Abstract
The light-dependent decrease in cyclic guanosine monophosphate (cGMP) in the rod outer segment is produced by a phosphodiesterase (PDE6), consisting of catalytic α and β subunits and two inhibitory γ subunits. The molecular mechanism of PDE6γ regulation of the catalytic subunits is uncertain. To study this mechanism in vivo, we introduced a modified Pde6g gene for PDE6γ into a line of Pde6gtm1/Pde6gtm1 mice that does not express PDE6γ. The resulting ILE86TER mice have a PDE6γ that lacks the two final carboxyl-terminal Ile86 and Ile87 residues, a mutation previously shown in vitro to reduce inhibition by PDE6γ. ILE86TER rods showed a decreased sensitivity and rate of activation, probably the result of a decreased level of expression of PDE6 in ILE86TER rods. More importantly, they showed a decreased rate of decay of the photoresponse, consistent with decreased inhibition of PDE6 α and β by PDE6γ. Furthermore, ILE86TER rods had a higher rate of spontaneous activation of PDE6 than WT rods. Circulating current in ILE86TER rods that also lacked both guanylyl cyclase activating proteins (GCAPs) could be increased several fold by perfusion with 100 µM of the PDE6 inhibitor 3-isobutyl-1-methylxanthine (IBMX), consistent with a higher rate of dark PDE6 activity in the mutant photoreceptors. In contrast, IBMX had little effect on the circulating current of WT rods, unlike previous results from amphibians. Our results show for the first time that the Ile86 and Ile87 residues are necessary for normal inhibition of PDE6 catalytic activity in vivo, and that increased basal activity of PDE can be partially compensated by GCAP-dependent regulation of guanylyl cyclase.
Keywords: rod, photoreceptor, phosphodiesterase, transduction, G-protein, retina, vision
1. Introduction
The absorption of a photon in the outer segment of a rod photoreceptor [see 1] produces an excited form of rhodopsin (Rh*), which binds a heterotrimeric G-protein called transducin and catalyzes the exchange of GTP for GDP on the transducin alpha subunit (Tα). The TαGTP then binds rod phosphodiesterase6 (PDE6), an enzyme complex that consists of catalytic PDE6α and PDE6β subunits and two regulatory PDE6γ subunits. In the dark, PDE6γ is bound to PDE6 α and β and inhibits catalytic activity. Upon light exposure, the newly formed TαGTP binds to PDE6γ, causing the inhibitory subunit to be displaced from the active site of a catalytic subunit. The PDE6 is then free to hydrolyze cGMP, and this hydrolysis decreases the outer segment cGMP concentration and produces a closing of cGMP-gated ion channels, which alters the rod membrane potential.
Because the PDE6γ subunit acts as the control point for regulating cGMP hydrolysis, it plays a key role in the transduction cascade. Little is known, however, about the molecular mechanism by which PDE6γ regulates PDE6 catalytic activity, though some information has been obtained from reconstituted systems. The PDE6γ contains a central lysine-rich region, in which 10 of 13 amino acids are basic [2]. These residues apparently contain one site for interaction with Tα [3] and are essential for binding of PDE6γ to the PDE6 α and β catalytic core [4]. The region involved in inhibiting PDE catalytic activity is thought to lie near the carboxyl terminus; deletions and point mutations in the carboxyl terminus have been shown in vitro to decrease inhibition of PDE activity [5–7]. Furthermore, the corresponding peptides with a mutated carboxyl terminus of PDE6γ fail to inhibit trypsin-activated PDE6 in vitro [8].
In order to test the function of specific amino acids or protein domains of PDE6γ in vivo, we constructed mutant PDE6γ cDNA under the control of the opsin promoter and generated transgenic mice by conventional means [9, 10]. The transgenes were then transferred by breeding to Pde6gtm1/Pde6gtm1 mice, homozygous for a targeted disruption of the endogenous PDE6 gene [11].
In this study, we examined the ILE86TER mutation lacking Ile86 and Ile87, the last two amino acids in PDE6γ. These amino acids have been previously shown in vitro to play an essential role in PDE6 function [2, 12]. We show that the light responses of mutant rods have a dramatically slower time course of decay, and that PDE6 in the mutant photoreceptors has a higher level of spontaneous activity in darkness. We conclude that Ile86 and Ile87 are essential for controlling PDE6γ inhibition of PDE6αβ in vivo.
2. Materials and Methods
2.1 Generation of ILE86TER animals
Experiments were performed in accordance with the rules and regulations of the NIH guidelines for research animals, as approved by the institutional animal care and use committees (IACUCs) of Columbia University, University of California, Los Angeles and University of Southern California. Animals were kept in cyclic 12-on/12-off lighting in approved cages and supplied with ample food and water. Animals in all experiments were sacrificed before tissue extraction by an approved procedure, usually decerebration or with an intraperitoneal injection of Nembutal.
The ILE86TER DNA construct for expression of Pde6g [13], together with the polyadenylation signal of the mouse protamine gene [14], was injected into the male pronucleus of oocytes. The ILE86TER point mutation was introduced by a standard PCR-based site-specific mutagenesis strategy [11]. The entire Pde6g cDNA coding region in the transgenic construct was sequenced to confirm the introduction of the point mutation and to verify that no other changes had been created inadvertently. KpnI and XbaI were used to excise vector sequences from the constructs. Fertilized oocytes were obtained from superovulated F1(DBA X C57BL6) females mated with homozygous Pde6gtm1/Pde6gtm1 males. The construct was injected into the male pronuclei of oocytes under a depression slide chamber. These microinjected oocytes were cultured overnight in M16 and transferred into the oviducts of 0.5-day post coitum pseudopregnant F1 females. The resulting transgenic mice were then backcrossed to Pde6gtm1/Pde6gtm1 mice to place the transgene into the knockout background. The mice were also tested for the absence of the rd1 mutation [15].
2.2 Identification of Transgenic Mice
DNA was isolated from tail tips or liver samples by homogenizing the tissue, digesting extensively with proteinase K and extracting with phenol. DNAs were analyzed by the polymerase chain reaction (PCR). The DNAs were also digested by SacI and analyzed by Southern blot hybridization with a Pde6g cDNA probe. Additional restriction digests were performed to analyze the structure of the integrated sequences, and to ensure that the DNA flanking the transgene was intact.
2.3 Immunoblot Analyses
Each retina was homogenized in 100 µl buffer (50 mM Tris, pH 7.4, 1 mM EDTA with protease inhibitor mixture from Roche Diagnostics, Indianapolis, IN), to which another 100 µl of sample loading buffer was added; the samples were boiled for 5 min. From this sample extract, different amounts were loaded onto a 4%–12% gradient gel (1, 2 and 3 µl for WT and 8, 12 and 16 µl for ILE86ter). For the detection of PDE6γ, blots were incubated with a 1:2,000 dilution of a polyclonal antibody directed against amino acid residues 2–16. Other antibodies were: PDE6α(PA1-720, 1:2,000, Thermo Scientific), RGS9 (from M.I. Simon, 1:5,000), Gαt (K60006R, 1:5,000, Meridian Life Science), Gβ1 (sc-379, 1:2,000, Santa Cruz Biotechnology). The secondary antibody was IRDye-labeled (1:10,000, LI-COR Biosciences), and the bands were detected and the fluorescence intensities were quantified with the Odyssey infrared imaging system (LI-COR Biosciences). In additional control experiments not shown in Fig. 1, we used the following primary antibodies to other phototransduction enzymes: GUCY2E, a polyclonal antibody to guanylyl cyclase 2E (gift of Prof Alexander M. Dizhoor, Pennsylvania College of Optometry, USA); GRK1 (rhodopsin kinase), polyclonal antibody sc-13078 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) to GRK1; rhodopsin, 1D4 monoclonal antibody to opsin (gift R. S. Molday of the University of British Columbia, Vancouver, Canada); the saryl hydrocarbon receptor-interacting protein-like 1, a polyclonal antibody (gift of Visvanathan Ramamurthy, Morgantown, WV, USA). In some experiments Western blots were visualized with the DuoLux Chemiluminescence substrate kit (Vector Laboratories, Inc., Burlingame, CA, USA) with a goat-anti-rabbit IgG-alkaline phosphatase conjugate. Blots were exposed to Hyperfilm-MP (Amersham Pharmacia Biotech, Piscataway, NJ, USA) and were preflashed to increase sensitivity and linearity according to the Sensitize™ protocol (Amersham Pharmacia Biotech).
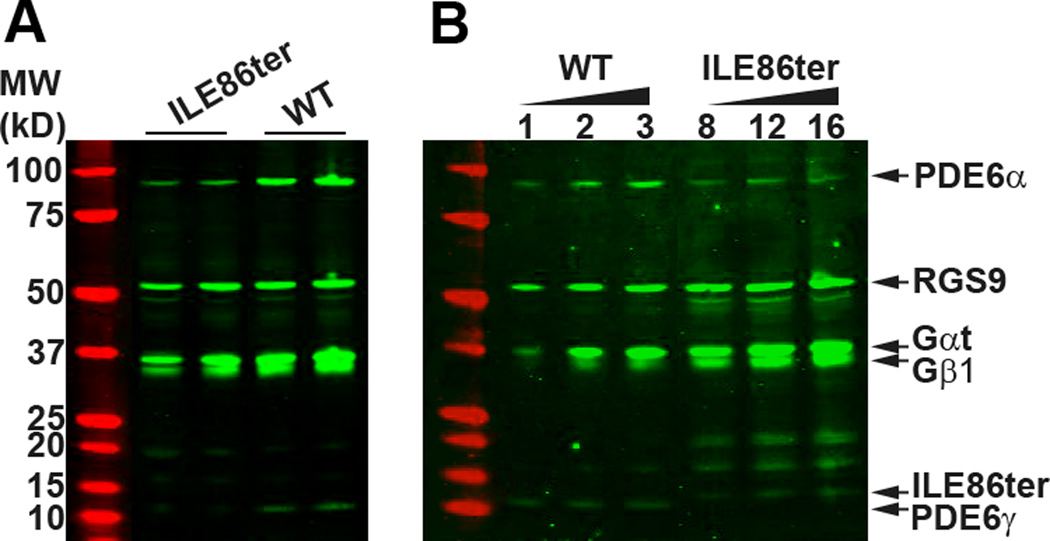
Fig. 1. Immunoblot analysis of the expression of PDE and other rod transduction proteins.
(A). The levels of RGS9, Gαt and Gβ1 were comparable between WT and ILE86ter retinas. However, PDE6 α and γ subunits were noticeably lowered in the ILE86ter retinas. Equal fraction of a retina (1/50) from an individual mouse was loaded onto each lane. (B). Quantification of PDE expression levels in ILE86ter and WT retinas. Representative blot of retinal extract prepared from WT and ILE86TER/GCAPs−/− mice. Each lane represent the amount loaded (µl) per retina (200 µl total sample volume). Based on the fluorescence signal quantified from each sample, the amount of PDE6α and PDE6γ in ILE86ter was 10 ± 3% of WT (N=3). Control experiments revealed no difference in PDE6 subunit expression levels between ILE86TER and ILE86TER/GCAPs−/− mice. Levels of other transduction proteins (RGS9, Gαt and Gβ1) were similar between WT and ILE86ter.
2.4 Histology
Mice were euthanized with an intraperitoneal injection of Nembutal. Each eye was rapidly removed, punctured at 12:00 along the limbus, and placed in a separate solution of 3% glutaraldehyde in phosphate buffered saline. After fixation for 1–2 days, the eyes were washed with saline and the 12:00 limbal puncture was used to orient the right and left eyes, which were kept in separate buffer so that the posterior segment containing the retina could be sectioned along the vertical meridian. A rectangular piece spanning the entire retina from superior to inferior orae serratae, including the optic nerve, was prepared for post fixing, dehydration, and embedding. A corner was cut out at the superior ora to allow identification of the upper retinal half of the segment. Sectioning proceeded along the long axis of the segment so that each section contained both upper and lower retina as well as the posterior pole. Eyes were dehydrated and embedded in paraffin. Hematoxylin-eosin (H&E) staining of paraffin sections was conducted as described [16]. Outer segment length was measured from thin sections examined in the electron microscope as previously described [17].
2.5 Suction-electrode recordings
Methods for recording responses of mouse rods have been given previously [18, 19]. Rods were perfused at 37°C with Dulbecco modified Eagle medium (D-2902, Sigma), supplemented with 15 mM NaHCO3, 2 mM Na succinate, 0.5 mM Na glutamate, 2 mM Na gluconate, and 5 mM NaCl, bubbled with 5% CO2 (pH 7.4). Data were filtered at 30 Hz (8 pole, Bessel) and sampled at 100 Hz. Flashes of 500 nm light 20 ms in duration were attenuated to different light levels by absorptive neutral density filters. At dim intensities, 10–20 individual responses presented at 5 s intervals were averaged to obtain the mean flash responses. At medium intensities, 5–10 responses were averaged, and the interflash interval was increased to 10 s. At bright intensities above saturation for the rods, only 3–5 responses were averaged, and the inter-flash interval was increased to 15–20 sec. Recordings always proceeded from dim intensities to brighter intensities, and the complete response-intensity data for an individual rod took about 20 min and bleached less than 0.5% of the visual pigment. The time course of PDE6 activity for Fig. 3D was calculated from Eqn. (24) of Pugh and Lamb [20]; the rate of change of activity was then computed by fitting a straight line to the initial rising phase as in Tsang et al. [11]. Unless otherwise stated, errors are given as standard errors of the mean (SE). Curve fitting and plotting of data were done with the program Origin (OriginLab Inc., Northampton, MA, USA).
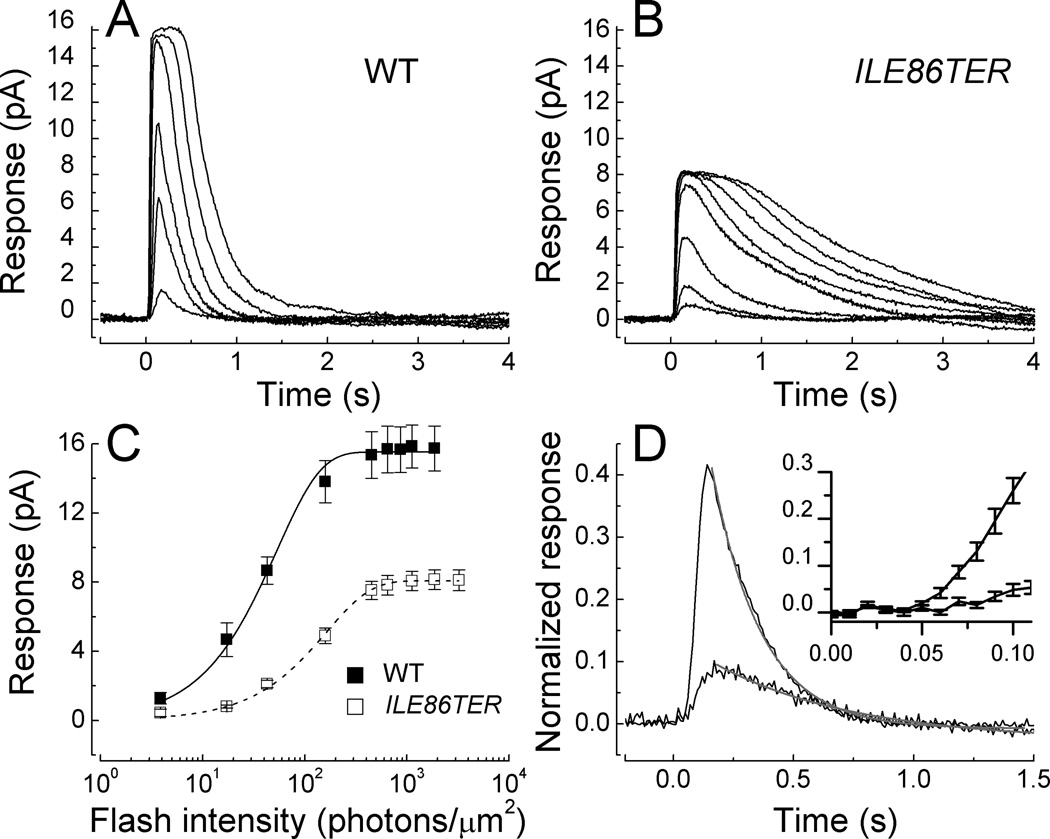
Fig. 3. Waveform and amplitude of WT and ILE86TER rods.
(A). Mean responses averaged from 10 WT rods to responses of 20 ms flashes of the following intensities (in photons µm−2): 3.9, 17, 43, 159, 453, and 1120. (B). Mean responses averaged from 18 ILE86TER rods to responses of 20 ms flashes of the following intensities (in photons µm2): 17, 43, 159, 453, 646, 1120, 1870, and 3250. (C). Peak amplitude of responses with SE as a function of intensity from 10 WT rods and 18 ILE86TER rods; same rods as in Figs. 3A and 3B. Means have been fitted with the exponential saturation function [46] of the form r = rmax [1 − exp(−kI)] where r is the peak amplitude of the response, rmax is the maximum value of the peak response amplitude at bright flash intensities, I is the flash intensity, and k is a constant. Best-fitting values were k = 0.019 photons−1 µm2 and rmax of 15.5 pA for WT rods, and k = 0.006 photons−1 µm2 and rmax of 8.1 pA for ILE86TER rods. (D). Mean responses from the same rods as in Figs. 3A and 3B to the same flash of intensity 17 photons µm−2. Declining waveforms of responses have been fitted with Equation (1); see text. Insert. Initial time courses of responses (with SE) are given on a faster time base to illustrate much slower rate of rise of ILE86TER response.
3. Results
3.1 Expression of ILE86TER Mutation in PDE6γ-Deficient Mice
To study the effect of the carboxyl terminus of PDE6γ on inhibition of the PDE6αβ catalytic core [4, 21], we produced transgenic mouse lines expressing the ILE86TER mutant allele, which lacks the two terminal isoleucines of PDE6γ, Ile86 and Ile87. ILE86TER mice were generated and crossed with Pde6gtm1/Pde6gtm1 to obtain animals that expressed only the mutant PDE6γ protein (see Methods). Immunoblots with retinal extracts from the transgenic line revealed that the levels of ILE86TER-mutant PDE6α and PDEγ were less than in WT (Fig. 1A), as in some previous mutant protein expression studies [22, 23]. We could however detect no difference in the levels of PDE6 subunits between ILE86TER mice and mice that were ILE86TER/GCAPs−/−, which we use in experiments described below (see Fig. 5).
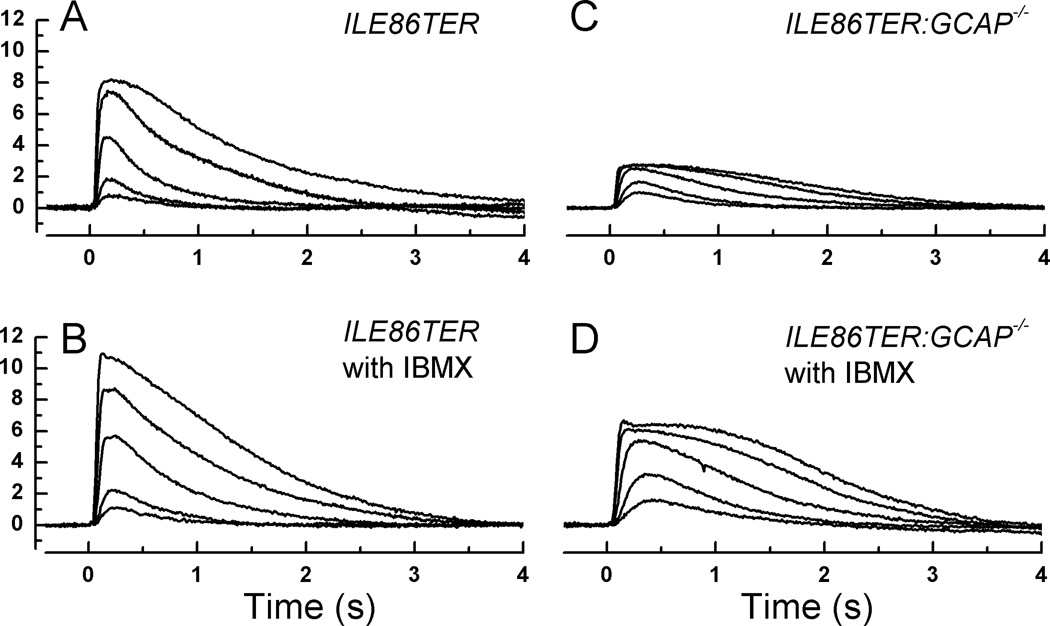
Fig. 5. Effect of IBMX on ILE86TER and ILE86TER/GCAPko rods.
A, Mean responses averaged from 18 ILE86TER rods to responses of 20 ms flashes of the following intensities (in photons µm2): 17, 43, 159, 453, and 1120. Same traces as in Fig. 4B. B, Mean responses averaged from 9 ILE86TER rods in presence of 100 µM IBMX to responses of 20 ms flashes of same intensities as in A. C, Mean responses averaged from 14 ILE86TER/GCAP−/− rods to responses of 20 ms flashes of same intensities as in A and B. D, Mean responses averaged from 11 ILE86TER/GCAP−/− rods in presence of 100 µM IBMX to responses of 20 ms flashes of the following intensities (in photons µm2): 17, 43, 159, 453, and 646.
To assess the expression levels more quantitatively, we loaded serial dilutions of samples from both WT and ILE86TER retinas (Fig. 1B). These experiments showed that both PDE6α and PDE6γ are expressed at a level of 10 ± 3% of WT (n=3). As both Figs. 1A and 1B demonstrate, the level of other transduction proteins such as RGS9 and transducin alpha and beta (Gαt and Gβ1) are unchanged in the mutant animals. In further control experiments not shown, we also found that levels of rhodopsin, rhodopsin kinase (GRK1), guanylyl cyclase 2D (GUCY2E), and aryl-hydrocarbon-interacting protein-like 1(AIPL1) were not significantly different in ILE86TER mice, compared to +/Pde6gtm1 and WT controls.
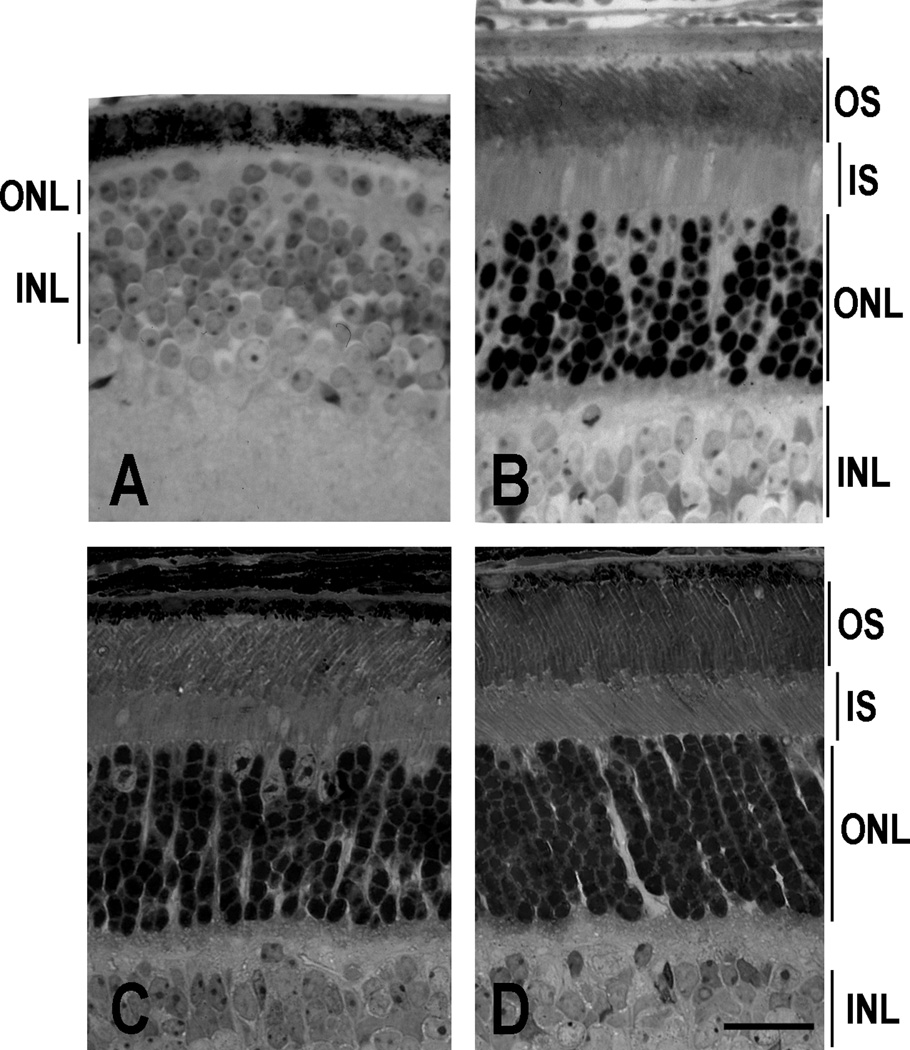
Retinal histology of sections prepared from 4-month-old mice showed 10–12 rows of photoreceptor nuclei in both heterozygous Pde6gtm1/+ mice with the ILE86TER transgene (not shown) and in homozygous Pde6gtm1/Pde6gtm1 mice with the ILE86TER transgene (Fig. 2B), similar to that of control C57/B6 mice (Fig. 2D). However, the parental homozygous Pde6gtm1/Pde6gtm1 mice without the transgene (Fig. 2A) showed complete degeneration [24]. Thus degeneration of the retina of the parental mutant mice was rescued by the mutant transgene, as was previously shown also to occur with the WT PDEγ transgene [11].
Fig. 2. ILE86TER transgene rescues retinal degeneration. Retinal light micrographs.
(A). An adult Pde6gtm1/Pde6gtm1 homozygote; (B). Pde6gtm1/Pde6gtm1 homozygote with the ILE86TER transgene; (C). ILE86TER/GCAPs−/− retina; and (D). C57B6 control. OS, outer segments; ONL, outer nuclear layer; INL, inner nuclear layer. The retina of the adult Pde6gtm1/Pde6gtm1 mouse lost all rod photoreceptors, and only a single layer of cones remains. Scale bars, 25 µm.
3.2 Suction-electrode recording from ILE86TER rods
Figs. 3A and 3B compare the average waveform of responses of 10 WT and 18 ILE86TER rods to 20 ms flashes of light of increasing intensity. Even though the level of PDE6 was reduced ten-fold in the mutant retinas, the rods still responded to light though with altered sensitivity, amplitude and time course. The mean peak amplitude (rmax) of the response from the ILE86TER rods was smaller than that of the WT (see Table 1). This indicates that a smaller number of the cGMP-gated channels in the ILE86TER rods were in the open state in darkness than in WT rods. This difference in peak response amplitude is also apparent in Fig. 3C, which plots the mean peak amplitude (with SE) to flashes of increasing intensity of WT responses from Fig. 3A, and ILE86TER responses from Fig. 3B, averaged rod by rod. This figure shows that the ILE86TER rods were less sensitive than WT rods, such that their mean response-intensity curve was shifted along the intensity axis to higher intensities (see also Table 1).
Table 1.
Kinetic and Sensitivity Parameters of Rods
| Animal line | rmax (pA) |
(pA photon−1 µm2) |
I1/2 (photons µm−2) |
ti (ms) |
τREC (ms) |
|---|---|---|---|---|---|
| WT (21) | 14.5±0.7 | 0.34±0.13 | 27±1 | 262±15 | 253±31 |
| WT IBMX (9) | 14.2±0.9 | 0.29±0.04 | 37±8 | 489±91 | 293±67 |
| ILE86TER (18) | 8.2±0.6 | 0.051±0.01 | 131±14 | 403±57 | 428±73 |
| ILE86TER IBMX (17) | 10.5±1.1 | 0.062±0.01 | 265±59 | 535±67 | 518±48 |
| ILE86TER GCAPko (20) | 2.0±0.3 | 0.025±0.004 | 44±7 | 568±38 | 480±45 |
| ILE86TER GCAPko IBMX (9) | 7.3±1.0 | 0.075±0.01 | 61±9 | 755±66 | 570±46 |
All values are means ± SE. Values of rmax (maximum response amplitude) were determined cell by cell from responses to saturating flashes; (dark-adapted flash sensitivity), by dividing the peak amplitude of the mean dim-flash response for each cell by the flash intensity; I1/2 (the intensity required to produce a half-maximal response), from the fit of response-intensity data for each cell to a Boltzmann function in the program Origin; ti (the integration time), from the time integral of the mean dim-flash response for each cell divided by the peak amplitude of the response; and τREC from the best-fitting exponential to the declining phase of the small-amplitude response.
Part of the difference in peak response amplitude and sensitivity between WT and ILE86TER rods can be attributed to a difference in outer segment length. WT rod outer segments averaged 25.7 ± 0.5 µm, as previously reported [17]; whereas the outer segments of ILE86TER rods averaged only 18.7 ± 0.5 µm in length and were thus about 0.73 as long as WT rods. Provided the density of outer segment cGMP-gated channels scales with the length of the outer segment [see 25], the decreased outer segment length by itself would predict a peak response amplitude of 10.6 pA, somewhat larger than our measured value of 8.2 pA (see Table 1). The decrease in outer segment length would reduce collecting area, decreasing sensitivity to 0.25 pA photon−1 µm2; but the actual measured mean of sensitivity was 0.051 pA photon−1 µm2. We conclude that much of the change in peak response amplitude but only a small fraction of the sensitivity change is the result of a decrease in outer segment length.
Figs. 3A and 3B also show that ILE86TER rods decay much more slowly than WT rods after exposure to brief flashes. Waveforms are explicitly compared in Fig. 3D, where we have plotted mean responses from the 10 WT and 18 ILE86TER rods of Figs. 3A and 3B to the same light intensity of 17 photons µm−2. Responses have been normalized to the mean peak amplitude of the response for the rods of each type to bright illumination (rmax); the ordinate in Fig. 3D therefore corresponds to the fraction of channels open in the dark which is closed by illumination. The larger response is from the WT rods. The time courses of decay of the mean responses have been fitted by a single exponential decay function of the form
| (1) |
where is the normalized rod response; is the normalized rod response at the beginning of the fit to the decay function, near the peak of the response; t is time; and τREC is the exponential time constant of response recovery. The fits with Eqn. (1) are indicated in Fig. 3D by the gray lines and give values of τREC of 206 ms for WT rods and 589 ms for ILE86TER rods. Individual measurements for dim light responses gave mean values of 253 ± 31 ms for 21 WT rods and 428 ± 73 ms for 18 ILE86TER rods, consistent with the fits in Fig. 3D. As a consequence of the slower decay, the integration time of ILE86TER rods was greater than that of WT rods (see Table 1).
In addition to their slower decay time, the responses in Fig. 3D can also be seen to rise more slowly after the presentation of the light flash. This can be observed more clearly in the insert to Fig. 3D, where we have plotted on a more rapid time base the mean response (with SE) of both WT and ILE86TER rods. The larger and more rapidly rising response is again from the WT rods. We estimated the rate of change of light-activated PDE6 activity from the slope of the initial time course of the response as in Pugh and Lamb [20] and Tsang et al. [11]. For the responses at the intensities used in Fig. 3D, the rate of change of PDE6 activity was about a factor of 10 smaller in ILE86TER rods than in WT (3.5 s−2 vs. 45 s−2). This decrease indicates a slower rate of activation of the enzyme and is consistent with the lower expression level of the PDE6 catalytic subunits in ILE86TER rods (Fig. 1). It may also explain the lower sensitivity of ILE86TER rods as compared to WT rods (see Table 1).
3.3 Effect of IBMX on WT rods
Deletion of the two terminal amino acids of PDE6γ has been shown in vitro to increase basal (spontaneous) activity of the PDE6 [2, 12], and in preliminary experiments we detected a nearly two-fold increase of basal PDE6 activity from extracts of ILE86TER retinas. We therefore investigated the possibility that basal PDE6 activity was higher in ILE86TER rods in vivo by perfusing the rods with 3-isobutyl-1-methylxanthine (IBMX), a partial competitive inhibitor that blocks PDE6 [26]. In amphibians, IBMX has been shown to produce large increases in the amplitude of the circulating current and a pronounced slowing of the rate of rise and decay of the photoreceptor response [27–29]. Electroretinogram (ERG) recordings from isolated cat eye suggest that similar effects may be produced by IBMX in mammals [30, 31], but because no previous recordings had been done from single mammalian rods perfused with IBMX, we first investigated the effect of this PDE6 blocker on WT mouse rods.
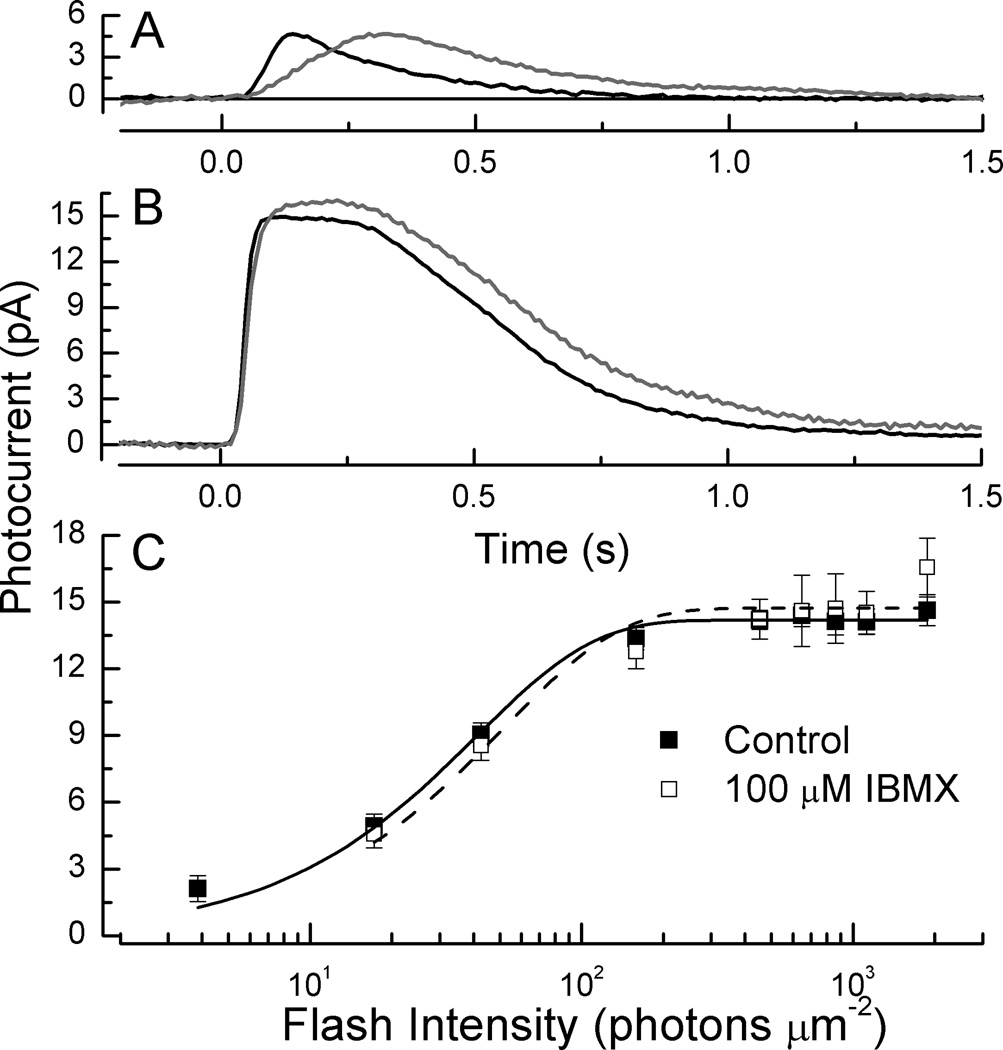
In Fig. 4A and 4B, we compare response waveforms of WT rods before (black traces) and during (gray traces) perfusion with 100 µM IBMX. As in previous studies in salamander [see especially 28], responses rose and decayed more slowly. From the initial time courses of the response at the dimmer intensity in part A, we estimated the rate of change of light-activated PDE6 activity, again as in Pugh and Lamb [20] and Tsang et al. [11]. We obtained a value of 37 s−2, in reasonable agreement with 45 s−2 obtained for the different group of WT rods in Fig. 3 at this same light intensity. In the presence of 100 µM IBMX, however, the value was no greater than 4–5 s−2, a factor of at least 7 smaller. We were surprised to discover, however, that we could detect little change in the maximum amplitude of the response to saturating light, indicating that circulating current was little affected (see Fig. 4B). In salamander, in contrast, this concentration of IBMX produces a several-fold increase in rmax [28, 29]. Part of this difference may result from the necessity of our perfusing the inner segment rather than the outer segment [28], but the site of perfusion should be less important in mouse than in salamander because of the much smaller volume of the mouse rod inner and outer segments. Furthermore, when we increased the IBMX concentration to 500 µM, we still observed little or no change in peak response amplitude, though this higher concentration gradually killed the rod from which we were making the recording, and subsequent attempts to record from other cells in the chamber perfused with this high concentration of IBMX were unsuccessful.
Fig. 4. Effect of IBMX on single WT mouse rods.
(A). Mean responses of 9 WT control rods (black) and 9 WT rods from the same retinas exposed to 100 µM IBMX, to flashes given at t = 0 of intensity 17 photons µm−2. (B). Same as in A but for saturating flashes of intensity 450 photons µm−2. (C). Response intensity curves of 9 rods before (■) and during (□) exposure to 100 µM IBMX. Means have been fitted with r = rmax [1 − exp(−kI)] as in Fig. 3D, for WT rods before exposure (continuous curve) with k = 0.0244 photons−1 µm2 and rmax of 14.2 pA, and for WT rods in IBMX (dashed curve) with k = 0.0194 photons−1 µm2 and rmax of 14.7 pA.
In Fig. 4C and in Table 1, we summarize the results of our experiments. After perfusion with 100 µM IBMX, we recorded a nearly two-fold increase in the integration time of the rod which was statistically significantly different from control responses (p<0.05, Student’s T), produced by the slowing of both the rising and falling phases of the response (Fig. 4A). There were, however, no significant changes in any of the other response parameters we measured, including sensitivity and circulating current.
3.4 Effect of IBMX on ILE86TER rods
When we then applied 100 µM IBMX to ILE86TER rods (Figs. 5A and 5B), we also observed an increase in integration time and modest slowing of the decay time of the response (Table 1), indicating inhibition of the PDE6 just as with WT rods. We were surprised, however, that IBMX again produced only a small increase in the maximum amplitude of the response, since previous biochemical measurements indicate that the deletion of the last two isoleucines of PDEγ should increase basal PDE activity [2, 12]. When rmax was averaged rod by rod (Table 1), the difference between ILE86TER rods with and without 100 µM IBMX was not significant (p>0.059, Student’s T). Even increasing the IBMX concentration to 500 µM produced little or no change in rmax (data not shown).
One possible explanation for the small effect of IBMX on circulating current in ILE86TER rods is that the blocking of the PDE6 by the drug was rapidly compensated by some other mechanism, such as a decrease in guanylyl cyclase activity, so that cGMP concentration (and the probability of outer segment channels being open) changed by only a small amount. We tested this notion by applying IBMX to ILE86TER/GCAPs−/− rods that also lacked the GCAP1 and GCAP2 molecules and were therefore unable to regulate guanylyl cyclase [32]. These rods had levels of expression of PDE subunits similar to those of ILE86TER rods (data not given), and their morphology is shown in Fig. 2C. The length of the outer segments of ILE86TER/GCAPs−/− rods, measured as for WT and ILE86TER rods in the electron microscope, averaged 16.9 ± 0.4 and was significantly smaller than the mean outer segment length of 18.7 ± 0.5 of ILE86TER rods (Student’s T, p<0.01). The difference in outer segment length was however not large enough to explain the four-fold decrease in maximum response amplitude of ILE86TER/GCAPs−/− rods compared to ILE86TER rods (Fig. 5C and Table 1), which we attribute to constitutive activation of basal PDE6 with no compensating change in guanylyl cyclase activity.
We tested our hypothesis of increased activation of PDE6 by perfusing the ILE86TER/GCAPs−/− rods with IBMX. At a concentration of 100 µM, IBMX produced an increase of peak response amplitude (and hence circulating current) by over three fold (Table 1), nearly to the value observed in ILE86TER rods (Fig. 5D). Thus in ILE86TER/GCAPs−/− rods, IBMX can inhibit an enhanced basal activity of the PDE6, consistent with previous biochemical measurements showing incomplete inhibition of PDE6 by this mutant form of PDEγ [2, 12].
4. Discussion
The PDE6 of photoreceptors catalyzes the light-induced decrease in cGMP that is ultimately responsible for gating the channels and generating the rod light response. The activity of PDE6 is regulated by the binding of inhibitory γ subunits to catalytic α and β subunits, but the molecular mechanism of this regulation is presently unknown [21]. In order to study this mechanism in rods in vivo, we have introduced a mutant form of PDE6γ into the mouse genome, in which the last two C-terminal amino acids Ile86 and Ile87 have been deleted. Previous work with reconstituted rod outer segments has indicated that the carboxyl-terminal tail of PDE6γ may influence γ subunit inhibition of PDE6 catalytic activity [6, 21, 33], and that the carboxyl-terminal Ile86 and Ile87 may play a particularly important role in this inhibition [4–6, 12, 21, 34–36].
Our studies demonstrate for the first time the essential role of the C-terminus of PDE6γ in the regulation of PDE activity in vivo. Rod responses decay more slowly than in WT animals. Although the expression of PDE6 subunits is less than in WT, this decrease in expression level should not by itself alter the time course of decay of the response because the ratio of expression of the α and γ subunits was unaltered (Fig. 1). Furthermore, the rate of decay was slower than WT over the whole range of light intensities we examined (Fig. 3) and even when responses were compared whose initial time courses (and rates of PDE activation) were chosen to be nearly equivalent (data not shown). Thus even for comparable rates of PDE activation, the decay of ILE86TER rods is slower.
Since previous experiments have demonstrated that the rate of photoreceptor decay is determined by the rate of hydrolysis of transducin alpha GTP [37] and shut off of activated PDE6 [38], a decrease in PDE6 expression should not by itself alter the decay time. The slower decay is unlikely to be caused by a pool of TαGTP unbound to PDE6 in the ILE86TER rods, because at the dim intensities we used to stimulate these photoreceptors, the number of activated transducins formed will still be much less than the number of PDE molecules in the rod outer segment. Instead, the slow time course of decay must occur because of less efficient shut off of light-activated PDE6 in the mutant photoreceptors, as the result of weaker re-association of PDE6γ to the PDE catalytic subunits [3, 4] and/or reduced rate of hydrolysis of transducin alpha GTP [39].
Our results with the blocker IBMX also demonstrate that the ILE86TER rods have a higher than normal basal activity of PDE6 [2, 12], once again reflecting decreased efficiency of inhibition of the PDE6 catalytic subunits by PDEγ. We first showed that perfusion of 100 µM IBMX on WT rods produced of the order of a 7-fold inhibition of the light-induced increase in PDE6 rate, but that the dark current of neither WT nor ILE86TER rods was significantly increased by IBMX (Table 1), unlike the effect of this inhibitor on rods in salamander [27–29]. One possible explanation for the small effect of IBMX on circulating current is that the blocking of PDE6 by the drug is rapidly compensated by some other mechanism such as a change in cyclase activity, so that cGMP concentration (and the number of outer segment channels open) remains relatively constant. We then tested this notion by recording from ILE86TER rods that also lacked the GCAP molecules and were therefore unable to modulate guanylyl cyclase [32]. These ILE86TER/GCAPs−/− rods have much lower circulating currents than ILE86TER rods, which by itself suggests that regulation by GCAPs may compensate for some part of the increase in basal PDE6 activity produced by the ILE86TER mutation. Furthermore, when ILE86TER/GCAPs−/− rods were perfused with IBMX, peak response amplitude (and therefore circulating current) was significantly increased, in support of our hypothesis that the increased basal PDE6 activity in ILE86TER rods is at least partially compensated by GCAP-mediated cyclase feedback. It is of some interest that the outer segments of ILE86TER rods are shorter than WT, and those of ILE86TER/GCAPs−/− rods are even shorter. This outer segment shortening may reflect the increased basal activity of the PDE6, which may act as an equivalent light [40].
In summary, our results show that the two terminal amino acids of PDE6γ play an essential role in the control of the PDE6αβ catalytic core. Our results substantiate biochemical measurements showing less efficient shutoff of PDE6 and increased basal activity of PDE6 lacking the two terminal isoleucines of PDEγ [2], and they provide a new appreciation of the role of the C-terminus of PDE6γ in the function of the rod. They also show that pathological changes in the spontaneous rate of the rod phosphodiesterase can be compensated at least partially by GCAP-dependent modulation of guanylyl cyclase.
Abnormally low cGMP phosphodiesterase activity is responsible for approximately 36,000 worldwide cases of retinal degeneration [41–45]. As there is no cure for these patients, the development of effective therapies to increase PDE6 activity requires an understanding of how PDE6 is regulated and integrated with other signal transduction pathways. As PDE6γ is an important regulatory component of PDE6, investigating the mechanisms by which PDE6γ regulates PDE6 is likely to improve our ability to control the progression of PDE6-related degenerations.
HIGHLIGHTS.
We made PDE6 ILE86TER mice lacking the two C-terminal PDE6γ Ile86 and Ile87 residues.
This mutation in vitro reduces the efficiency of PDE6γ inhibition of PDE6α and PDE6β.
Light responses of ILE86TER rods decayed more slowly than those of WT rods.
Experiments with IBMX showed that mutant rods had more spontaneous PDE6 activity.
The C-terminus of PDE6γ plays an essential role in control of PDE6 activity in vivo.
Acknowledgments
We are grateful to members of the Bernard & Shirlee Brown Glaucoma Laboratory for support, especially J. Mie Kasanuki, Yao Li, and Jean J. Pak. Work was supported by NIH grants EY01844 to GLF, EY12155 to JC, and EY018213 to SHT, and by additional assistance to SHT from NIH P30EY019007 (Core Support for Vision Research), and TS080017 from Department of Defense, as well as from the Foundation Fighting Blindness and Schneeweiss Stargardt Fund, the Bernard Becker-Association of University Professors in Ophthalmology-Research to Prevent Blindness Award, the Dennis W. Jahnigen Award of the American Geriatrics Society, the Joel Hoffman Fund, the Gale and Richard Siegel Stem Cell Fund, the Crowley Family Fund, the Charles Culpeper Scholarship, the Schneeweiss Stem Cell Fund, the Irma T. Hirschl Charitable Trust, the Bernard and Anne Spitzer Stem Cell Fund, and the Barbara & Donald Jonas Family Fund. SHT has also been a Fellow of the Burroughs-Wellcome Program in Biomedical Sciences.
Abbreviations
- BSA
bovine serum albumin
- cGMP
cyclic guanosine monophosphate
- ES
embryonic stem cell
- GAP
GTPase accelerating protein
- GCAPs
guanylyl cyclase activating proteins
- GDP
guanosine diphosphate
- GTP
guanosine triphosphate
- IBMX
isobutylmethylxanthine
- IDV
integral density value
- OS
outer segment
- PDE
cGMP phosphodiesterase
- PDE6
cGMP phosphodiesterase 6
- Rh*
active form of bleached rhodopsin (metarhodopsin II)
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- T
transducin
- Tα
alpha subunit of transducin
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Experiments were performed in New York, New York, and in Los Angeles, California. The conception and design of the experiments were done by S. H. Tsang, J. Chen, M. L. Woodruff, and G. L. Fain; the collection, analysis and interpretation of data were done by C.-S. Lin, M. L. Woodruff, B. D. Jacobson, M. C. Naumann, C. W. Hsu, R. J. Davis, Marianne Cilluffo, S. H. Tsang, and G. L. Fain; and the drafting of the article or revising it critically for important intellectual content was done by J. Chen, S. H. Tsang, M. L. Woodruff and G. L. Fain.
Contributor Information
Stephen H. Tsang, Email: sht2@columbia.edu.
Michael L. Woodruff, Email: michaelw@physci.ucla.edu.
Chyuan-Sheng Lin, Email: csl5@columbia.edu.
Barry D. Jacobson, Email: bdj@alum.mit.edu.
Matthew C. Naumann, Email: mn2400@columbia.edu.
Chun Wei Hsu, Email: cwh2118@columbia.edu.
Richard J. Davis, Email: rd2224@columbia.edu.
Marianne C. Cilluffo, Email: mariannc@ucla.edu.
Jeannie Chen, Email: jeannie.chen@keck.usc.edu.
Gordon L. Fain, Email: gfain@ucla.edu.
References
- 1.Fain GL. Sensory Transduction. Sunderland, MA: Sinauer, Inc.; 2003. [Google Scholar]
- 2.Granovsky AE, Artemyev NO. Biochemistry. 2001;40(44):13209–13215. doi: 10.1021/bi011127j. [DOI] [PubMed] [Google Scholar]
- 3.Lipkin VM, Dumler IL, Muradov KG, Artemyev NO, Etingof RN. FEBS Lett. 1988;234(2):287–290. doi: 10.1016/0014-5793(88)80100-5. [DOI] [PubMed] [Google Scholar]
- 4.Artemyev NO, Hamm HE. Biochem J. 1992;283(Pt 1):273–279. doi: 10.1042/bj2830273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez K, Cunnick J, Takemoto D. Biochem Biophys Res Commun. 1991;181(3):1094–1096. doi: 10.1016/0006-291x(91)92050-t. [DOI] [PubMed] [Google Scholar]
- 6.Takemoto DJ, Hurt D, Oppert B, Cunnick J. Biochem J. 1992;281(Pt 3):637–643. doi: 10.1042/bj2810637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger AL, Cerione RA, Erickson JW. J Biol Chem. 1997;272(5):2714–2721. doi: 10.1074/jbc.272.5.2714. [DOI] [PubMed] [Google Scholar]
- 8.Granovsky AE, Natochin M, Artemyev NO. J Biol Chem. 1997;272(18):11686–11689. doi: 10.1074/jbc.272.18.11686. [DOI] [PubMed] [Google Scholar]
- 9.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, N.Y.: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 10.Tsang SH, Yamashita CK, Lee WH, Lin CS, Goff SP, Gouras P, Farber DB. Vision Res. 2002;42(4):439–445. doi: 10.1016/s0042-6989(01)00213-9. [DOI] [PubMed] [Google Scholar]
- 11.Tsang SH, Burns ME, Calvert PD, Gouras P, Baylor DA, Goff SP, Arshavsky VY. Science. 1998;282:117–121. doi: 10.1126/science.282.5386.117. [DOI] [PubMed] [Google Scholar]
- 12.Skiba NP, Artemyev NO, Hamm HE. J Biol Chem. 1995;270(22):13210–13215. doi: 10.1074/jbc.270.22.13210. [DOI] [PubMed] [Google Scholar]
- 13.Tuteja N, Farber DB. FEBS Lett. 1988;232(1):182–186. doi: 10.1016/0014-5793(88)80413-7. [DOI] [PubMed] [Google Scholar]
- 14.Lem J, Applebury ML, Falk JD, Flannery JG, Simon MI. Neuron. 1991;6(2):201–210. doi: 10.1016/0896-6273(91)90356-5. [DOI] [PubMed] [Google Scholar]
- 15.Pittler SJ, Baehr W. Proc Natl Acad Sci U S A. 1991;88(19):8322–8326. doi: 10.1073/pnas.88.19.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsang SH, Woodruff ML, Jun L, Mahajan V, Yamashita CK, Pedersen R, Lin CS, Goff SP, Rosenberg T, Larsen M, Farber DB, Nusinowitz S. Hum Mutat. 2007;28(3):243–254. doi: 10.1002/humu.20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsang SH, Woodruff ML, Janisch KM, Cilluffo MC, Farber DB, Fain GL. J Physiol. 2007;579(Pt 2):303–312. doi: 10.1113/jphysiol.2006.121772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodruff ML, Janisch KM, Peshenko IV, Dizhoor AM, Tsang SH, Fain GL. J Neurosci. 2008;28(9):2064–2074. doi: 10.1523/JNEUROSCI.2973-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CK, Woodruff ML, Chen FS, Chen D, Fain GL. J Neurosci. 2010;30(4):1213–1220. doi: 10.1523/JNEUROSCI.4353-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugh EN, Jr, Lamb TD. Biochim Biophys Acta. 1993;1141(2–3):111–149. doi: 10.1016/0005-2728(93)90038-h. [DOI] [PubMed] [Google Scholar]
- 21.Artemyev NO, Natochin M, Busman M, Schey KL, Hamm HE. Proc Natl Acad Sci U S A. 1996;93(11):5407–5412. doi: 10.1073/pnas.93.11.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raport CJ, Lem J, Makino C, Chen CK, Fitch CL, Hobson A, Baylor D, Simon MI, Hurley JB. Invest Ophthalmol Vis Sci. 1994;35(7):2932–2947. [PubMed] [Google Scholar]
- 23.Kerov V, Chen D, Moussaif M, Chen YJ, Chen CK, Artemyev NO. J Biol Chem. 2005;280(49):41069–41076. doi: 10.1074/jbc.M508849200. [DOI] [PubMed] [Google Scholar]
- 24.Tsang SH, Gouras P, Yamashita CK, Kjeldbye H, Fisher J, Farber DB, Goff SP. Science. 1996;272(5264):1026–1029. doi: 10.1126/science.272.5264.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodruff ML, Olshevskaya EV, Savchenko AB, Peshenko IV, Barrett R, Bush RA, Sieving PA, Fain GL, Dizhoor AM. J Neurosci. 2007;27(33):8805–8815. doi: 10.1523/JNEUROSCI.2751-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beavo JA, Rogers NL, Crofford OB, Hardman JG, Sutherland EW, Newman EV. Mol Pharmacol. 1970;6(6):597–603. [PubMed] [Google Scholar]
- 27.Capovilla M, Caretta A, Cervetto L, Torre V. J Physiol (Lond) 1983;343:295–310. doi: 10.1113/jphysiol.1983.sp014893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cervetto L, McNaughton PA. J Physiol (Lond) 1986;370:91–109. doi: 10.1113/jphysiol.1986.sp015924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornwall MC, Fain GL. J Physiol (Lond) 1994;480(Pt 2):261–279. doi: 10.1113/jphysiol.1994.sp020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandberg MA, Pawlyk BS, Crane WG, Schmidt SY, Berson EL. Vision Res. 1987;27(9):1421–1430. doi: 10.1016/0042-6989(87)90152-0. [DOI] [PubMed] [Google Scholar]
- 31.Pawlyk BS, Sandberg MA, Berson EL. Vision Res. 1991;31(7–8):1093–1097. doi: 10.1016/0042-6989(91)90035-4. [DOI] [PubMed] [Google Scholar]
- 32.Mendez A, Burns ME, Sokal I, Dizhoor AM, Baehr W, Palczewski K, Baylor DA, Chen J. Proc Natl Acad Sci U S A. 2001;98(17):9948–9953. doi: 10.1073/pnas.171308998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stryer L. J Biol Chem. 1991;266(17):10711–10714. [PubMed] [Google Scholar]
- 34.Natochin M, Artemyev NO. J Biol Chem. 1996;271(33):19964–19969. doi: 10.1074/jbc.271.33.19964. [DOI] [PubMed] [Google Scholar]
- 35.Lipkin VM, Bondarenko VA, Zagranichny VE, Dobrynina LN, Muradov KG, Natochin M. Biochim Biophys Acta. 1993;1176(3):250–256. doi: 10.1016/0167-4889(93)90052-q. [DOI] [PubMed] [Google Scholar]
- 36.Berger AL, Cerione RA, Erickson JW. Biochemistry. 1999;38(4):1293–1299. doi: 10.1021/bi981683m. [DOI] [PubMed] [Google Scholar]
- 37.Krispel CM, Chen D, Melling N, Chen YJ, Martemyanov KA, Quillinan N, Arshavsky VY, Wensel TG, Chen CK, Burns ME. Neuron. 2006;51(4):409–416. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 38.Tsang SH, Woodruff ML, Chen CK, Yamashita CY, Cilluffo MC, Rao AL, Farber DB, Fain GL. J Neurosci. 2006;26:4472–4480. doi: 10.1523/JNEUROSCI.4775-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slep KC, Kercher MA, He W, Cowan CW, Wensel TG, Sigler PB. Nature. 2001;409(6823):1071–1077. doi: 10.1038/35059138. [DOI] [PubMed] [Google Scholar]
- 40.Fain GL. Bioessays. 2006;28(4):344–354. doi: 10.1002/bies.20382. [DOI] [PubMed] [Google Scholar]
- 41.McLaughlin ME, Sandberg MA, Berson EL, Dryja TP. Nat Genet. 1993;4(2):130–134. doi: 10.1038/ng0693-130. [DOI] [PubMed] [Google Scholar]
- 42.McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP. Proc Natl Acad Sci U S A. 1995;92(8):3249–3253. doi: 10.1073/pnas.92.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang SH, Pittler SJ, Huang X, Oliveira L, Berson EL, Dryja TP. Nat Genet. 1995;11(4):468–471. doi: 10.1038/ng1295-468. [DOI] [PubMed] [Google Scholar]
- 44.Dryja TP, Rucinski DE, Chen SH, Berson EL. Invest Ophthalmol Vis Sci. 1999;40(8):1859–1865. [PubMed] [Google Scholar]
- 45.Dvir L, Srour G, Abu-Ras R, Miller B, Shalev SA, Ben-Yosef T. Am J Hum Genet. 2010;87(2):258–264. doi: 10.1016/j.ajhg.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamb TD, McNaughton PA, Yau KW. J Physiol. 1981;319:463–496. doi: 10.1113/jphysiol.1981.sp013921. [DOI] [PMC free article] [PubMed] [Google Scholar]