Abstract
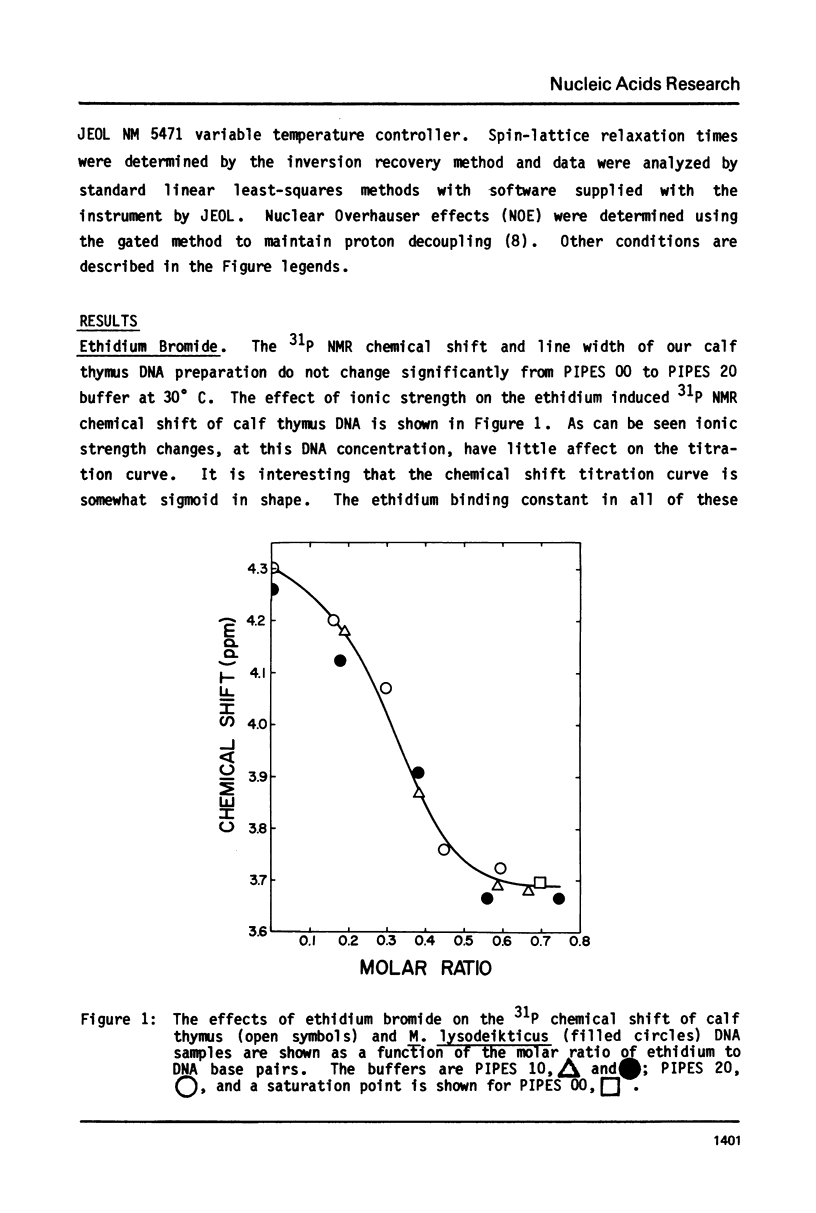
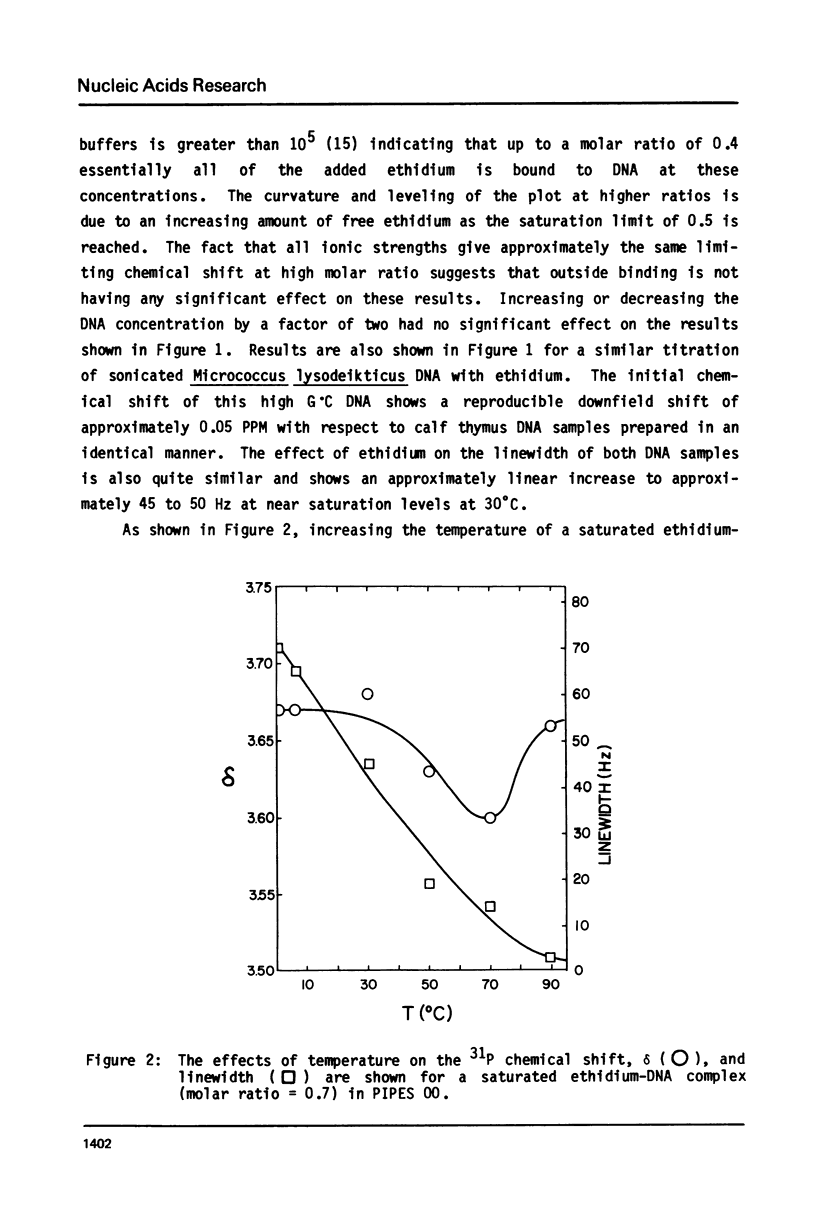
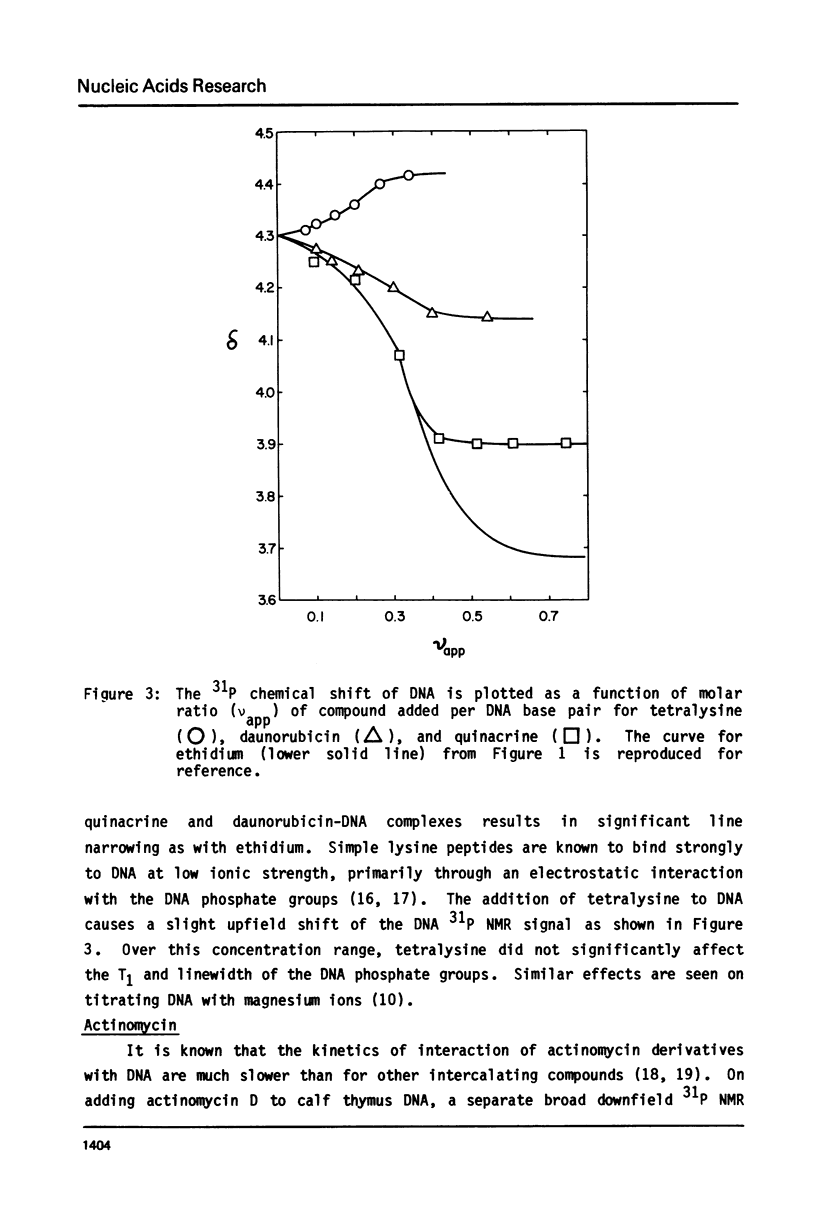
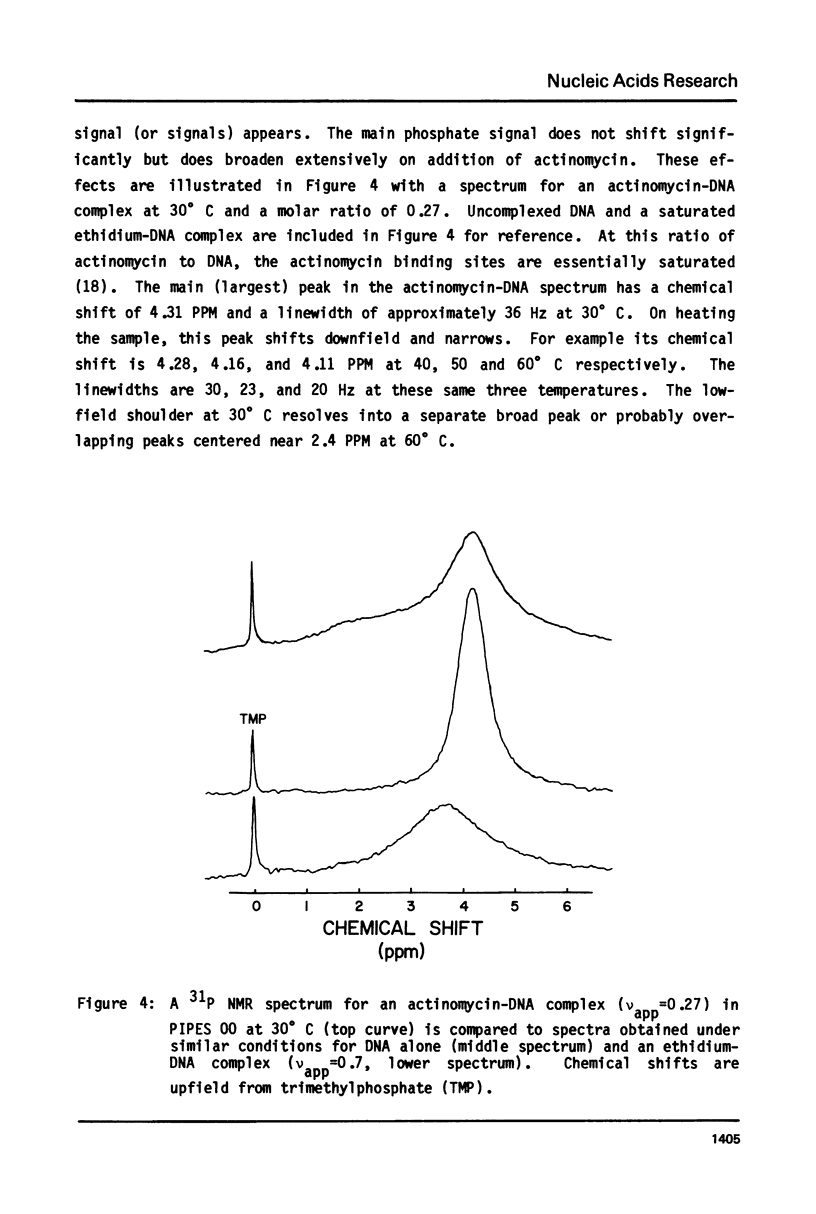
The binding of actinomycin D, ethidium, quinacrine, daunorubicin, and tetralysine to DNA has been investigated using 31P NMR. Titration of DNA with actinomycin yields a new downfield peak or overlapping peaks as would be expected from the slow dissociation kinetics of this compound. The other intercalators shift the DNA 31P signal downfield as a single exchange averaged peak. Tetralysine causes a slight upfield shift. The chemical shift titration curves for the intercalators are sigmoid curves suggesting that cooperative processes or competing effects on the chemical shift are being observed. The magnitude of the chemical shift change at saturation of DNA with the compounds is found to vary significantly and to be linearly related to the DNA base pair unwinding angle for the compounds. Analysis of 31P spin lattice relaxation times (T1) and linewidths as a function of temperature (below Tm) and titration with the above compounds indicates that T1 does not change significantly while linewidth increases with decreasing temperature and increasing bound intercalator. One interpretation of these results is that in both cases the overall motion of DNA becomes slower while the internal motion is not greatly affected.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baase W. A., Johnson W. C., Jr Circular dichroism and DNA secondary structure. Nucleic Acids Res. 1979 Feb;6(2):797–814. doi: 10.1093/nar/6.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresloff J. L., Crothers D. M. DNA-ethidium reaction kinetics: demonstration of direct ligand transfer between DNA binding sites. J Mol Biol. 1975 Jun 15;95(1):103–123. doi: 10.1016/0022-2836(75)90339-3. [DOI] [PubMed] [Google Scholar]
- Dattagupta N., Crothers D. M. Solution structural studies of the Ag(I)-DNA complex. Nucleic Acids Res. 1981 Jun 25;9(12):2971–2985. doi: 10.1093/nar/9.12.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dattagupta N., Hogan M., Crothers D. M. Interaction of netropsin and distamycin with deoxyribonucleic acid: electric dichroism study. Biochemistry. 1980 Dec 23;19(26):5998–6005. doi: 10.1021/bi00567a009. [DOI] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R., Hillen W., Wells R. D. A 300-MHz proton nuclear magnetic resonance investigation of deoxyribonucleic acid restriction fragments: dynamic properties. Biochemistry. 1981 Jun 23;20(13):3764–3769. doi: 10.1021/bi00516a015. [DOI] [PubMed] [Google Scholar]
- Gorenstein D. G. Nucleotide conformational analysis by 31P nuclear magnetic resonance spectroscopy. Annu Rev Biophys Bioeng. 1981;10:355–386. doi: 10.1146/annurev.bb.10.060181.002035. [DOI] [PubMed] [Google Scholar]
- Jones R. L., Lanier A. C., Keel R. A., Wilson W. D. The effect of ionic strength on DNA-ligand unwinding angles for acridine and quinoline derivatives. Nucleic Acids Res. 1980 Apr 11;8(7):1613–1624. doi: 10.1093/nar/8.7.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns D. R. High-resolution nuclear magnetic resonance studies of double helical polynucleotides. Annu Rev Biophys Bioeng. 1977;6:477–523. doi: 10.1146/annurev.bb.06.060177.002401. [DOI] [PubMed] [Google Scholar]
- Levy G. C., Hilliard P. R., Jr, Levy L. F., Rill R. L., Inners R. Carbon 13 spin-lattice relaxation, linewidth, and nuclear Overhauser enhancement measurements of nucleosome length DNA. J Biol Chem. 1981 Oct 10;256(19):9986–9989. [PubMed] [Google Scholar]
- Lipari G., Szabo A. Nuclear magnetic resonance relaxation in nucleic acid fragments: models for internal motion. Biochemistry. 1981 Oct 13;20(21):6250–6256. doi: 10.1021/bi00524a053. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., deHaseth P. L., Record M. T., Jr Pentalysine-deoxyribonucleic acid interactions: a model for the general effects of ion concentrations on the interactions of proteins with nucleic acids. Biochemistry. 1980 Jul 22;19(15):3522–3530. doi: 10.1021/bi00556a017. [DOI] [PubMed] [Google Scholar]
- Mariam Y. H., Wilson W. D. A 31P NMR analysis of the helix to coil transition of natural DNA samples: evidence for the existence of different conformational states. Biochem Biophys Res Commun. 1979 Jun 13;88(3):861–866. doi: 10.1016/0006-291x(79)91488-8. [DOI] [PubMed] [Google Scholar]
- McDonald C. C., Phillips W. D., Lazar J. Nuclear magnetic resonance determination of thymine nearest neighbor base frequency ratios in deoxyribonucleic acid. J Am Chem Soc. 1967 Aug 2;89(16):4166–4170. doi: 10.1021/ja00992a035. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M. Studies of the binding of actinomycin and related compounds to DNA. J Mol Biol. 1968 Jul 28;35(2):251–290. doi: 10.1016/s0022-2836(68)80024-5. [DOI] [PubMed] [Google Scholar]
- Patel D. J. Proton and phosphorus NMR studies of d-CpG(pCpG)n duplexes in solution. Helix-coil transition and complex formation with actinomycin-D. Biopolymers. 1976 Mar;15(3):533–558. doi: 10.1002/bip.1976.360150310. [DOI] [PubMed] [Google Scholar]
- Saucier J. M., Festy B., Le Pecq J. B. The change of the torsion of the DNA helix caused by intercalation. II. Measurement of the relative change of torsion induced by various intercalating drugs. Biochimie. 1971;53(9):973–980. doi: 10.1016/s0300-9084(71)80065-2. [DOI] [PubMed] [Google Scholar]
- Shafer R. H., Burnette R. R., Mirau P. A. Spectroscopic analysis of the equilibrium and kinetic DNA binding properties of several actinomycin analogs. Nucleic Acids Res. 1980 Mar 11;8(5):1121–1132. doi: 10.1093/nar/8.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo H. NMR relaxation processes of 31P in macromolecules. Biopolymers. 1980 Mar;19(3):509–522. doi: 10.1002/bip.1980.360190306. [DOI] [PubMed] [Google Scholar]
- Wakelin L. P., Waring M. J. Kinetics of drug-DNA interaction. Dependence of the binding mechanism on structure of the ligand. J Mol Biol. 1980 Dec 5;144(2):183–214. doi: 10.1016/0022-2836(80)90032-7. [DOI] [PubMed] [Google Scholar]
- Wilson D. W., Grier D., Reimer R., Bauman J. D., Preston J. F., Gabbay E. J. Sturcture-activity relationship of daunorubicin and its peptide derivatives. J Med Chem. 1976 Mar;19(3):381–384. doi: 10.1021/jm00225a008. [DOI] [PubMed] [Google Scholar]
- Wilson D. W., Grier D., Reimer R., Bauman J. D., Preston J. F., Gabbay E. J. Sturcture-activity relationship of daunorubicin and its peptide derivatives. J Med Chem. 1976 Mar;19(3):381–384. doi: 10.1021/jm00225a008. [DOI] [PubMed] [Google Scholar]
- Wilson W. D., Jones R. L. Intercalating drugs: DNA binding and molecular pharmacology. Adv Pharmacol Chemother. 1981;18:177–222. doi: 10.1016/s1054-3589(08)60255-0. [DOI] [PubMed] [Google Scholar]
- Wilson W. D., Lopp I. G. Analysis of cooperativity and ion effects in the interaction of quinacrine with DNA. Biopolymers. 1979 Dec;18(12):3025–3041. doi: 10.1002/bip.1979.360181210. [DOI] [PubMed] [Google Scholar]
- Winkle S. A., Krugh T. R. Equilibrium binding of carcinogens and antitumor antibiotics to DNA: site selectivity, cooperativity, allosterism. Nucleic Acids Res. 1981 Jul 10;9(13):3175–3186. doi: 10.1093/nar/9.13.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]


