Abstract
Cell-based impedance biosensing is an emerging technology that can be used to non-invasively and instantaneously detect and analyze cell responses to chemical and biological agents. This article highlights the fabrication and measurement technologies of cell impedance sensors, and their application in toxin detection and anti-cancer drug screening. We start with an introduction that describes the capability and advantages of cell-based sensors over conventional sensing technology, followed by a discussion of the influence of cell adhesion, spreading and viability during cell patterning on the subsequent impedance measurements and sensing applications. We then present an electronic circuit that models the cell-electrode system, by which the cellular changes can be detected in terms of impedance changes of the circuit. Finally, we discuss the current status on using cell impedance sensors for toxin detection and anti-cancer drug screening.
1. Introduction
Cell-based biosensors, often referred to as “cytosensors”, are new hybrid systems that utilize living biological cells as sensing elements to monitor physiological changes caused by internal or external stimuli.1–6 Cells have multiple, complex analyte-recognition and signal-transduction/amplification mechanisms.7 Their diverse recognition activities make this emerging technology an ideal tool for the detection of chemical and biological toxins or mutagens and the screening of pharmacologically active compounds, offering new opportunities for biomedical applications such as drug evaluation, biothreat detection, and environmental pollutant identification.1–5, 8 Conventional molecular biosensors are principally chemical-, antibody-, or nucleic acid-based assays using specific chemical properties or molecular recognition mechanisms to identify a particular, singular agent, and thus are limited to detecting only known biothreats.5, 9 Compared to conventional molecular sensors, cell-based sensors have a number of advantages, including: (1) the ability to detect many forms of substances, including chemical toxins, bacteria, and viruses2, 10–14 due to the many types of receptors presented on cell membranes; (2) high sensing efficiency and rapid response since sensing receptors are embedded in cell membranes and therefore remain non-denatured; (3) providing insights into the physiological effect of an analyte and thus having the ability to detect and identify known and unknown biothreats;3–5 and (4) low cost, since there is no need to separate and purify proteins that are embedded in a cell membrane.
Cell-based biosensors are addressable optically or electronically.5 Typical applications of cell-based sensors include pH detection to monitor metabolism-activity, fluorescence imaging of protein-expression related to cell signaling, electrical probing and analysis of intra- and extra-cellular membrane potential using microelectrodes, and impedance detection for monitoring cell adhesion and spreading.1, 2, 5 Among electronically addressable cell-based biosensors, cellular impedance biosensors are capable of determining the events of cell adhesion, spreading, growth, motility, and death for any adherent cell types (both excitable and nonelectrically active).15, 16 By monitoring electric alternations at the contact between the cell and electrode, cellular impedance biosensors detect, analyze, and determine the physical effects of external analytes on cells non-invasively, quantitatively, and instantaneously. The measured output signal indicates the average change of cell properties over the cell population on each electrode, as a result of cellular proliferation or motility, cell-cell separation, or subtle intracellular changes induced by external stimuli.
Cellular impedance biosensors are typically developed by immobilizing cells on an array of electrodes on an insulative substrate with each electrode hosting a group of cells. The electrode microarray is integrated with microelectronics for real-time data acquisition, analysis, and display. The primary challenge in the development of microarrays of cytosensors is the ability to immobilize cells on designated areas of a substrate without compromising their viability and functionality.
This article describes the fabrication and measurement principles of cellular impedance sensor microarrays and highlights recent advances and challenges in toxin detection and drug screening applications using this advanced technology. An overview of cell patterning techniques is given, and the mechanisms of cell adhesion, spreading, and viability are also discussed as they are related to the efficacy of cellular biosensors.
2. Cell Patterning, Adhesion, and Viability
Microelectrode Arrays for Cell Patterning
In order to monitor biological responses of cells, a sensing device must accommodate cells in designated areas addressable electronically or optically. The process that places live cells on these areas is commonly known as cell patterning. In a typical electronically addressable sensing device, cells are patterned on an array of electrodes. The primary requirement for cell patterning is cellular selectivity and viability, which refer, respectively, to the ability to precisely place cells only on the designated areas (e.g. electrodes) and retain the patterned cells alive and functional. Successful development of such a technique depends on the mechanism governing cell-material interactions and on the ability to tailor the material surface to favor cell immobilization.
A number of techniques have been developed to fabricate micron-scale cellular patterns, including metallic and elastomeric stenciling,17, 18 microcontact printing,19–21 elastomeric membranes,22, 23 and microfluidic channels.24–26 Most of these techniques use a mechanical cell delivery device in contact with the substrate to guide selective cell adhesion.27, 28 The metallic and elastomeric stencil techniques apply a micromachined thin sheet (stencil) containing holes over a substrate during the cell-seeding process.17, 18 The holes are sized and positioned so that cells are patterned in replica of the patterned stencil; the stencil is peeled off after cell attachment. These stenciling techniques can create cellular micropatterns on multiple substrate materials including polymers, gels, and metals. Microcontact printing utilizes a poly-dimethylsiloxane (PDMS) stamp to create cell patterns on surfaces through a protein-mediated process in which the protein is deposited on the substrate through contact-transfer to guide cell adhesion.20, 25 This technique can pattern cells on both planar and non-planar surfaces. Elastomeric membranes are polymer films that have circular or square holes through which proteins and cells can be patterned on a substrate of a variety of material types, such as glass and plastic.22, 23 The membrane is peeled off after cell attachment, which allows cells to spread from the initial pattern. Microfluidic systems pattern cells by the use of laminar flow through microchannels29 that are created by a stamp similar to that used in microcontact printing, and an elastomeric film defining the desired pattern is sealed within the channels to create a protein template for cell adhesion.27 Cells from the microflow adsorb to the protein template on the substrate or can be seeded directly on the substrate by size-restricting microchannels.24
In all of these techniques the patterns are formed either by generation of heterogeneous chemistry on a single material or by deposition of a second material of certain geometry followed by surface modification to form heterogeneous chemistry. Other methods for directly positioning cell-array structures without using proteins or peptides for guidance include dielectrophoretic fields30 and hydrodynamic flow.31 Ink jet printing has recently been introduced,26 which can pattern multiple cells on one substrate. In contrary to the direct positioning methods, an alternative approach, which is of particular interest for patterning cells on heterogeneous materials (e.g. metal electrodes on an insulting substrate), patterns cells by surface engineering and mediated cell adhesion with no positioning device involved. In this approach, cell adhesive proteins or peptides are first coated on the sites on which the cells adhere while the rest of the surface is passivated with a polymer coating to resist protein adsorption and cell adhesion.32, 33
Patterning cells on a cell impedance biosensor device requires a substrate comprising both conductive and insulative materials. A basic design includes an array of conducting microelectrodes and interconnecting conduits (e.g. gold) on insulating substrate (e.g. glass or dry silicon dioxide), where the electrodes are exposed while the interconnects are covered by an insulative passivation material (e.g. silicon dioxide).34 Figure 1 illustrates a pattern design (left) for patterning single cells on gold electrodes over a silicon dioxide substrate and the layout of material architecture for a single electrode (right). Electrode size and geometry can vary depending on the cells’ physical characteristics and the strategy to pattern cells on the electrode array. The cell patterning technique must be capable of generating highly selective and reproducible cell patterns and retaining cell functionality during and after the cell adhesion to the electrodes.35
Figure 1.
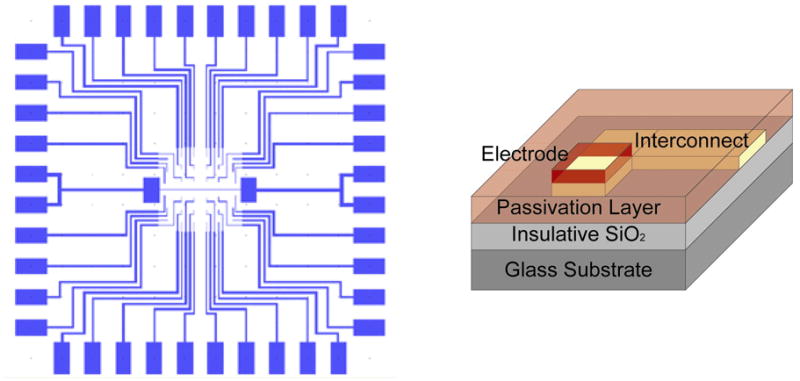
Design of a microelectrode array for cell impedance sensing: Gold-patterned substrate (left) and the material architecture of a single electrode (right).
The success of cell impedance biosensors hinges on the ability to pattern cells on electrodes in a tightly-bound, well-spread conformation. Such a conformation would reduce the electric signal loss due to current leaking along undesired pathways when there is a direct electrode-to-electrolyte exposure, thereby improving detection sensitivity.36 Thus, the mechanisms governing cell attachment and spreading are of significant importance in choosing the cell patterning strategy.
Cell Adhesion and Spreading on Substrates
Cell adhesion triggers signals that regulate cell survival, growth, cell-cycle progression and differentiation, and motility in multiple cell systems.37 Each stage of adhesion, i.e., attachment, spreading, migration, and immobilization, involves changes in cell morphology and cytoskeletal structure that are regulated by incompletely defined protein kinase and lipid second messenger pathways.38–41 Thus, it is preferred to pattern cells onto microelectrodes through natural cell attachment, which is commonly accomplished by immobilizing adhesive proteins or peptides on the electrodes to mediate the subsequent cell adhesion, promote cell spreading, and retain cell viability.42
Cells bind to adhesive extracellular matrix (ECM) components through integrin–ligand interaction, most commonly by the attraction of specific peptide regions in the ECM proteins to integrin transmembrane receptors on the plasma membrane. Integrin receptors are αβ heterodimers with a large extracellular domain and two short cytoplasmic tails, where 18 β and 8 β subunits have been identified to dimerize into 24 distinct receptors.43 Most integrins are expressed on a wide variety of cell types, and most cells express several integrin receptors. Ligand specificity is determined by the subunits of a given integrin, but integrins often bind to more than one type of ligands.
Integrin–ligand mediated adhesion activates receptor integrins to undergo conformational changes that provide spatial and temporal control of the binding activity between the cell and the extracellular ligand.43 Following this coupling, bound receptors rapidly associate with the actin cytoskeleton and cluster together to form focal adhesions, discrete supermolecular complexes that are the structural links between the cytoskeleton and ECM.44 Focal adhesions generate mechanical forces mediating stable adhesion, spreading, and migration; and cell spreading on the surface is driven by the traction force set by adhesion molecules peripheral to the initial focal adhesion contacts between the cell and the surface.45, 46 The availability, conformation, and distribution of cell binding domains dictate the morphology of adhered cells and the extent of cell spreading.46 Cell spreading can minimize the distance between an adhered cell and its substratum surface, which is typically on the order of 10 to 15 nm.47
An approach based on integrin-mediated cell adhesion strategy to produce single-cell patterns has been demonstrated using a small peptide Lys-Arg-Glu-Asp-Val-Tyr (KREDVY).46 The process starts with creating an array of gold electrodes on an insulative silicon oxide substrate by photolithography. The KREDVY peptide is then selectively immobilized on the electrodes through a pre-coated alkanethiol self-assembled monolayer (SAM) by covalent bonding with amine-terminated functional groups, while the silicon dioxide background is coated with a layer of polyethylene glycol-silane to resist cell adhesion (Figure 2A). The selective single-cell patterning onto the electrodes is accomplished by exposure of this chemically modified substrate to cells in suspension. With this approach, single cells can be patterned on electrodes in a tightly and well-spread fashion (Figure 2B). This cell patterning technique holds great promise to improve the signal-to-noise ratio for cell impedance measurements.
Figure 2.
(A) Schematic representation of a SAM-modified gold electrode coated with covalently-bound adhesion ligands for mediated cell adhesion. (B) Optical DIC image of a single 9L (rat glioma) cell adhered on the gold electrode coated with covalently-bound chlorotoxin ligand, revealing the well-spread cell morphology.
Cell Viability
Retaining the viability of cells attached to the substrate is essential for the cells to function as sensing elements. When studying viability of patterned cells, it is often important to determine the manner by which a cell dies, which can be either pathological or programmed death. Pathological cell death, known as necrosis, occurs when a cell is compromised by toxins or other external factors. Necrosis is characterized by the swelling and rupture of the plasma membrane, resulting in the release of intracellular debris. Programmed cell death is a naturally occurring, strictly regulated process that eliminates damaged cells and maintains constant cell numbers in developed organisms. This process is characterized by a distinct morphological change known as apoptosis, which can be readily detected by a cellular impedance sensor.
The apoptosis mechanism is the desired response of a cancerous tumor to effective chemotherapies or radiation treatments.48 Resistance of tumors to specific therapies is characterized by a failure to induce apoptosis.49 Impedance characterization can provide a simple and efficient method to measure the concentration of chemotherapeutics and the response-time functions of patterned cells to the chemotherapeutics.
3. Cellular Electrical Impedance Detection
Cellular Impedance Model
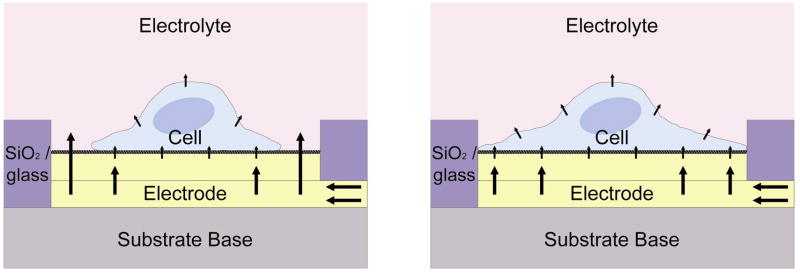
When a cell is immobilized on an electrode, it impedes electrical current flowing directly from the electrode into the bulk electrolyte due to the interference of its anchored plasma membrane above the electrode surface.50 Thus, as current passes from an electrode to an attached cell, it is dispersed through the narrow cell-substrate spaces available, as well as any intercellular junctions in large, multicellular electrode systems.16 Figure 3 illustrates a comparison in electrode-cell-electrolyte current flow between a cell poorly-spread (which exposes part of electrode surface area to the electrolyte) and a cell well-spread over the entire electrode surface. Because cells restrict electrical current flow, a cell can be modeled by basic impedance elements comprising both resistors and capacitors. However, differing cell impedance models exist as this phenomenon is not thoroughly understood.
Figure 3.
Illustration of current passing from an electrode through an adhered single cell that does not cover the entire electrode surface area (left) and one that fully spreads across the electrode (right). In the former, the current passes through the least resistive path between the electrode and electrolyte, resulting in cell signal loss.
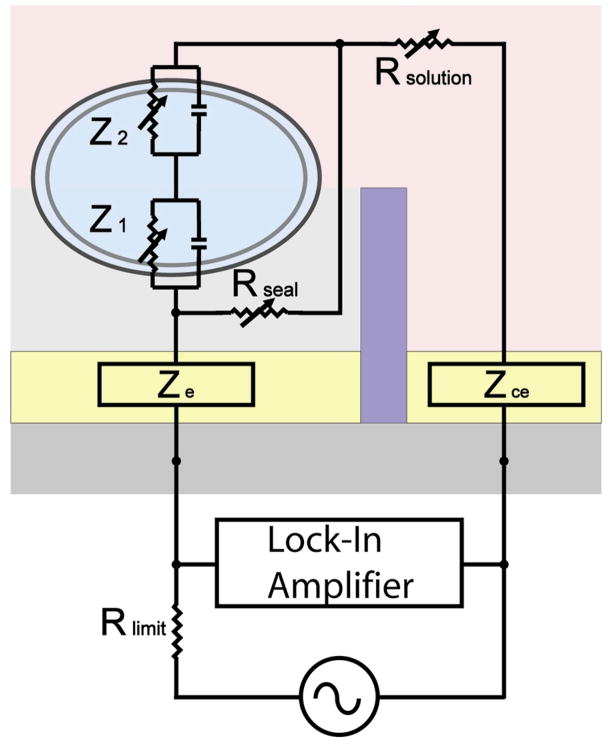
The impedance characteristics of a cell may be best described as a capacitive plasma membrane, due to its insulative properties, in parallel with a variable resistor that represents the parallel combination of all the ion channels open for transport of specific ions across the membrane.34 Cellular impedance is thus represented by the series sum of the apical and basal plasma membrane impedances, each denoted by a variable resistor and a capacitor in parallel, where the variable resistance depends on the state of the ion channels.34 Figure 4 depicts the total measurable impedance of a cell attached to an electrode, which consists of the electrode impedance (ZE), the basal and apical cell membrane impedances (Z1, Z2), the variable resistance (Rsolution) of the electrolyte solution, the counter electrode impedance (ZCE), and the variable “sealing” resistance (Rseal) along the thin channel layer of medium between the cell, electrode, and the passivation layer.
Figure 4.
Impedance model of a cell adhered to an electrode. Z1 and Z2 represent the basal and apical plasma membrane impedances, respectively, each with a resistive and a capacitive element in parallel. Rseal is the variable resistance between the cell and the substrate, increasing with the extent of cell spreading and focal adhesions. Rsolution is the variable resistance of the electrolyte solution, determined by ion concentration in the solution. ZE and ZCE represent the characteristic impedances of the electrode and counter electrode and their interfacial impedance with the electrolyte solution. ZCE is usually negligible in comparison to ZE and Zcell due to the large geometric size of counter electrode.
In order for cell impedance Zcell (= Z1 + Z2) to be quantified accurately from the measured impedance Zmeasurement, the sealing resistance Rseal must be on the same order of magnitude or greater than the membrane impedance, as indicated in Equation 1.
| (1) |
Depending on the manner in which cells are immobilized on the electrode, the lipid core of the plasma membrane is kept at a certain distance from the electrode by weak bonding forces between the membrane and the metal electrode and by molecules that protrude from the membrane, i.e. protein molecules (e.g. integrins, glycocalix) that face the outer leaflet of the cell membrane and those that are deposited onto the substrate (e.g. ECM proteins). These forces and proteins keep the lipid core of the membrane at a certain distance from the substrate, stabilizing a cleft between cell and electrode surface that is filled with electrolyte.51 The resulting shunt resistance between the electrode and the electrolyte may seriously degrade the actual impedance characterization of the cell membrane. If Rseal is significantly less than ZE + Zcell, it dominates the measured total impedance rather than Zcell. A tight coupling between the cell and the electrode will decrease the signal loss through shunt pathways in the cleft and thereby increase the signal-to-noise ratio (SNR), improving detection sensitivity. Thus, cell-surface distance and cell spreading are the most crucial variables to be controlled for accurate and reliable impedance measurements.
Cellular Impedance Measurement
Cellular impedance measurements require low noise voltage source instrumentation, transimpedance and/or “lock-in” voltage amplification, and subsequent signal processing to obtain cell impedance data from the measured voltage signal. A challenge in acquiring the desired signal is the removal of noise, which includes biological noise, electrode noise, thermal (“white”) noise due to electronic instrumentation, and interference from external sources.34 Environment noise is caused by fluctuations in the pH and temperature, and biological noise is often attributed to poor seals at the cell-electrode interface.
In measuring the small signal from any cellular action or response, lock-in amplifiers are utilized because they can detect small AC signals, even among noise sources many thousands of times larger. Lock-in amplifiers use a technique known as phase-sensitive detection to single out the component of a signal at a specific reference frequency and phase, rejecting noise at frequencies other than that of the reference frequency. The resultant resolution of the electrical measurement will therefore be high enough to pick up the fractional changes of the membrane conductance of a single cell. In addition to reducing electronic noise, the environmental conditions must also be maintained in a meticulous manner while allowing for the induction of known or unknown analytes to be screened. The cells must also be maintained at constant pH, osmolarity, and temperature, otherwise risk detecting false positive biothreat signals.5
Typically, cell impedance sensing is conducted in the following manner. A constant AC voltage is applied to the system and the resulting electric potential (voltage) across the electrode-cell(s)-electrolyte-counter electrode is monitored. The amplitude of current passed through the cell is held in the nanoampere (nA) range, usually controlled by a current-limiting resistor in a voltage divider configuration, which creates a negligible electrical stimulation to the cell during the impedance measurement. Baseline measurements of the electrode-electrolyte combination without cells are recorded prior to cell seeding and the baseline voltage would remain constant as long as the frequency of the input signal is constant.36 Cellular impedance is calculated using the difference between the baseline voltage and the voltage measured after cells are attached to the electrode, which removes the contribution of the electrode-electrolyte combination and parasitic elements. Thus, any impedance changes are attributed to the changes in cellular properties, whether due to micromotion or in response to environmental substances.
Cellular Impedance Sensing Applications: Cytotoxicity
The subject of environmental toxins from the air, water, and soil has stirred serious debate about their true impact on human health and the global environment. There are a multitude of views on testing protocols (e.g. conditions of exposure) and data interpretation which evoke a need for a compelling technological approach to provide trusted information and logical solutions to environmental issues. For nearly a decade, cellular impedance biosensors have been widely researched to evaluate their capabilities in monitoring the effects of cytotoxicity on cells. For these devices to be effective, they must be capable of detecting low doses of any toxic substance introduced to the cellular environment in a manner that meets the requirements of rapid and sensitive analysis, stability, reproducibility, and portability.
The most commonly utilized impedance cytosensor is the electric cell-substrate impedance sensing (ECIS) system, which can be used for characterization of cell attachment, micromotion,52–55 and cytotoxicity.56–59 ECIS sensors were pioneered by Ivar Giaever and Charles Keese, and since have become commercially available.52 These sensors exhibit great sensitivity in monitoring cellular response to the changes of external conditions such as temperature, pH, and a number of biological and chemical compounds.50, 53, 60 To date, ECIS has been able to provide quantitative information in real-time about cell movement, morphological changes, and functional alteration when exposed to various harmful substances. There have been many studies using ECIS to detect toxic or noxious agents, such as Tween 20, benzalkonium chloride (BAK), Triton X100, sodium lauryl sulfate, cadmium chloride (CdCl2), sodium arsenate (Na2HAsO4), mercury chloride (HgCl2), 1,3,5-trinitrobenzene (TNB), and cycloheximide (CHX), with various types of cells.56–59, 61 These studies examined how cell attachment and spreading are effected by different toxin concentrations, varying inoculation and exposure time.57 A pronounced resistance change corresponding to differing toxin concentrations is observed with great reproducibility.
These experiments generally begin by administering the toxic compound to the cell suspension immediately prior to cell seeding over the electrode wells. In doing so, plotting the impedance as a function of time (for varying concentrations) consistently shows that a lethal dosage of the toxicant causes a majority of cells to die instantly, resulting in a slight or no increase in the maximum measured resistance.58 This data indicates that an immediate cell death response prohibits cell attachment and subsequent spreading. If toxin concentration is not as acute, cells might still be able to attach and spread on the detecting electrode, leading to an initial increase in impedance, followed by a steady-state decrease if the cells are continuously subjected to the toxin.58
Despite the recognized success of ECIS sensors, there is a limitation on the sensitivity of cell response due to the manner of cell adherence. Each detecting electrode on an ECIS system adheres thousands of cells, and the average cellular impedance is determined from this monolayer of cells. Cells naturally associate with each other for the interest of survival, working for both themselves and each other. This close proximity and association between the plasma membranes of neighboring cells within the monolayer undergo alterations at the nanometer scale, which can cause significant changes in individual cellular impedance.62, 63 Individual alterations cannot be disseminated for each cell present, only a characteristic response of aggregate cells. Thus, vital information can go undetected across a layer of cells because ECIS devices are not capable of resolving the morphological changes of individual cells within an adhered cell monolayer. Additionally, ECIS sensors are incapable of addressing the need to examine the cell membrane properties, particularly the effects of different compounds on the ionic channels.5
Cellular Impedance Sensing Applications: Anticancer Drug Screening
Reliable assessment of cell death is a pivotal issue in developing effective and safe anticancer drug therapies. Many current anticancer drugs kill cancer cells by inducing apoptosis, and thus, a cost-effective platform to screen such drugs based on detection of cellular apoptosis may prove to be particularly beneficial to the development process. Traditional biochemical assays and techniques determine cell viability by monitoring cellular metabolic rate, activity of cytoplasmic enzymes, and the release of artificial labels. However, none of them are suitable for high throughput analysis in real-time under continuous, automated conditions. Apoptosis is characterized with dramatic changes in cell morphology, ionic channel conductance, and extracellular membrane integrity, as well as altered intracellular structure (e.g., the observed condensation and fragmentation of organelles and the cell nucleus).64 Cell impedance biosensors are capable of detecting these electrical property changes at significantly reduced cost and expedited assessment procedure, and thus offer an optimal approach in anticancer drug screening.
Unlike cellular impedance toxin sensor applications, impedance sensing to detect apoptosis is slow to catch on. ECIS has been shown to monitor apoptosis-induced changes in cell shape in real-time, differing from detection tools such as fluorescence microscopy and flow cytometry (for suspended cells) with caspase-3 or DNA fragmentation assay kits.61 It has been shown that a response to an apoptotic-inducing substance, such as CHX, resulted in a rapid and monotonic decrease in impedance.61 The time course of the impedance change was verified when compared to the assay results of caspase-3 activity, a major protease that is responsible for apoptosis-induced proteolysis of various intracellular target proteins. Recently, the ECIS technique was demonstrated to analyze aspirin-induced changes in colon cancer cells (HT-29) and deduced that impedance alterations were consistent with apoptosis, verified through transmission electron microscope (TEM) images showing apoptotic morphologies of these cells.65
The limited use of cell impedance biosensors in drug screening is mainly due to design constraints with these traditional microelectrode biosensors. Measuring the impedance from a large cell population attached on one electrode results in complex and even questionable interpretations of the acquired data. While averages of collected signals resulted from cellular activities, such as proliferation, motility, and cell-cell separation, can be achieved over the cell population, it is difficult, if not impossible, to precisely monitor changes in cell membrane properties of individual cells with current ECIS systems. Thus, the response of a single cell to a given dose of a pharmacological drug or harmful toxin may prove to be more significant than that of multiple cells. In a single-cell sensing system, cell response to drugs alone, without cell-cell interaction, can be accurately recorded.
4. Concluding Remarks
Cellular impedance biosensors show tremendous promise in quantifying cellular events such as adhesion, spreading, growth, motility, and in serving as a predictor of in vivo responses to environmental toxins, pharmacological drugs, and other chemicals and substances. These sensors offer an alternative to conventional analytical techniques with advantages of high speed, accuracy, sensitivity, and an ability to detect large numbers of potential threats.
Future success in this field lies in the ability to immobilize single cells on microelectrode arrays in a manner that minimizes the cell-surface distance and promotes full cell spreading over the electrode. Single-cell impedance sensors make it possible to experimentally study cellular pathways without interference from other cells, thereby eliminating the uncertainty incurred by cell-cell interaction. Electrical signals measured from an individual cell-electrode construct can be many times larger than a cluster of unordered cells placed on a common electrode. Statistical analysis of the behavior of individual cells is not possible without a single-cell based system, since only highly identical targets may generate meaningful statistical data.
Acknowledgments
The authors would like to acknowledge the funding support from the National Institutes of Health (NIH/GMS R01GM075095) for the project of “Microelectrode arrays of single cell biosensors”.
References
- 1.Bousse L. Sensors And Actuators B-Chemical. 1996;34:270. [Google Scholar]
- 2.Pancrazio JJ, Whelan JP, Borkholder DA, Ma W, Stenger DA. Annals Of Biomedical Engineering. 1999;27:697. doi: 10.1114/1.225. [DOI] [PubMed] [Google Scholar]
- 3.Stenger DA, Gross GW, Keefer EW, Shaffer KM, Andreadis JD, Ma W, Pancrazio JJ. Trends In Biotechnology. 2001;19:304. doi: 10.1016/s0167-7799(01)01690-0. [DOI] [PubMed] [Google Scholar]
- 4.McFadden P. Science. 2002;297:2075. doi: 10.1126/science.297.5589.2075. [DOI] [PubMed] [Google Scholar]
- 5.Kovacs GTA. Proceedings Of The Ieee. 2003;91:915. [Google Scholar]
- 6.Veiseh M, Wickes BT, Castner DG, Zhang MQ. Biomaterials. 2004;25:3315. doi: 10.1016/j.biomaterials.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Schultz JS. Scientific American. 1991;265:64. doi: 10.1038/scientificamerican0891-64. [DOI] [PubMed] [Google Scholar]
- 8.Ziegler C. Fresenius Journal of Analytical Chemistry. 2000;366:552. doi: 10.1007/s002160051550. [DOI] [PubMed] [Google Scholar]
- 9.Lin HJ, Charles PT, Andreadis JD, Churilla AM, Stenger DA, Pancrazio JJ. Analytica Chimica Acta. 2002;457:97. [Google Scholar]
- 10.Gilchrist KH, Barker VN, Fletcher LE, DeBusschere BD, Ghanouni P, Giovangrandi L, Kovacs GTA. Biosensors & Bioelectronics. 2001;16:557. doi: 10.1016/s0956-5663(01)00169-5. [DOI] [PubMed] [Google Scholar]
- 11.Hanein Y, Bohringer KF, Wyeth RC, Willows AO. Towards MEMS Probes for Intracellular Recording. 2002 [Google Scholar]
- 12.Martinoiz S, Rosso N, Grattarola M, Lorenzelli L, Margesin B, Zen M. Biosensors & Bioelectronics. 2001;16:1043. doi: 10.1016/s0956-5663(01)00202-0. [DOI] [PubMed] [Google Scholar]
- 13.Yicong W, Ping W, Xuesong Y, Gaoyan Z, Huiqi H, Weimin Y, Xiaoxiang Z, Jinghong H, Dafu C. Drug evaluations using a novel microphysiometer based on cell-based biosensors. 2001. [DOI] [PubMed] [Google Scholar]
- 14.Bloom FR, Price P, Lao G, Xia J, Crowe JH, Battista JR, Helm RF, Slaughter S, Potts M. Biosensors & Bioelectronics. 2001;16:603. doi: 10.1016/s0956-5663(01)00175-0. [DOI] [PubMed] [Google Scholar]
- 15.Ehret R, Baumann W, Brischwein M, Schwinde A, Stegbauer K, Wolf B. Biosensors & Bioelectronics. 1997;12:29. doi: 10.1016/0956-5663(96)89087-7. [DOI] [PubMed] [Google Scholar]
- 16.Luong JHT. Analytical Letters. 2003;36:3147. [Google Scholar]
- 17.Jimbo Y, Robinson HPC, Kawana A. Ieee Transactions on Biomedical Engineering. 1993;40:804. doi: 10.1109/10.238465. [DOI] [PubMed] [Google Scholar]
- 18.Folch A, Jo BH, Hurtado O, Beebe DJ, Toner M. Journal of Biomedical Materials Research. 2000;52:346. doi: 10.1002/1097-4636(200011)52:2<346::aid-jbm14>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 19.Kam L, Boxer SG. Journal of Biomedical Materials Research. 2001;55:487. doi: 10.1002/1097-4636(20010615)55:4<487::aid-jbm1041>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 20.Mrksich M, Dike LE, Tien J, Ingber DE, Whitesides GM. Experimental Cell Research. 1997;235:305. doi: 10.1006/excr.1997.3668. [DOI] [PubMed] [Google Scholar]
- 21.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Biotechnology Progress. 1998;14:356. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- 22.Ostuni E, Kane R, Chen CS, Ingber DE, Whitesides GM. Langmuir. 2000;16:7811. [Google Scholar]
- 23.Wang N, Ostuni E, Whitesides GM, Ingber DE. Cell Motility and the Cytoskeleton. 2002;52:97. doi: 10.1002/cm.10037. [DOI] [PubMed] [Google Scholar]
- 24.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu HK, Schueller OJA, Whitesides GM. Electrophoresis. 2000;21:27. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 25.Folch A, Toner M. Annual Review of Biomedical Engineering. 2000;2:227. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 26.Whitesides GM, Ostuni E, Takayama S, Jiang XY, Ingber DE. Annual Review of Biomedical Engineering. 2001;3:335. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 27.Folch A, Toner M. Biotechnology Progress. 1998;14:388. doi: 10.1021/bp980037b. [DOI] [PubMed] [Google Scholar]
- 28.Ostuni E, Chen CS, Ingber DE, Whitesides GM. Langmuir. 2001;17:2828. [Google Scholar]
- 29.Takayama S, McDonald JC, Ostuni E, Liang MN, Kenis PJA, Ismagilov RF, Whitesides GM. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5545. doi: 10.1073/pnas.96.10.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuhr G, Shirley SG. Journal of Micromechanics and Microengineering. 1995;5:77. [Google Scholar]
- 31.Thielecke H, Stieglitz T, Beutel H, Matthies T, Heinrich H, Meyer JU. Ieee Engineering in Medicine and Biology Magazine. 1999;18:48. doi: 10.1109/51.805144. [DOI] [PubMed] [Google Scholar]
- 32.Veiseh M, Zhang Y, Hinkley K, Zhang MQ. Biomedical Microdevices. 2001;3:45. [Google Scholar]
- 33.Veiseh M, Zareie MH, Zhang MQ. Langmuir. 2002;18:6671. [Google Scholar]
- 34.Borkholder DA. Cell based biosensors using microelectrodes. Stanford University; Palo Alto: 1998. [Google Scholar]
- 35.Lan S, Veiseh M, Zhang MQ. Biosensors & Bioelectronics. 2005;20:1697. doi: 10.1016/j.bios.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 36.Huang XQ, Nguyen D, Greve DW, Domach MM. Ieee Sensors Journal. 2004;4:576. [Google Scholar]
- 37.Hynes RO. Cell. 1992;69:11. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 38.Huttenlocher A, Sandborg RR, Horwitz AF. Current Opinion In Cell Biology. 1995;7:697. doi: 10.1016/0955-0674(95)80112-x. [DOI] [PubMed] [Google Scholar]
- 39.Heidemann SR, Buxbaum RE. Journal Of Cell Biology. 1998;141:1. doi: 10.1083/jcb.141.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz MA, Baron V. Current Opinion in Cell Biology. 1999;11:197. doi: 10.1016/s0955-0674(99)80026-x. [DOI] [PubMed] [Google Scholar]
- 41.Stockton RA, Jacobson BS. Molecular Biology Of The Cell. 2001;12:1937. doi: 10.1091/mbc.12.7.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veiseh M, Zhang MQ. Journal Of The American Chemical Society. 2006;128:1197. doi: 10.1021/ja055473q. [DOI] [PubMed] [Google Scholar]
- 43.Garcia AJ. Interfaces to control cell-biomaterial adhesive interactions. 2006. [Google Scholar]
- 44.Geiger B, Bershadsky A, Pankov R, Yamada KM. Nature Reviews Molecular Cell Biology. 2001;2:793. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 45.Lehnert D, Wehrle-Haller B, David C, Weiland U, Ballestrem C, Imhof BA, Bastmeyer M. Journal Of Cell Science. 2004;117:41. doi: 10.1242/jcs.00836. [DOI] [PubMed] [Google Scholar]
- 46.Veiseh M, Veiseh O, Martin MC, Asphahani F, Zhang M. Langmuir. 2007;23:4472. doi: 10.1021/la062849k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truskey GA, Burmeister JS, Grapa E, Reichert WM. Journal Of Cell Science. 1992;103:491. doi: 10.1242/jcs.103.2.491. [DOI] [PubMed] [Google Scholar]
- 48.Schmitt CA. Nature Reviews Cancer. 2003;3:286. doi: 10.1038/nrc1044. [DOI] [PubMed] [Google Scholar]
- 49.Igney FH, Krammer PH. Nature Reviews Cancer. 2002;2:277. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 50.Giaever I, Keese CR. Nature. 1993;366:591. doi: 10.1038/366591a0. [DOI] [PubMed] [Google Scholar]
- 51.Fromherz P. Physica E-Low-Dimensional Systems & Nanostructures. 2003;16:24. [Google Scholar]
- 52.Giaever I, Keese CR. Proceedings Of The National Academy Of Sciences Of The United States Of America-Biological Sciences. 1984;81:3761. doi: 10.1073/pnas.81.12.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giaever I, Keese CR. Proceedings Of The National Academy Of Sciences Of The United States Of America. 1991;88:7896. doi: 10.1073/pnas.88.17.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lo CM, Keese CR, Giaever I. Biophysical Journal. 1995;69:2800. doi: 10.1016/S0006-3495(95)80153-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao C, Lachance B, Sunahara G, Luong JHT. Analytical Chemistry. 2002;74:1333. doi: 10.1021/ac011104a. [DOI] [PubMed] [Google Scholar]
- 56.Keese CR, Karra N, Dillon B, Goldberg AM, Giaever I. In Vitro & Molecular Toxicology-A Journal Of Basic And Applied Research. 1998;11:183. [Google Scholar]
- 57.Xiao CD, Lachance B, Sunahara G, Luong JHT. Analytical Chemistry. 2002;74:5748. doi: 10.1021/ac025848f. [DOI] [PubMed] [Google Scholar]
- 58.Xiao C, Luong JHT. Biotechnology Progress. 2003;19:1000. doi: 10.1021/bp025733x. [DOI] [PubMed] [Google Scholar]
- 59.Xiao C, Luong JHT. Toxicology And Applied Pharmacology. 2005;206:102. doi: 10.1016/j.taap.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 60.Tiruppathi C, Malik AB, Delvecchio PJ, Keese CR, Giaever I. Proceedings Of The National Academy Of Sciences Of The United States Of America. 1992;89:7919. doi: 10.1073/pnas.89.17.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arndt S, Seebach J, Psathaki K, Galla HJ, Wegener J. Biosensors & Bioelectronics. 2004;19:583. doi: 10.1016/s0956-5663(03)00269-0. [DOI] [PubMed] [Google Scholar]
- 62.Lo CM, Ferrier J. Physical Review E. 1998;57:6982. [Google Scholar]
- 63.Liu HB, Plopper G, Earley S, Chen YQ, Ferguson B, Zhang XC. Biosensors & Bioelectronics. 2007;22:1075. doi: 10.1016/j.bios.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 64.van der Meer FJ, Faber DJ, Aalders MCG, Van Leeuwen TG. European Heart Journal. 2003;24:152. [Google Scholar]
- 65.Yin HY, Wang FL, Wang AL, Cheng J, Zhou YX. Analytical Letters. 2007;40:85. [Google Scholar]