold or greater under fasted/re-fed conditions. These peptides constitute the top 5% largest changes in acetylation levels among the 733 acetylated peptides, based on two standard deviations from the center of the observed log10 (re-fed/fasted) distribution as a cut off. Thirty-one acetylated peptides of these 58 are from metabolic proteins or chaperones. Many of these protein acetylation events may be relevant metabolic changes that accompany fuel switching and will serve as targets for future investigation. Our results show that acetylation levels in ATP-generating or utilizing metabolic processes such as glycolysis, Krebs cycle, gluconeogenesis, lipid synthesis and oxidation likely serve as control points or play a role in modulation of these pathways. In fact, acetylation is a classic candidate for such critical regulation, owing to its requirement of acetyl CoA, an important intermediate molecule linking such major pathways, as its acetyl group donor. The results presented here are first to demonstrate that there variation exists in the global quantitative profiles of acetylated metabolic proteins in all these organs, that are dramatically affected by comparison of the fasted to re-fed state.
Materials and Methods
1. Materials
Deacetylase inhibitors (TSA, Nicotinamide, Butyric acid), 2,2,2-Trifluoroethanol (TFE), Ammonium bicarbonate (ABC), Iodoacetamide (IAA) and Tergitol solution (70% NP40 in water) were purchased from Sigma-Aldrich (St. Louis, MO) and used without purification.
Affinity-purified anti-acetyl lysine antibody immobilized onto protein A-conjugated agarose beads was purchased from ImmunoChem Pharmaceuticals Inc. (Burnaby, British Columbia, Canada). TCEP, BupH™ Tris Buffered Saline Pack (IP buffer), BCA Protein Assay Kit and mass spectrometry-grade Trypsin endoproteinase were purchased from Pierce (Rockford, IL, USA). Protease inhibitor cocktail tablets were purchased from Roche Diagnostics (Indianapolis, IN, USA).
2. Animals
Four to five months old male FVB/N background mice studied were housed in a full-barrier facility with a 12-hour light/dark cycle (7:00 am/7:00 pm) throughout the study. Animals had full access to food (standard chow from Harlan Teklad 4% Mouse/Rat Diet cat # 7001) and water unless otherwise stated. All animal experiments were performed in accordance with National Institutes of Health guidelines and with the approval of the Animal Care and Use Committee of Albert Einstein College of Medicine. For the fasting/re-feeding studies, mice were allowed to eat for an hour and half after the beginning of the dark cycle (7:00 pm) and fasting was initiated by removing the food at 8:30 pm. Animals (n=5) were sacrificed the next day at the end of: a) 18 hours fast (2:30 pm) for fasting studies, b) 13 hours fast (9:30 am) followed by 5 hours re-feeding (2:30 pm) for re-feeding studies. Harvested tissues (liver, brown adipose, white adipose, heart muscle, skeletal muscle, kidney and brain) were snap-frozen in liquid N2 and stored at −80° C until further analysis.
3. Sample preparation
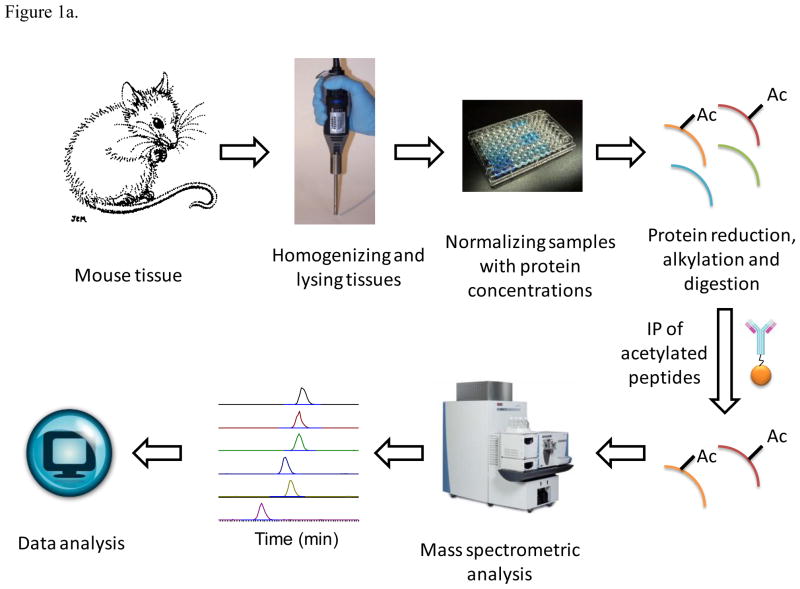
Matching tissues or organs from five mice were pooled together. Pooling the samples from different biological sources has been shown to reduce the biological variation, as one might anticipate, and in many cases, increases the ability to study the differences caused by treatment as described in several references 46–48. Because of limited sample sizes available in a single animal, pooled samples were required to study all seven tissues presented here. Sample preparation was divided into: 1) Tissue protein extraction, 2) normalization and tryptic digestion and 3) the enrichment of the acetylated peptides. A general experimental scheme is shown in Figure 1. Tissues were homogenized in the lysis buffer, which contains 50% TFE, 0.1M ABC, 5mM TCEP, 10μM TSA, 10 mM nicotinamide, 50mM butyric acid and protease inhibitor cocktail. The pH of the lysis buffer was adjusted to 7.4. Minimal volume of the lysis buffer was used in the homogenizing step to get highly concentrated protein solutions. The samples were sonicated to reduce the viscosity and heated at 99 °C for 5 min. The protein solutions were centrifuged at 16,000 × G for 20 min to separate solids. Protein concentration in the supernatant portions was measured with a BCA protein quantification kit. All samples were normalized by using an aliquot containing the same amount of total protein. Thirty milligrams of protein from each sample were reduced and alkylated with a final concentration of 50mM TCEP and IAA, respectively. The volume of each sample was diluted 5 times with 100 mM ABC buffer; and the pH was adjusted to 8–8.2. The samples were then subjected to digestion with trypsin with a 500:1 protein-to-enzyme ratio and were incubated overnight at 37 °C. Then the mixtures were heated at 99 °C for 5 min to denature trypsin and reduce activity. The mixtures were centrifuged at 16,000 × G for 2 min; and the supernatants were separated from the insoluble fractions. TFE was evaporated from the supernatants which were placed under vacuum (Thermo Speed Vacuum SPD131DDA). We previously found 0.1% concentration of the non-ionic detergent NP40 to be optimal for immunoprecipitation23 so this was the final concentration of NP40 used here, and the pH was adjusted to 7.4. For each 30 mg of digested protein sample, a 200 μL slurry aliquot of the anti-acetylated lysine antibody beads was used for immunoprecipitation. The aliquots of the antibody slurry were washed with phosphate buffered saline (PBS) 3 times, and then added into the peptide solutions. The mixtures were then incubated 3 hours at 4 °C, centrifuged and the antibody beads were separated from the supernatants. The beads were washed 4 times with IP buffer and eluted with a 100 μL of the elution buffer 3 times (0.1% formic acid and 50% acetonitrile in DI water). Then the eluents were dried under vacuum and re-dissolved in a 20 μL buffer of 0.1% formic acid in DI water.
Figure 1.
Experimental Scheme. Seven tissues or organs from the same 5 fasted or re-fed mice were pooled, homogenized and lysed. Different samples were normalized using the same starting amount of total protein. Thirty milligrams of protein from each sample were reduced and alkylated, trypsin digested, and anti-acetylated lysine antibody beads were used for immunoprecipitation from the digested protein sample supernatants. The enriched acetylation products were fractionated with UPLC (nanoAcquity Waters, Milford, MA) and analyzed with a built-in-house Velos-FT mass spectrometer24 (see Methods).
4. Mass spectrometric analyses
The enriched acetylation products were fractionated with UPLC (nanoAcquity Waters, Milford, MA) and analyzed with a built-in-house Velos-FT mass spectrometer24 (Submitted to Analytical Chemistry). The FT analyzer enables high resolution and accurate mass detection and the Velos mass spectrometer allows fast scan speed and better sensitivity, which is important for the identification of the low abundance acetylated peptides. A 35 cm long C18 column was made in house by packing fused silica capillary (360 μm × 75 μm) with MAGIC C18AQ 100A 5U beads (Michrom Bioresources, Inc., Auburn, CA). The electrospray ionization (ESI) tip was made by pulling the end of the column with a laser puller (Model P-2000, Sutter Instrument Co.). A 2 cm long trap column was prepared similarly by packing a frit fused silica capillary (360 μm × 100 μm) with MAGIC C18AQ 200A 5U beads. The following LC gradient was used: 0–60 min 5%–35% buffer B, 60–75 min flushing with 80% buffer B and 75–100 min equilibrating with 5% buffer B. Buffer A and buffer B consisted of 0.1% formic acid, 0.1% formic acid and 95% acetonitrile in DI water, respectively. One MS measurement in the FT analyzer with 25K resolving power was followed by 10 MSMS measurements in DDA mode in the Velos instrument. The dynamic exclusion repeat and the exclusion duration were 15 sec. Each sample was analyzed in triplicate and in each analysis 5 μL of sample solution was loaded onto the column. Two LC-MSMS measurements of a mixture of 0.5 pmol angiotension I and neurotensin were run after the triplicate analyses of each sample. The first run was used to clean the column, and the second run was used to verify the column performance. A total of 42 LC-MSMS experiments with mouse tissue and organ samples were acquired, and used to quantify acetylated peptides.
5. Data analyses
Peptide identification
The obtained LC-MSMS data were searched against the mouse database (IPI_mouse_3.26) with Mascot (version: 2.3.01). The parameters in database search were: missed cleavage = 3, precursor error tolerance = 15 ppm, fragmentation error tolerance = 0.6 Da, fixed modification of carbamidomethyl on cysteine, variable modifications of acetylation on lysine and oxidation on methionine. The false discovery rate (FDR) was determined in Mascot by enabling decoy database search 44, 45. The acetylated sites are assigned both by Mascot search and manual verification. In most cases, only a single possible modification site (internal lysine residue) exists in the peptide sequence, yielding unambiguous acetylation site assignment. In cases where multiple possible acetylation sites were assigned by Mascot, manual verification of the MS/MS fragmentation patterns was used to determine acetylation sites. The mass error distribution between the observed and the theoretical masses of all identified peptides from the fasted and re-fed samples are plotted in Supplementary Figure 1. The center and standard deviation of the acetylated peptide mass error distribution is at 3.5 ppm and 5 ppm, respectively. Many non-specific binding peptides were identified even after immunoprecipitation, which indicates mass spectrometric fragmentation and database search identification of the peptides are necessary to distinguish the acetylated and non-acetylated peptides. Importantly, protease digestion of total cellular extracts prior to IP, and LC-MSMS analysis of acetylated peptides, presents an additional level of specificity in the analysis of global acetylation as compared to the IP of acetylated proteins, since non-specific binders are readily distinguished during the database search (reviewed in Guan and Xiong32). For example, the use of variable acetyl modification on lysine side chains during database search allows differentiation of nonspecific binding peptides and bona fide acetylated lysine-containing peptides. On the other hand, in the analysis of immunoprecipitated acetylated proteins, non-acetylated peptides could originate from either specific or non-specific binders which can complicate the analysis. Non-specific binding proteins could show different abundances after immunoprecipitation and mistakenly be considered as differentially acetylated proteins without mass spectrometry identification of the actual acetylated peptides. In addition, protease digestion of total cellular extracts prior to IP exposes a greater amount of acetylated lysine residues and significantly helps to enrich the acetylated peptides. This was seen by our increased identification of organ specific acetylated proteins in comparison to studies where spots identified as having increased acetylation are cut out of 1-D or 2-D gels, and trypsin cleavage is done in gel, followed by extraction of the digested tryptic peptides10, 25.
Label free quantification of the acetylated peptides
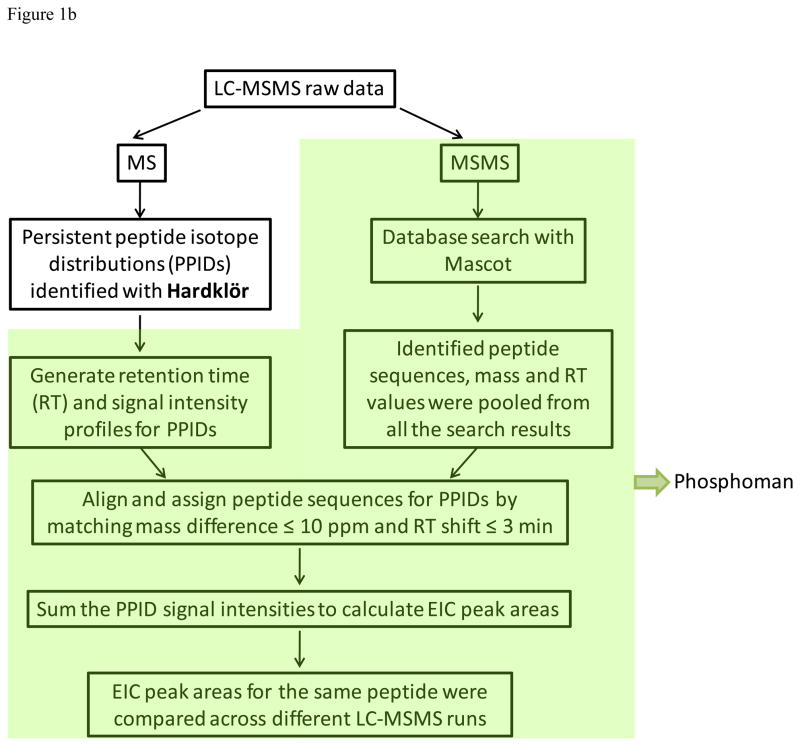
Custom software written in-house called Phosphoman was used for quantification of the acetylated peptides from multiple samples. Phosphoman was written to enable quantification of peptide results with Mascot, the existing database search algorithm used in our lab. The data analysis workflow consists of three major functions including: 1) chromatographic alignment of multiple sample injections, 2) assignment of peptide sequence identifications to their respective precursor ion signals, and 3) quantitative analysis of the peptide signals across all data sets. Each of these functions is described in detail below.
Chromatographic alignment of multiple sample injections
Peptide isotope distributions from the primary spectra of each run were identified using the feature detection software, Hardklör26, with a correlation cutoff of 0.9. Persistent peptide isotope distributions (PPIDs) were acquired for each run by tracing the peptide signals identified with Hardklör that persisted over at least three consecutive spectra (allowing for a single gap) with a 10 ppm monoisotopic mass tolerance. For each scan event in which a persistent signal was observed, the retention time and signal intensity were recorded to create a chromatographic profile for that PPID. The list of PPIDs for each run could then be aligned with each other by matching mass within 10 ppm and retention time overlap with a 3 minute tolerance.
Assignment of peptide sequence identifications
Because the mass spectra were acquired using shotgun methods, a single peptide sequence database was assembled from the pooled database searching results for all the samples being compared. For a peptide sequence to be accepted into the database, it must be observed in at least one run with a consistent chromatographic elution profile. PPIDs were assigned a peptide sequence from the database if the monoisotopic masses matched within 10 ppm and were within the range of observed retention times with a 3 minutes tolerance.
Quantitative analysis of the peptide signals
Differences in peptide signal abundance levels were compared across the different samples analyzed with Phosphoman. The peptide signal intensity at each scan event of a PPID was summed together to obtain a measurement of the extracted ion chromatographic (EIC) peak area for that PPID. All the peptides were used for alignment, however only the modified one were analyzed quantitatively. A scheme of data analysis is shown in Figure 1b.
Study of the deviation with an internal standard
The deviation caused by sample preparation and mass spectrometric analyses was carefully studied. An acetylated BSA sample was prepared as described in a previous report27. An equal amount (15 picomoles) of the acetylated BSA protein was spiked into each mouse liver sample before the samples were subjected to trypsin digestion. Therefore, equal amounts of the acetylated BSA peptides were used as internal standards. Supplementary Figure 2 shows the quantification results of all the identified acetylated BSA peptides, with sequences indicated. Good overall reproducibility of the experiment is observed. None of the acetylated BSA peptide log10 ratios fall outside ± 2σ of the distribution of log10 (re-fed/fasted) of liver samples. The averaged EIC area observed from the re-fed sample is 1.1-fold of that from the fasted sample and the averaged standard deviation in measured BSA peptide PPID areas is 18%.
Results
1. Label free quantification
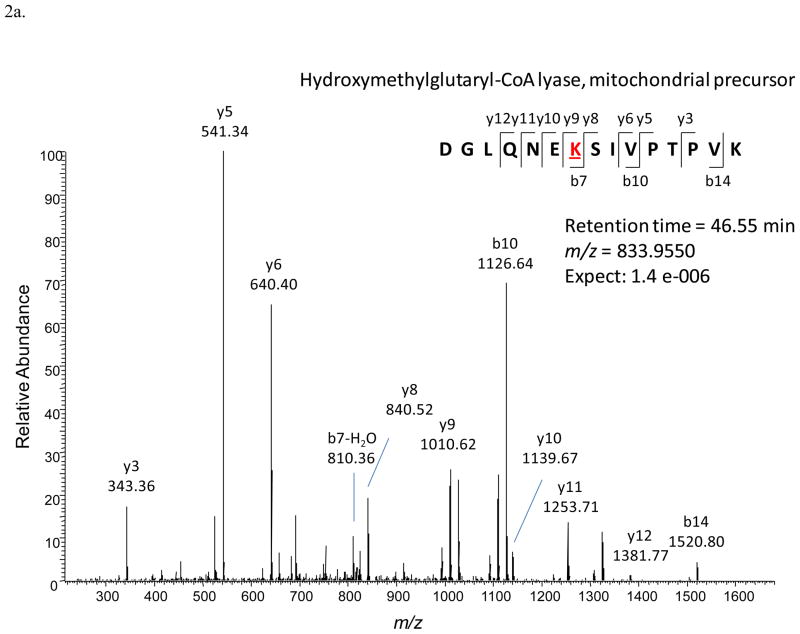
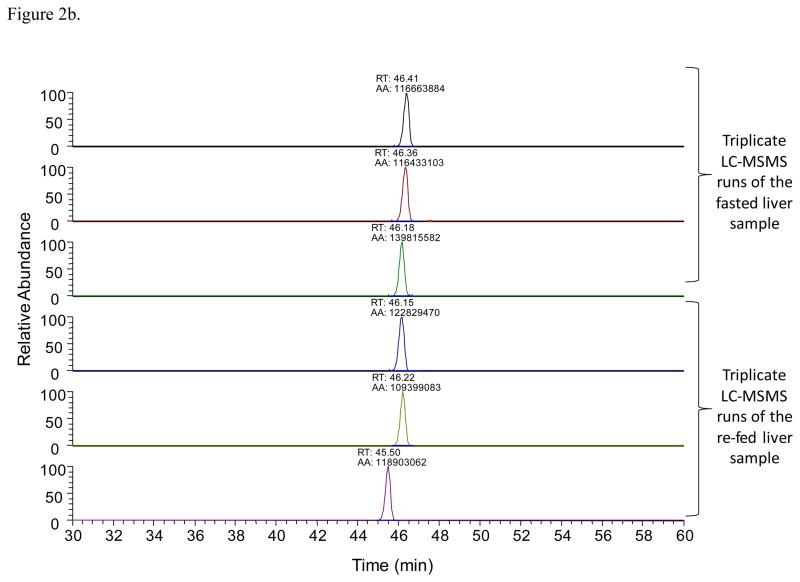
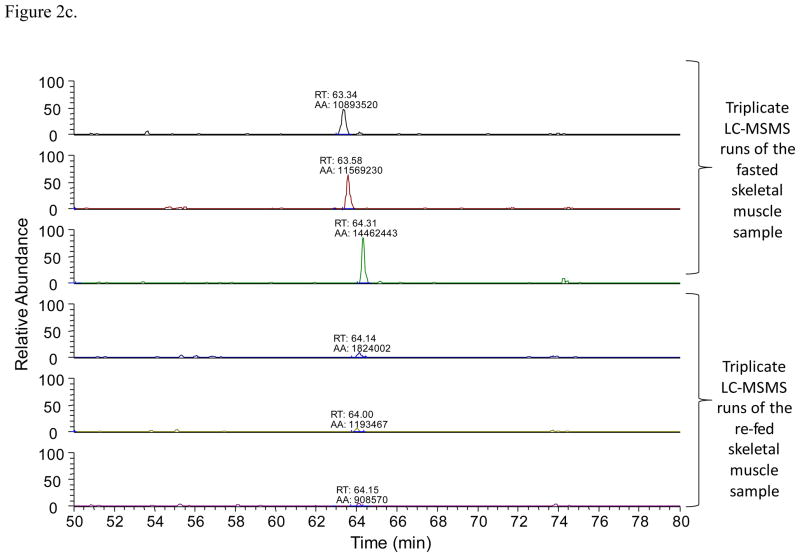
Once an acetylated peptide was identified in one LC-MSMS run, a mass-and-time tag was given to the peptide to allow its identification in the other LC-MSMS measurements with the same accurate mass (≤ 10 ppm) and retention time (± 1.5 min)28. For LFQ analyses, EIC peak areas of each acetylated peptide were compared between different LC-MSMS experiments. Figure 2 illustrates an example of the acetylated peptide from hydroxymethylglutaryl-CoA lyase that was identified. The acetylated peptide was identified by using the LC-MSMS data from the fasted liver sample (Figure 2a). Accurate mass and retention time was used to extract the specific peptide’s EICs from the other LC-MSMS runs. Figure 2b was generated with the software Xcalibur with a mass error tolerance of 0.01 Da (< 10 ppm) to illustrate how quantification with Phosphoman was achieved. Phosphoman does not yield graphical data, but reports only numerical quantification results. Between different runs, the observed retention time variance is less than 1 minute, which demonstrates good reproducibility of the LC performance. Integrated EIC peak areas are listed as “AA” values on top of each EIC peaks. Six EIC traces are from the triplicate runs of the fasted and re-fed liver tissues respectively. The average areas and standard deviations were calculated from each group of the triplicate runs. In this case, the average of the fasted liver sample area is 1.24 × 108 (in arbitrary units) and the standard deviation is 1.36 × 107; while the average of the re-fed liver sample is 1.17 × 108 and the standard deviation is 0.72 × 107. In addition, Figure 2c shows the quantification results of an acetylated peptide (FCVGLQK@IEEIFKK) that was found to be altered in abundance (3-fold or greater) with feeding status. The average peak area of the peptide under the fasting condition is 1.23 × 107 with a standard deviation of 1.91 × 106, and that under the re-feeding condition is 1.31 × 106 with a standard deviation of 4.66 × 105. In this study, 733 acetylated peptides like this example were identified and automatically quantified in the LFQ approach with the software Phosphoman. For comparison with existing methods, data obtained from fasted liver samples were also analyzed with the published software MaxQuant29. The relative peak areas (area of a specific peptide/the most abundant peptide’s area) and the standard deviations were calculated with the two software tools respectively (Supplementary Table 1). For most peptides, good agreement in calculated peak areas and standard deviation values was observed between Phosphoman and MaxQuant results.
Figure 2.
An example of LFQ analysis of the acetylated peptides
2a. Identification of the acetylated peptide DGLQNEK@SIVPTPVK with the LC-MSMS data and Mascot database search. The modified lysine residue is marked with @.
2b. EIC peaks of the acetylated peptide DGLQNEK@SIVPTPVK with the retention time and peak areas labeled. The retention time shift is less than 1 min. The peak areas are used in the LFQ analysis.
2c. An example of a peptide with altered abundance observed during fasted-to-fed transition: FCVGLQK@IEEIFKK.
2. Mouse acetylome
Here, the acetylome of seven vital mouse tissues harvested under different physiological conditions (fasting and re-feeding) are reported. Unique peptides identified from protein groups were clustered together and treated as a single entry. Peptides that have the same sequence and modification but were reported by Mascot to be modified at different residues were validated by manual inspection of the specific MSMS fragments that are unique to the modification positions. Validated peptides with a common sequence but modifications at different sites are treated as different entries in the table. An index number was given to each entry. Multiple acetylated peptides from the same protein were grouped in the list, so they have neighboring index numbers. A total of 733 non-redundant acetylated peptides from 337 mouse proteins were identified with a false discovery rate (FDR) less than 1.5% (Supplementary Table 2), which constitutes the largest mouse acetylome dataset produced to date. Previously, Kim et al. and Schwer et al. reported 195 and 287 acetylated proteins from mouse liver tissues8, 11 and our results have 31 and 86 proteins in common with these previous reports, respectively.
3. Tissue specific acetylome profiles
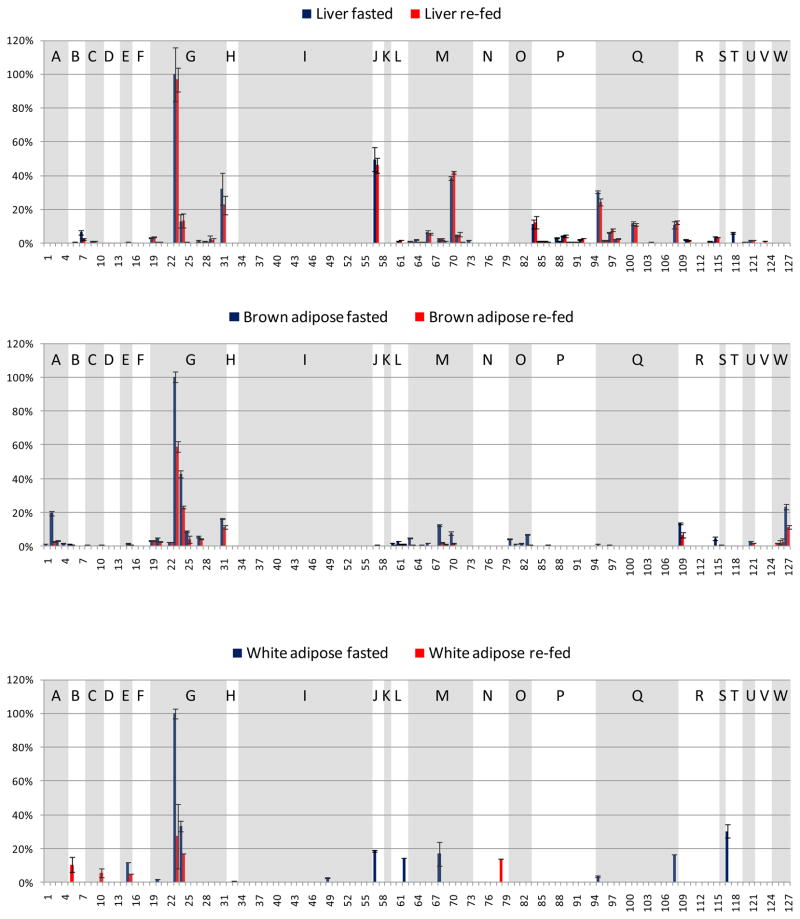
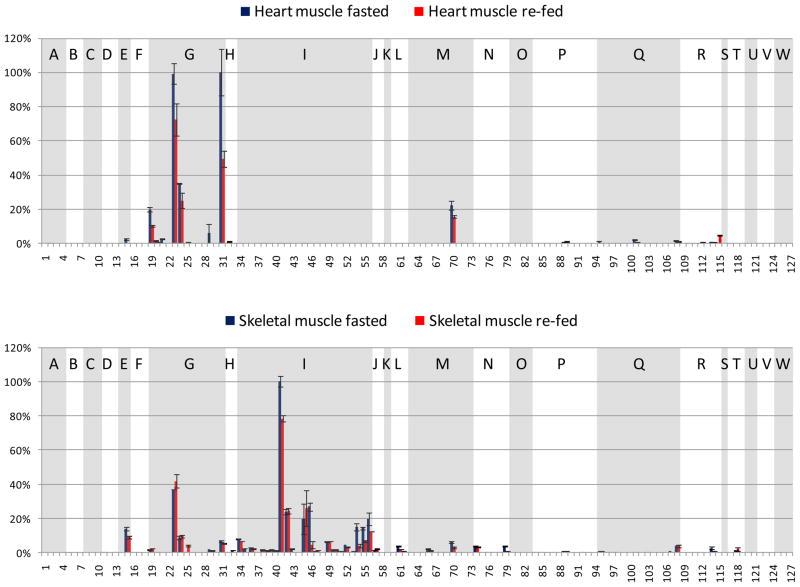
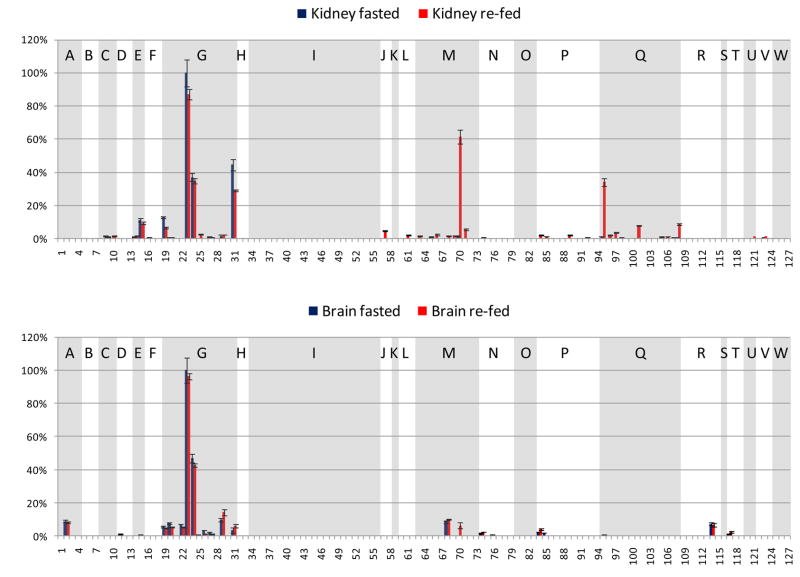
A goal of this study was to investigate the acetylation sites observed in multiple mouse tissues and organs and to quantify acetylation level changes upon fasting to re-fed transition. Thus, a challenge in the analysis was to visualize a large number of acetylation sites, how they changed with feeding status, and how they did so across tissue types or organs. To achieve a comparison that can be more readily visualized, a reduced number of identified acetylated peptides are displayed from 23 metabolic and chaperone proteins. Since this work involved a fasting to re-fed transition, metabolic proteins were considered most relevant for the comparison across multiple organs, and these 21 metabolic and 2 chaperone proteins were selected based on prior knowledge of function. Selection of peptides from this set of proteins resulted in 127 acetylated peptides (Table 1). Each panel in Figure 3 illustrates one organ-specific acetylome profile of the 23 selected proteins identified in these experiments, where extracted ion chromatographic peak areas from each peptide were derived through analysis with Phosphoman and were used for quantification. In each panel, all signals were normalized to the most abundant acetylated peptide from this subset observed in that tissue or organ, and standard deviations were calculated from triplicate LC-MSMS experiments. Peptides from the same protein are grouped and shaded with white or gray background. The selected 23 proteins are indicated with single letters on top of each group of peptides and the protein names are listed in Figure 3. Acetylation sites on several peptides were observed to be common across different tissues. For example, peptides 23 and 24 from ATP synthase coupling factor 6 (F6) have high abundance in all the samples, regardless of tissue types and feeding status. However in many other cases, the same acetylated peptide shows remarkably different abundance levels in different tissues (see Discussion). For example, these include acetylated peptides 57 (from protein Hydroxymethylglutaryl-CoA lyase) and 127 (from Glycine cleavage system H protein) that show abundant, but opposing trends in liver and brown adipose tissues and yet appear absent or less abundant in other tissues. Global measurements on protein levels including the substrates for acetylation identified here will enable determination of the extent of differential protein acetylation that exists in each tissue. Nonetheless, measurements on protein acetylation patterns themselves may be important for improved understanding of tissues-specific energy sensing mechanisms and energy homeostasis.
Table 1.
Identified acetylated peptides from the selected metabolic and chaperone proteins
| Index | Protein accession number | Protein name | Peptide sequence |
|---|---|---|---|
| 1 | IPI00116074 | Aconitate hydratase, mitochondrial precursor | AIITK@SFAR |
| 2 | IPI00116074 | Aconitate hydratase, mitochondrial precursor | YDLLEK@NINIVR |
| 3 | IPI00116074 | Aconitate hydratase, mitochondrial precursor | FK@LEAPDADELPR |
| 4 | IPI00116074 | Aconitate hydratase, mitochondrial precursor | CTTDHISAAGPWLK@FR |
| 5 | IPI00117978 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial precursor | WDYDK@NEWK |
| 6 | IPI00117978 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial precursor | AHGSVVK@SEDYAFPTYADR |
| 7 | IPI00117978 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial precursor | AHGSVVK@SEDYAFPTYADRR |
| 8 | IPI00315794 | Cytochrome b5 type B precursor | QYYIGDVHPSDLK@PK |
| 9 | IPI00315794 | Cytochrome b5 type B precursor | EM#LK@QYYIGDVHPSDLKPK |
| 10 | IPI00315794 | Cytochrome b5 type B precursor | ATPEASGSGEK@VEGSEPSVTYYR |
| 11 | IPI00121440 | Electron transfer flavoprotein subunit beta | VLAK@LAEK |
| 12 | IPI00121440 | Electron transfer flavoprotein subunit beta | VLAK@LAEKEK |
| 13 | IPI00121440 | Electron transfer flavoprotein subunit beta | LAEKEK@VDLLFLGK |
| 14 | IPI00122549 | Isoform Pl-VDAC1 of Voltage-dependent anion-selective channel protein 1 | LTLSALLDGK@NVNAGGHK |
| 15 | IPI00122549 | Isoform Pl-VDAC1 of Voltage-dependent anion-selective channel protein 1 | FGIAAK@YQVDPDACFSAK |
| 16 | IPI00118986 | ATP synthase O subunit, mitochondrial precursor | IGEK@YVDMSAK |
| 17 | IPI00118986 | ATP synthase O subunit, mitochondrial precursor | TVKVK@SLNDITK |
| 18 | IPI00118986 | ATP synthase O subunit, mitochondrial precursor | TVK@VKSLNDITK |
| 19 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | LFVDK@IR |
| 20 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | FEVIDK@PQS |
| 21 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | LFVDK@IREYK |
| 22 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | ELDPVQK@LFVDK |
| 23 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | FDDPKFEVIDK@PQS |
| 24 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | FDDPK@FEVIDKPQS |
| 25 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | FDDPK@FEVIDK@PQS |
| 26 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | GEMDTFPTFK@FDDPK |
| 27 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | GEM#DTFPTFK@FDDPK |
| 28 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | QM#YGK@GEM#DTFPTFK |
| 29 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | GEMDTFPTFKFDDPK@FEVIDKPQS |
| 30 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | GEMDTFPTFK@FDDPKFEVIDKPQS |
| 31 | IPI00125460 | ATP synthase coupling factor 6, mitochondrial precursor | GEMDTFPTFKFDDPK@FEVIDKPQS |
| 32 | IPI00120076 | Creatine kinase, sarcomeric mitochondrial precursor | FSK@ILENLR |
| 33 | IPI00120076, IPI00127596 | Creatine kinase, sarcomeric mitochondrial precursor | VISM#EK@GGNM#K |
| 34 | IPI00127596 | Creatine kinase M-type | GK@YYPLK |
| 35 | IPI00127596 | Creatine kinase M-type | IEEIFK@K |
| 36 | IPI00127596 | Creatine kinase M-type | HGGYK@PTDK |
| 37 | IPI00127596 | Creatine kinase M-type | LANLSK@HPK |
| 38 | IPI00127596 | Creatine kinase M-type | LM#VEM#EK@K |
| 39 | IPI00127596 | Creatine kinase M-type | PFGNTHNK@FK |
| 40 | IPI00127596 | Creatine kinase M-type | GGVHVK@LANLSK |
| 41 | IPI00127596 | Creatine kinase M-type | HPK@FEEILTR |
| 42 | IPI00127596 | Creatine kinase M-type | VLTPDLYNK@LR |
| 43 | IPI00127596 | Creatine kinase M-type | FCVGLQK@IEEIFK |
| 44 | IPI00127596 | Creatine kinase M-type | LSVEALNSLTGEFK@GK |
| 45 | IPI00127596 | Creatine kinase M-type | LNYK@PQEEYPDLSK |
| 46 | IPI00127596 | Creatine kinase M-type | FCVGLQK@IEEIFKK |
| 47 | IPI00127596 | Creatine kinase M-type | HNNHM#AK@VLTPDLYNK |
| 48 | IPI00127596 | Creatine kinase M-type | AVEK@LSVEALNSLTGEFK |
| 49 | IPI00127596 | Creatine kinase M-type | FKLNYK@PQEEYPDLSK |
| 50 | IPI00127596 | Creatine kinase M-type | FK@LNYKPQEEYPDLSK |
| 51 | IPI00127596 | Creatine kinase M-type | LSVEALNSLTGEFKGK@YYPLK |
| 52 | IPI00127596 | Creatine kinase M-type | TDLNHENLK@GGDDLDPNYVLSSR |
| 53 | IPI00127596 | Creatine kinase M-type | HKTDLNHENLK@GGDDLDPNYVLSSR |
| 54 | IPI00127596 | Creatine kinase M-type | SMTEQEQQQLIDDHFLFDK@PVSPLLLASGMAR |
| 55 | IPI00127596, IPI00127596 | Creatine kinase M-type | SMTEQEQQQLIDDHFLFDK@PVSPLLLASGM#AR |
| 56 | IPI00127596 | Creatine kinase M-type | SM#TEQEQQQLIDDHFLFDK@PVSPLLLASGM#AR |
| 57 | IPI00127625 | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor | DGLQNEK@SIVPTPVK |
| 58 | IPI00127625 | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor | DGLQNEK@SIVPTPVKIR |
| 59 | IPI00130804 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial precursor | SITFSK@L |
| 60 | IPI00134809 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial precursor | HK@DAFLK |
| 61 | IPI00134809 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial precursor | HKDAFLK@K |
| 62 | IPI00134809 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial precursor | HNLK@LGFMSAFVK |
| 63 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | VLK@YAGLK |
| 64 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | VDFSK@VPK |
| 65 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | VLK@YAGLKK |
| 66 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | GATPYGGVK@LEDLIVK |
| 67 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | GKPDVVVK@EDEEYKR |
| 68 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | EAWDAGK@FASEITPITISVK |
| 69 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | DGLTDVYNK@IHM#GNCAENTAK |
| 70 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | SKEAWDAGK@FASEITPITISVK |
| 71 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | GATPYGGVK@LEDLIVKDGLTDVYNK |
| 72 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | AGIPK@EEVKEVYM#GNVIQGGEGQAPTR |
| 73 | IPI00154054 | Acetyl-CoA acetyltransferase, mitochondrial precursor | TVFQK@ENGTITAANASTLNDGAAALVLM#TAEAAQR |
| 74 | IPI00221402, IPI00119458, IPI00649880 | Fructose-bisphosphate aldolase A | DGADFAK@WR |
| 75 | IPI00221402, IPI00133580 | Fructose-bisphosphate aldolase A | GILAADESTGSIAK@R |
| 76 | IPI00221402 | Fructose-bisphosphate aldolase A | K@ELSDIAHR |
| 77 | IPI00221402, IPI00649880 | Fructose-bisphosphate aldolase A | AAQEEYIK@R |
| 78 | IPI00221402, IPI00649880 | Fructose-bisphosphate aldolase A | CPLLK@PWALTFSYGR |
| 79 | IPI00221402 | Fructose-bisphosphate aldolase A | CVLK@IGEHTPSALAIMENANVLAR |
| 80 | IPI00223092 | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (Trifunctional protein), alpha subunit | M#FEK@LEK |
| 81 | IPI00223092 | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (Trifunctional protein), alpha subunit | GQQQVFK@GLNDK |
| 82 | IPI00223092 | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (Trifunctional protein), alpha subunit | DSIFSNLIGQLDYK@GFEK |
| 83 | IPI00223092 | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (Trifunctional protein), alpha subunit | AGLEQGSDAGYLAESQK@FGELALTK |
| 84 | IPI00133903 | Stress-70 protein, mitochondrial precursor | LK@EEISK |
| 85 | IPI00133903 | Stress-70 protein, mitochondrial precursor | HIVK@EFKR |
| 86 | IPI00133903 | Stress-70 protein, mitochondrial precursor | NAEK@YAEEDR |
| 87 | IPI00133903 | Stress-70 protein, mitochondrial precursor | YDDPEVQK@DTK |
| 88 | IPI00133903 | Stress-70 protein, mitochondrial precursor | NAEK@YAEEDRR |
| 89 | IPI00133903 | Stress-70 protein, mitochondrial precursor | AQFEGIVTDLIK@R |
| 90 | IPI00133903 | Stress-70 protein, mitochondrial precursor | RYDDPEVQK@DTK |
| 91 | IPI00133903 | Stress-70 protein, mitochondrial precursor | QATK@DAGQISGLNVLR |
| 92 | IPI00133903 | Stress-70 protein, mitochondrial precursor | ETGVDLTK@DNM#ALQR |
| 93 | IPI00133903 | Stress-70 protein, mitochondrial precursor | YDDPEVQKDTK@NVPFK |
| 94 | IPI00133903 | Stress-70 protein, mitochondrial precursor | ASNGDAWVEAHGK@LYSPSQIGAFVLM#K |
| 95 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | EGFEK@ISK |
| 96 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | IGIEIIK@R |
| 97 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | SIAK@EGFEK |
| 98 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | SIDLK@DKYK |
| 99 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | SIDLKDK@YK |
| 100 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | LAK@LSDGVAVLK |
| 101 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | SIAKEGFEK@ISK |
| 102 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | GVM#LAVDAVIAELK@K |
| 103 | IPI00308885, IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | VGGTSDVEVNEKK@DR |
| 104 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | VGEVIVTK@DDAM#LLK |
| 105 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | CIPALDSLK@PANEDQK |
| 106 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | TLNDELEIIEGM#K@FDR |
| 107 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | K@PLVIIAEDVDGEALSTLVLNR |
| 108 | IPI00308885 | 60 kDa heat shock protein, mitochondrial precursor | IQEITEQLDITTSEYEKEK@LNER |
| 109 | IPI00323592 | Malate dehydrogenase, mitochondrial precursor | GEDFVK@NM#K |
| 110 | IPI00323592 | Malate dehydrogenase, mitochondrial precursor | KGEDFVK@NM#K |
| 111 | IPI00323592 | Malate dehydrogenase, mitochondrial precursor | ASIKK@GEDFVK |
| 112 | IPI00323592 | Malate dehydrogenase, mitochondrial precursor | NLGIGK@ITPFEEK |
| 113 | IPI00323592 | Malate dehydrogenase, mitochondrial precursor | ANTFVAELK@GLDPAR |
| 114 | IPI00323592 | Malate dehydrogenase, mitochondrial precursor | ITPFEEK@MIAEAIPELK |
| 115 | IPI00323592 | Malate dehydrogenase, mitochondrial precursor | ITPFEEK@M#IAEAIPELK |
| 116 | IPI00330523 | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor | TFDK@ILIANR |
| 117 | IPI00466128 | Alcohol dehydrogenase | YDSTHYK@ETWK |
| 118 | IPI00466128 | Alcohol dehydrogenase | GDNPFPK@NADGTVR |
| 119 | IPI00466128 | Alcohol dehydrogenase | M#PLIGLGTWK@SEPGQVK |
| 120 | IPI00230034 | D-dopachrome decarboxylase | FFPLEAWQIGK@K |
| 121 | IPI00230034 | D-dopachrome decarboxylase | LCAATATILDK@PEDR |
| 122 | IPI00312058 | Catalase | DAQLFIQK@K |
| 123 | IPI00312058 | Catalase | TFYTK@VLNEEER |
| 124 | IPI00312058 | Catalase | IQALLDK@YNAEKPK |
| 125 | IPI00453724 | Glycine cleavage system H protein, mitochondrial precursor | MTLSDPSELDELM#SEEAYEK@YVK |
| 126 | IPI00453724 | Glycine cleavage system H protein, mitochondrial precursor | M#TLSDPSELDELMSEEAYEK@YVK |
| 127 | IPI00453724 | Glycine cleavage system H protein, mitochondrial precursor | M#TLSDPSELDELM#SEEAYEK@YVK |
Figure 3.
Fasted and re-fed tissue-specific mouse acetylome profiles of the selected 127 acetylated peptides from the 23 metabolic proteins and chaperones. x-axis: the index number of the selected 127 acetylated peptides, y-axis: relative EIC area (the EIC area of a specific acetylated peptide divided by that of the most abundant acetylated peptide from the selected proteins in that tissue). Blue bars indicate the fasted state, pink bars the re-fed state. Shown panels are: a. liver, b. brown adipose, c. white adipose, d. heart muscle, e. skeletal muscle, f. kidney, g. brain. The peptides from the same protein are grouped, and the proteins are indicated with single letters.
A: Aconitate hydratase, mitochondrial precursor (ACO2)
B: Cytochrome c oxidase subunit 4 isoform 1, mitochondrial precursor (COX4)
C: Cytochrome b5 type B precursor (Cyb5b)
D: Electron transfer flavoprotein subunit beta (β-ETF)
E: Isoform Pl-VDAC1 of Voltage-dependent anion-selective channel protein 1 (VDAC1)
F: ATP synthase O subunit, mitochondrial precursor (OSCP)
G: ATP synthase coupling factor 6, mitochondrial precursor (F6)
H: Creatine kinase, sarcomeric mitochondrial precursor (S-MtCK)
I: Creatine kinase M-type (M-CK)
J: Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (HMGCL)
K: Product of the ECH1 gene, Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial precursor (ECH1)
L: Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial precursor (α-KGDH)
M: Acetyl-CoA acetyltransferase, mitochondrial precursor (ACAT1)
N: Fructose-bisphosphate aldolase A (ALDOA)
O: Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (Trifunctional protein), alpha subunit (TP-α)
P: L: Stress-70 protein, mitochondrial precursor (GRP-75)
Q: 60 kDa heat shock protein, mitochondrial precursor (HSP-60)
R: Malate dehydrogenase, mitochondrial precursor (MDH2)
S: Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (PCCA-α)
T: Alcohol dehydrogenase (ADH)
U: D-dopachrome decarboxylase (DDT)
V: Catalase (CAT)
W: Glycine cleavage system H protein, mitochondrial precursor (GCSH)
4. Comparison of mouse metabolic acetylome under different feeding conditions
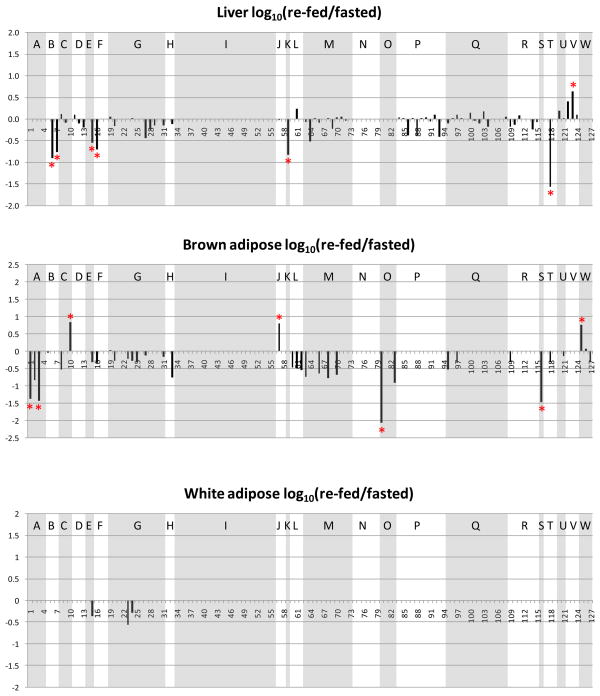
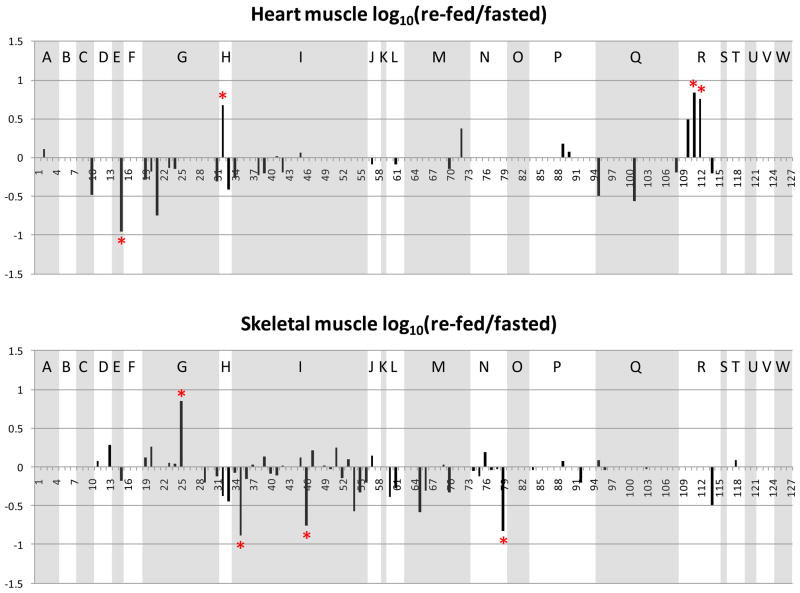
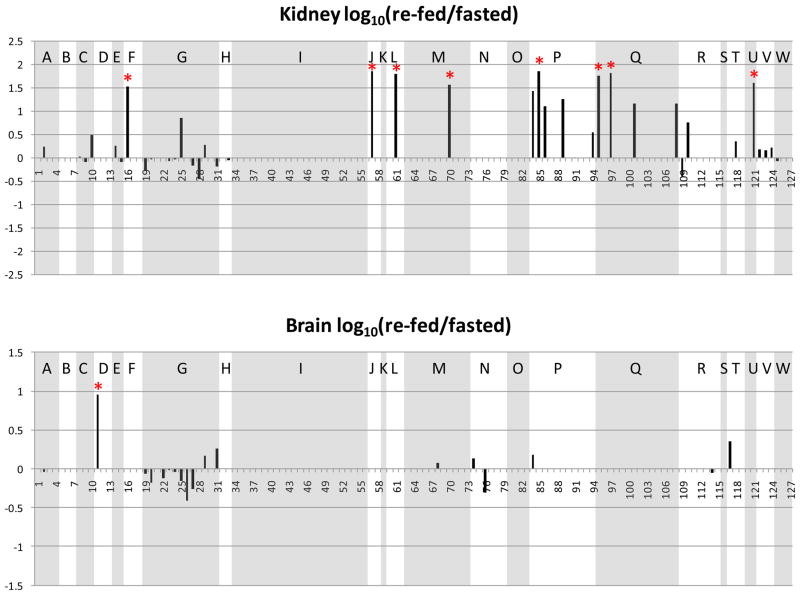
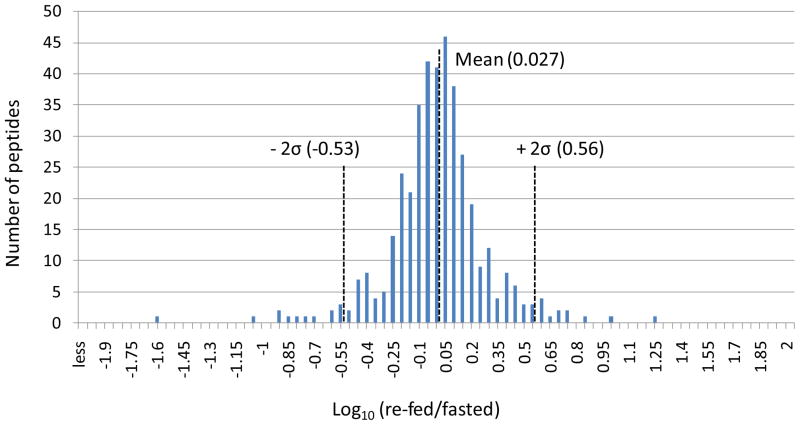
Changes of the observed levels of acetylated peptides under fasting and re-feeding conditions are visualized by plotting the log10 (re-fed/fasted) values of the areas of the acetylated peptides in Figure 4. The same acetylated peptides from those metabolic and chaperone proteins plotted in Figure 3 are used in generating Figure 4. The sequences and protein origins of these peptides are listed in Table 1, and their index values are used as the x-axis in Figure 4. For an identified acetylated peptide in a specific tissue, the quantified EIC area from the re-fed sample was divided by that from the fasted sample. Log10 (re-fed/fasted) higher than or lower than 0 indicates the acetylated peptide abundance is increased or decreased in the re-fed sample, respectively. Similar to Figure 3, the peptides from the same protein are grouped and shaded to allow visualization of multiple sites for each protein. Furthermore, to determine which acetylated peptides appeared with the highest altered abundance levels, measurements on the distributions of the log10 (re-fed/fasted) data were performed. For each tissue or organ, data from all the quantified peptides were used to plot the distribution of the log10 values. For example, Figure 5 illustrates the distribution derived from the liver data. Most log10 (re-fed/fasted) values are clustered at 0.027 (approximately 1:1 of re-fed: fasted), only a few are distant from the distribution center. The average and standard deviation were then calculated from the distribution plot. Acetylated peptides with ratios greater than ± two standard deviations from the mean were selected as those with the top 5% largest changed levels (> 3-fold change, more discussion in the supplementary file). In this way, a total of 58 acetylated peptides were found to show the greatest altered levels under fasted/re-feeding conditions (> 3-fold change) from seven tissues or organs, among which 31 peptides are from the 23 selected metabolic proteins and chaperones (listed in Table 2 and marked with * in Figure 4). Excitingly, many of the log10 (re-fed/fasted) values for insulin sensitive tissues, such as liver, brown adipose and skeletal muscle are negative, which indicates acetylation levels for a majority of proteins are decreased in these tissues under the re-feeding condition. On the other hand, insulin insensitive organs like kidney and brain show the opposite trend and the log10 (re-fed/fasted) values of many proteins are observed to be positive. This suggests that re-fed/fasted acetylation changes reflect tissue-specific fuel utilization changes (metabolic flexibility), rather than being an indicator of tissue or organ specific insulin effects. For example, if insulin-specific effects were the only cause for the shift in acetylation patterns between fasted and re-fed conditions, one might expect that insulin insensitive organs to exhibit little or no change upon re-feeding. The altered abundance levels of the acetylated peptides may be resultant from the altered acetylation levels in the fasted and re-fed mice, and/or altered protein levels. To investigate this, catalase and alcohol dehydrogenase protein levels in fasted and re-fed mouse liver samples were measured with western blot analyses (Supplementary Figure 3). The quantification results show that the protein levels are unaltered, while the acetylation levels detected on peptides from these proteins appear with fasted-to-fed ratios among the top 5% of all those measured (greater than 3-fold change). In addition, it should be noted that differential regulation of these proteins on a tissue-specific basis, whether that includes PTM levels, protein abundance levels or both implicates differing functional role of these species in each tissue under different feeding conditions. This in itself presents a novel and exciting result that can impact understanding of metabolic regulation in a new way. Furthermore in several cases, many peptides from a given protein were identified as acetylated; however only one or a couple were found to change more than 3-fold. For example, 2–10 acetylated peptides were quantified from protein G (ATP synthase F6) from each tissue or organ; however the re-fed/fasted ratios for most peptides hover around 1:1 in all the cases except for peptide 25 in skeletal muscle. Another example is protein I (creatine kinase M-type). In this case, the difference between muscle and the other tissues is dramatic. In skeletal and heart muscle, more than 10 acetylated peptides of creatine kinase were quantified; however none of those peptides could be quantified in any of the other tissues. Such observation illustrates excellent agreement with the known tissue-specific expression since this protein is found with expression in skeletal muscle much higher than in any other tissue and agrees with the finding of creatine kinase M acetylation in an earlier report25. On the peptide level, from the 19 quantified acetylated peptides from creatine kinase M-type in skeletal muscle, the level changes of only 2 were found to change greater than 3-fold, while the other 17 appear unchanged. Such observation highlights the significance of acetylation on those two specific sites. In another case, different subunits of a complex were acetylated in different tissues. For example, the F6 subunit of the peripheral arm of the ATP synthase complex49 was hyperacetylated in skeletal muscle, whereas the oligomycin sensitivity conferral protein (OSCP) of the peripheral arm was hyperacetylated in kidney, but hypoacetylated in liver. Further investigations of three-dimensional structures of these acetylated lysine sites may reveal key mechanisms relevant to protein function and how they can be modulated by acetylation under different physiological conditions.
Figure 4.
Log10 (re-fed/fasted) values of the acetylated peptide EIC areas in different tissues. Shown panels are: a. liver, b. brown adipose, c. white adipose, d. heart muscle, e. skeletal muscle, f. kidney, g. brain. The peptides from the same protein are grouped, and the proteins are indicated the same as in Figure 3.
Figure 5.
Distribution plot of the liver log10 (re-fed/fasted) data. The center of the distribution is 0.027, and the standard deviation is 0.267.
Table 2.
Acetylated peptides from metabolic proteins that appear altered in abundance (3-fold or higher) in fasted-to-fed comparison. Note: acetylated peptides from metabolic proteins in white adipose samples were not observed with altered abundance levels of 3-fold or higher and thus, are not included in this table.
| liver
| ||||
|---|---|---|---|---|
| Protein accession number | Peptide sequence | log10(re-fed/fasted) | Index | Protein name |
| IPI00466128 | R.GDNPFPK@NADGTVR.Y | −1.56 | 118 | Alcohol dehydrogenase |
| IPI00117978 | R.AHGSVVK@SEDYAFPTYADR.R | −0.90 | 6 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial precursor |
| IPI00130804 | K.SITFSK@L.- | −0.82 | 59 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial precursor |
| IPI00117978 | R.AHGSVVK@SEDYAFPTYADRR.D | −0.76 | 7 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial precursor |
| IPI00118986 | R.IGEK@YVDM#SAK.S | −0.70 | 16 | ATP synthase O subunit, mitochondrial precursor |
| IPI00122549 | R.FGIAAK@YQVDPDACFSAK.V | −0.55 | 15 | Isoform Pl-VDAC1 of Voltage-dependent anion-selective channel protein 1 |
| IPI00312058 | R.TFYTK@VLNEEER.K | 0.64 | 123 | Catalase |
|
| ||||
|
Brown adipose
| ||||
| Protein accession number | Peptide sequence | log10(re-fed/fasted) | Index | Protein name |
|
| ||||
| IPI00223092 | R.M#FEK@LEK.S | −2.08 | 80 | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (Trifunctional protein), alpha subunit |
| IPI00330523 | K.TFDK@ILIANR.G | −1.47 | 116 | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor |
| IPI00116074 | K.FK@LEAPDADELPR.S | −1.43 | 3 | Aconitate hydratase, mitochondrial precursor |
| IPI00116074 | R.AIITK@SFAR.I | −1.37 | 1 | Aconitate hydratase, mitochondrial precursor |
| IPI00453724 | K.MTLSDPSELDELM#SEEAYEK@YVK.S | 0.76 | 125 | Glycine cleavage system H protein, mitochondrial precursor |
| IPI00127625 | R.DGLQNEK@SIVPTPVK.I | 0.81 | 57 | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor |
| IPI00315794 | M.ATPEASGSGEK@VEGSEPSVTYYR.L | 0.83 | 10 | Cytochrome b5 type B precursor |
|
| ||||
|
Heart muscle
| ||||
| Protein accession number | Peptide sequence | log10(re-fed/fasted) | Index | Protein name |
|
| ||||
| IPI00122549 | R.FGIAAK@YQVDPDACFSAK.V | −0.95 | 15 | Isoform Pl-VDAC1 of Voltage-dependent anion-selective channel protein 1 |
| IPI00120076 | R.FSK@ILENLR.L | 0.68 | 32 | Creatine kinase, sarcomeric mitochondrial precursor |
| IPI00323592 | K.NLGIGK@ITPFEEK.M | 0.75 | 112 | Malate dehydrogenase, mitochondrial precursor |
| IPI00323592 | K.ASIKK@GEDFVK.N | 0.84 | 111 | Malate dehydrogenase, mitochondrial precursor |
|
| ||||
|
Skeletal muscle
| ||||
| Protein accession number | Peptide sequence | log10(re-fed/fasted) | Index | Protein name |
|
| ||||
| IPI00127596 | K.IEEIFK@K.A | −0.89 | 35 | Creatine kinase M-type |
| IPI00221402 | R.CVLK@IGEHTPSALAIMENANVLAR.Y | −0.83 | 79 | Fructose-bisphosphate aldolase A |
| IPI00127596 | R.FCVGLQK@IEEIFKK.A | −0.76 | 46 | Creatine kinase M-type |
| IPI00125460 | K.FDDPK@FEVIDK@PQS.- | 0.85 | 25 | ATP synthase coupling factor 6, mitochondrial precursor |
|
| ||||
|
Kidney
| ||||
| Protein accession number | Peptide sequence | log10(re-fed/fasted) | Index | Protein name |
|
| ||||
| IPI00118986 | R.IGEK@YVDM#SAK.S | 1.52 | 16 | ATP synthase O subunit, mitochondrial precursor |
| IPI00154054 | R.SK@EAWDAGKFASEITPITISVK.G | 1.56 | 70 | Acetyl-CoA acetyltransferase, mitochondrial precursor |
| IPI00230034 | R.LCAATATILDK@PEDR.V | 1.61 | 121 | D-dopachrome decarboxylase |
| IPI00308885 | K.EGFEK@ISK.G | 1.76 | 95 | 60 kDa heat shock protein, mitochondrial precursor |
| IPI00134809 | R.HKDAFLK@K.H | 1.80 | 61 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial precursor |
| IPI00308885 | R.SIAK@EGFEK.I | 1.83 | 97 | 60 kDa heat shock protein, mitochondrial precursor |
| IPI00133903 | R.HIVK@EFKR.E | 1.87 | 85 | Stress-70 protein, mitochondrial precursor |
| IPI00127625 | R.DGLQNEK@SIVPTPVK.I | 1.87 | 57 | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor |
|
| ||||
|
Brain
| ||||
| Protein accession number | Peptide sequence | log10(re-fed/fasted) | Index | Protein name |
|
| ||||
| IPI00121440 | R.VLAK@LAEK.E | 0.96 | 11 | Electron transfer flavoprotein subunit beta |
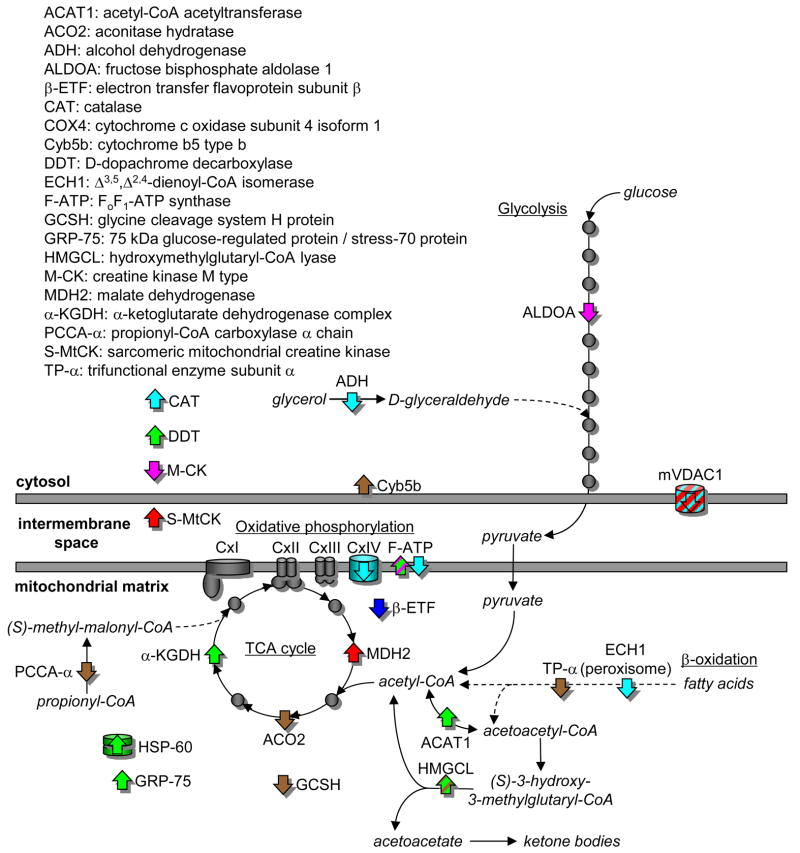
Figure 6 shows the proteins with greater than 3-fold differences in acetylation between the fasted and re-fed states in the context of metabolism, including fatty acid oxidation, glycolysis, the citric acid cycle and oxidative phosphorylation. Table 3 also shows that there were organ specific differences in these proteins with the biggest number of proteins detected in kidney, brown adipose and liver. From Table 3 and Figure 6 it appears that many of the enzymes with differences in the abundance of acetylated peptides between the fasted and re-fed state are involved in the production of acetyl CoA or its consumption in the TCA cycle and oxidative phosphorylation. The majority of the metabolic proteins with differences in the abundance of acetylated peptides are detected in one organ only. The exceptions are the OSCP of FoF1-ATP synthase (kidney and liver) and hydroxymethylglutaryl-CoA lyase (HMGCL) (kidney and brown adipose), and the mitochondrial voltage dependent anion channel 1 (mVDAC1) (liver and heart).
Figure 6.
Differences in tissue-specific acetylation of enzymes, transporters and chaperones of bioenergetic pathways in the cytoplasm and mitchondrial matrix between fasted and re-fed mice. Proteins showing a re-fed:fasted ratios of the extracted ion chromatographic peak areas for the persistent peptide isotope distributions greater than 3 are shown as an up arrow (⇑) and less than 0.33 as a down arrow (⇓), The proteins are color-coded according to tissue: brain (blue), brown fat (brown), heart muscle (red), kidney (green), liver (cyan) and skeletal muscle (magenta). Proteins showing no differential acetylation are shown as grey circles.
Table 3.
Enzymes, chaperones and carriers of bioenergetic metabolism showing differential acetylation between fasted and re-fed mice in different tissues. Proteins detected in more than one tissue are underlined.
| Tissue | Mitochondrial | Cytosolic | |||||
|---|---|---|---|---|---|---|---|
| FAO | TCA cycle | OxPh | Other | Transport | Glycolysis | Other | |
| Kidney | ACAT1 | α-KGDH | OSCP | HSP-60 | - | - | DDT |
| HMGCL | GRP-75 | ||||||
| Brown fat | TP-α | ACO2 | - | GCSH | - | - | CYB5B |
| HMGCL | |||||||
| PCCA-α | |||||||
| Liver | ECH1 | - | COX4 | - | mVDAC1 | ADH | CAT |
| OSCP | |||||||
| Muscle | - | - | F6 | - | - | ALDOA | M-CK |
| Heart | - | MDH2 | - | S-MtCK | mVDAC1 | - | - |
| Brain | - | - | β-ETF | - | - | - | - |
Abbreviations: Fatty acid oxidation (FAO), tricarboxylic acid cycle (TCA cycle), oxidative phosphorylation (OxPh), acetyl-CoA acetyltransferase (ACAT1), aconitase hydratase (ACO2), alcohol dehydrogenase (ADH), fructose-bisphosphate aldolase A (ALDOA), electron transfer flavoprotein subunit β (β-ETF), catalase (CAT), cytochrome c oxidase subunit 4 isoform 1 (COX4), cytochrome b5 type b (CYB5B), D-dopachrome decarboxylase (DDT), Δ3,5, Δ2,4-dienoyl-CoA isomerase (ECH1), ATP synthase coupling factor 6 (F6), glycine cleavage system H protein (GCSH), 75 kDa glucose-regulated protein/stress-70 protein (GRP-75), hydroxymethylglutaryl-CoA lyase (HMGCL), creatine kinase M type (M-CK), malate dehydrogenase (MDH2), α-ketoglutarate dehydrogenase complex (α-KGDH), oligomycin sensitivity conferral protein (OSCP), propionyl-CoA carboxylase α chain (PCCA-α), sarcomeric mitochondrial creatine kinase (S-MtCK), and trifunctional enzyme subunit α (TP-α).
Discussion
Approximately 2000 acetylated proteins have been previously identified in mammalian cells, and metabolic enzymes are highly represented7, 9, 30, 31. It has been postulated, mainly on the basis of liver proteomic studies, that acetylation serves to coordinate flux in the central metabolism network, as nearly all enzymes involved in glycolysis, gluconeogenesis, the TCA cycle, fatty acid oxidation, the urea cycle, glycogen metabolism, OXPHOS, and amino acid metabolism are acetylated (reviewed in Guan and Xiong32, and Patel et al30). The global tissue and organ specific metabolic acetylome data we have collected, summarized in Table 3 and Figure 6, is more extensive than has been done previously, and suggests something further, that changes in lysine acetylation may control tissue specific fuel switching between the fasted and fed states. Between the fasted and re-fed states, our results showed a large number of proteins with greater than 3-fold differences in the abundance of their acetylated peptides in metabolic pathways of the TCA cycle, β-oxidation, oxidative phosphorylation and glycolysis, as well as associated chaperones and transport proteins. Furthermore, these differences were organ specific, and heavily mitochondrial biased (Table 3), even though our global technique used whole tissue and did not enrich for mitochondria (Figure 1). Lysine acetylation has been associated with increases or decreases in protein activity9,31–33, and it will require metabolomic and fluxomic profiling to distinguish whether organs showed different metabolic responses to the fasted to re-fed transition that correlated positively or negatively with acetylation. Certainly, studies showing acetylation of malate dehydrogenase (MDH2) increases in response to glucose, and deacetylation of MDH2 decreases its activity9, support regulation of TCA cycle flux by acetylation in response to glucose changes that can occur with fasting and re-feeding. Considerations of TCA cycle flux regulation will require both metabolic profiling and fluxomic studies as while MDH2 activity is increased by acetylation, deacetylation of the SdhA subunit, key for coupling TCA cycle flux to the OXPHOS chain, increased the Complex II activity33.
One might speculate on the tissue specific effects of acetylation, based on our fasted/re-fed data (described in Fig. 4). For example, in skeletal muscle, glycolysis and creatine phosphorylation should be more active in the re-fed state34, 35, where we see decreases of acetylation levels in creatine kinase M-type and fructose-bisphosphate aldolase A. Thus, acetylation may be used to negatively regulate these enzyme activities in times when blood glucose levels drop, decreasing demands for glycolysis and energy storage. On the other hand, the increased re-fed/fasted acetylation of ATP synthase F6 may facilitate the synthesis of ATP for the fed state. In heart muscle, VDAC1, the voltage-dependent anion-selective channel, is hypoacetylated. VDACs behave as a general diffusion pore for small hydrophilic molecules, with a voltage-dependent switch between an anion-selective high-conductance state with high metabolite flux and a cation-selective low-conductance state with limited passage of metabolites. It may be that acetylation restricts the passage of metabolites, which would be increased for the fasted to fed transition. Heart MDH2 is hyperacetylated, in agreement with the increased TCA cycle flux expected for the fed vs. fasted state. For liver, the decreased re-fed/fasted acetylation for cytochrome c oxidase subunit 4, ATP synthase OSCP subunit, VDAC1, enoyl CoA hydratase 1 (ECH1), and alcohol dehydrogenase (ADH) may all serve to maximize fuel storage, and energy generation in the fasted to fed transition. As shown in Figure 6, ECH1 activity is key to unsaturated fatty acid oxidation, and it is known that acetylation increases the activity of another enzyme of fatty acid oxidation, enoyl-coenzyme A hydratase/3-hydroxyacyl-coenzyme A (EHHADH)9. Therefore, in the fed state, where fatty acid synthesis is active, it is logical that ECH1 is hypoacetylated, with presumably decreased activity. ADH may help funnel glycerol (increased from hydrolysis of triglycerides in the intestine after re-feeding) into the glycolytic pathway (Figure 6), and presumably hypoacetylation would increase ADH activity. Detailed discussion on the acetylation changes in many other proteins in different organs or tissues are included in the supplementary discussion. It is important to note however, that altered levels of acetylated peptides identified here include both biologically-significant level changes and those due to normal biological variation. Thus while not conclusive, the acetylation sites identified in this manuscript serve as useful guide for further biological studies and for the first time, show the variability in acetylation site levels that exists within different tissues from the same animals.
Conclusion
Lysine acetylation is emerging as a critical PTM found in many areas and may be part of a fasted/re-fed feedback control system for metabolic network regulation fuel switching. Thus, proteome studies are critical to elucidate and help understand the functional roles fulfilled by protein acetylation. This study presents the most comprehensive acetylome data from a set of mouse organs produced to date. Seven hundred and thirty-three acetylated peptides from 337 mouse proteins were identified and quantified, many of which are likely relevant to fasted/re-fed transition and fuel switching. From this comprehensive acetylome dataset, a new view is attained revealing previously unrecognized differential acetylation that is present in various organs or tissues derived from the same set of animals. Different organs such as liver, brown and white adipose, skeletal and heart muscle, kidney and brain show dramatic variation in quantitative profiles of acetylated peptides under differential feeding conditions. A total of 58 acetylated peptides changed 3-fold or greater under fasted/re-fed conditions, 31 of which are from metabolic proteins or chaperones. These acetylated sites represent a valuable set of targets to guide future research on fuel switching and molecular pathways relevant to metabolic disorders.
Supplementary Material
Acknowledgments
We thank Professor Derek LeRoith from Metabolism Institute, Mount Sinai Medical Centre for providing the FVB/N mice used. The present work was supported by NIH grants R01GM086688, R01RR023334 and S10RR025107 to J. E. Bruce; DK58132-01A2 grant to I. J. Kurland, and method development is additionally supported by Diabetes Research and Training Center (DRTC) NIH grant P60DK020541, and NIAID grant U19AI091175-01. A. J. Robinson is supported by the Medical Research Council, UK. Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Batta K, Das C, Gadad S, Shandilya J, Kundu TK. Reversible acetylation of non histone proteins: role in cellular function and disease. Subcell Biochem. 2007;41:193–212. [PubMed] [Google Scholar]
- 2.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41(1):185–98. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 3.Norris KL, Lee JY, Yao TP. Acetylation goes global: the emergence of acetylation biology. Sci Signal. 2009;2(97):pe76. doi: 10.1126/scisignal.297pe76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? Embo J. 2000;19(6):1176–9. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32(3):959–76. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mischerikow N, Heck AJ. Targeted large-scale analysis of protein acetylation. Proteomics. 2011;11(4):571–89. doi: 10.1002/pmic.201000397. [DOI] [PubMed] [Google Scholar]
- 7.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 8.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23(4):607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 9.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, Sack MN, Jing E, Kahn CR, Friedman JE, Jonscher KR. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2011;433(3):505–14. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, Lombard DB. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8(5):604–6. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClatchy DB, Liao L, Park SK, Xu T, Lu B, Yates Iii JR. Differential proteomic analysis of mammalian tissues using SILAM. PLoS One. 2011;6(1):e16039. doi: 10.1371/journal.pone.0016039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storlien LH, Baur LA, Kriketos AD, Pan DA, Cooney GJ, Jenkins AB, Calvert GD, Campbell LV. Dietary fats and insulin action. Diabetologia. 1996;39(6):621–31. doi: 10.1007/BF00418533. [DOI] [PubMed] [Google Scholar]
- 14.Chomentowski P, Coen PM, Radikova Z, Goodpaster BH, Toledo FG. Skeletal muscle mitochondria in insulin resistance: differences in intermyofibrillar versus subsarcolemmal subpopulations and relationship to metabolic flexibility. J Clin Endocrinol Metab. 2011;96(2):494–503. doi: 10.1210/jc.2010-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galgani JE, Moro C, Ravussin E. Metabolic flexibility and insulin resistance. Am J Physiol Endocrinol Metab. 2008;295(5):E1009–17. doi: 10.1152/ajpendo.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes. 2000;49(5):677–83. doi: 10.2337/diabetes.49.5.677. [DOI] [PubMed] [Google Scholar]
- 17.Mandarino LJ, Consoli A, Jain A, Kelley DE. Interaction of carbohydrate and fat fuels in human skeletal muscle: impact of obesity and NIDDM. Am J Physiol. 1996;270(3 Pt 1):E463–70. doi: 10.1152/ajpendo.1996.270.3.E463. [DOI] [PubMed] [Google Scholar]
- 18.Storlien L, Oakes ND, Kelley DE. Metabolic flexibility. Proc Nutr Soc. 2004;63(2):363–8. doi: 10.1079/PNS2004349. [DOI] [PubMed] [Google Scholar]
- 19.Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997;77(3):731–58. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 20.Blaxter K. Energy metabolism in animals and man. Cambridge University Press; 1989. [Google Scholar]
- 21.Cannon B, Nedergaard J. Metabolic consequences of the presence or absence of the thermogenic capacity of brown adipose tissue in mice (and probably in humans) Int J Obes (Lond) 2010;34 (Suppl 1):S7–16. doi: 10.1038/ijo.2010.177. [DOI] [PubMed] [Google Scholar]
- 22.Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, Eychmuller A, Gordts PL, Rinninger F, Bruegelmann K, Freund B, Nielsen P, Merkel M, Heeren J. Brown adipose tissue activity controls triglyceride clearance. Nat Med. 2011;17(2):200–5. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, Zhang H, Bruce JE. Optimizing the detergent concentration conditions for immunoprecipitation (IP) coupled with LC-MS/MS identification of interacting proteins. Analyst. 2009;134(4 ):755–62. doi: 10.1039/b813335b. [DOI] [PubMed] [Google Scholar]
- 24.Weisbrod CR, Hoopmann MR, Senko MW, Bruce JE. A New Dual Linear Ion Trap Fourier Transform Ion Cyclotron Resonance Mass Spectrometer: the Velos-FT. Anal Chem. 2011 Submitted 12/10. [Google Scholar]
- 25.Iwabata H, Yoshida M, Komatsu Y. Proteomic analysis of organ-specific post-translational lysine-acetylation and -methylation in mice by use of anti-acetyllysine and -methyllysine mouse monoclonal antibodies. Proteomics. 2005;5(18 ):4653–64. doi: 10.1002/pmic.200500042. [DOI] [PubMed] [Google Scholar]
- 26.Hoopmann MR, Finney GL, MacCoss MJ. High-speed data reduction, feature detection, and MS/MS spectrum quality assessment of shotgun proteomics data sets using high-resolution mass spectrometry. Anal Chem. 2007;79(15):5620–32. doi: 10.1021/ac0700833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonas A, Weber G. Presence of arginine residues at the strong, hydrophobic anion binding sites of bovine serum albumin. Biochemistry. 1971;10(8):1335–9. doi: 10.1021/bi00784a010. [DOI] [PubMed] [Google Scholar]
- 28.Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, Masselon C, Markillie LM, Moore RJ, Romine MF, Shen Y, Stritmatter E, Tolic N, Udseth HR, Venkateswaran A, Wong KK, Zhao R, Smith RD. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc Natl Acad Sci U S A. 2002;99(17):11049–54. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 30.Patel J, Pathak RR, Mujtaba S. The biology of lysine acetylation integrates transcriptional programming and metabolism. Nutr Metab (Lond) 2011;8:12. doi: 10.1186/1743-7075-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao YL, Yang WM. Beyond histone and deacetylase: an overview of cytoplasmic histone deacetylases and their nonhistone substrates. J Biomed Biotechnol. 2011;2011:146493. doi: 10.1155/2011/146493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2011;36(2 ):108–16. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49(2 ):304–11. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halias HM, Snittyns’kyi VV, Ianovych VH. The regulatory characteristics of creatine kinase activity in the skeletal muscle of piglets in the neonatal period. Fiziol Zh. 1995;41(1–2 ):25–9. [PubMed] [Google Scholar]
- 35.Dumas JF, Bielicki G, Renou JP, Roussel D, Ducluzeau PH, Malthiery Y, Simard G, Ritz P. Dexamethasone impairs muscle energetics, studied by (31)P NMR, in rats. Diabetologia. 2005;48(2 ):328–35. doi: 10.1007/s00125-004-1631-0. [DOI] [PubMed] [Google Scholar]
- 36.Wang SP, Robert MF, Gibson KM, Wanders RJ, Mitchell GA. 3-Hydroxy-3-methylglutaryl CoA lyase (HL): mouse and human HL gene (HMGCL) cloning and detection of large gene deletions in two unrelated HL-deficient patients. Genomics. 1996;33(1 ):99–104. doi: 10.1006/geno.1996.0164. [DOI] [PubMed] [Google Scholar]
- 37.Ross BD, Espinal J, Silva P. Glucose metabolism in renal tubular function. Kidney Int. 1986;29(1 ):54–67. doi: 10.1038/ki.1986.8. [DOI] [PubMed] [Google Scholar]
- 38.Cabiscol E, Belli G, Tamarit J, Echave P, Herrero E, Ros J. Mitochondrial Hsp60, resistance to oxidative stress, and the labile iron pool are closely connected in Saccharomyces cerevisiae. J Biol Chem. 2002;277(46 ):44531–8. doi: 10.1074/jbc.M206525200. [DOI] [PubMed] [Google Scholar]
- 39.Shan Y, Napoli E, Cortopassi G. Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum Mol Genet. 2007;16(8 ):929–41. doi: 10.1093/hmg/ddm038. [DOI] [PubMed] [Google Scholar]
- 40.Tsai CL, Barondeau DP. Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry. 2010;49(43 ):9132–9. doi: 10.1021/bi1013062. [DOI] [PubMed] [Google Scholar]
- 41.Rouault TA, Tong WH. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008;24(8 ):398–407. doi: 10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, Rosen N, Neckers L. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell. 2007;25(1 ):151–9. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Wang SY, Zhang XH, Zhao M, Hou CM, Xu YJ, Du ZY, Yu XD. FK228 inhibits Hsp90 chaperone function in K562 cells via hyperacetylation of Hsp70. Biochem Biophys Res Commun. 2007;356(4 ):998–1003. doi: 10.1016/j.bbrc.2007.03.076. [DOI] [PubMed] [Google Scholar]
- 44.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18 ):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 45.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2(9 ):667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 46.Karp NA, Lilley KS. Investigating sample pooling strategies for DIGE experiments to address biological variability. Proteomics. 2009;9(2 ):388–397. doi: 10.1002/pmic.200800485. [DOI] [PubMed] [Google Scholar]
- 47.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci U S A. 2005;102(12 ):4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diz AP, Truebano M, Skibinski DO. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis. 2009;30(17 ):2967–2975. doi: 10.1002/elps.200900210. [DOI] [PubMed] [Google Scholar]
- 49.Rees DM, Leslie AG, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci U S A. 2009;106(51):21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.