Abstract
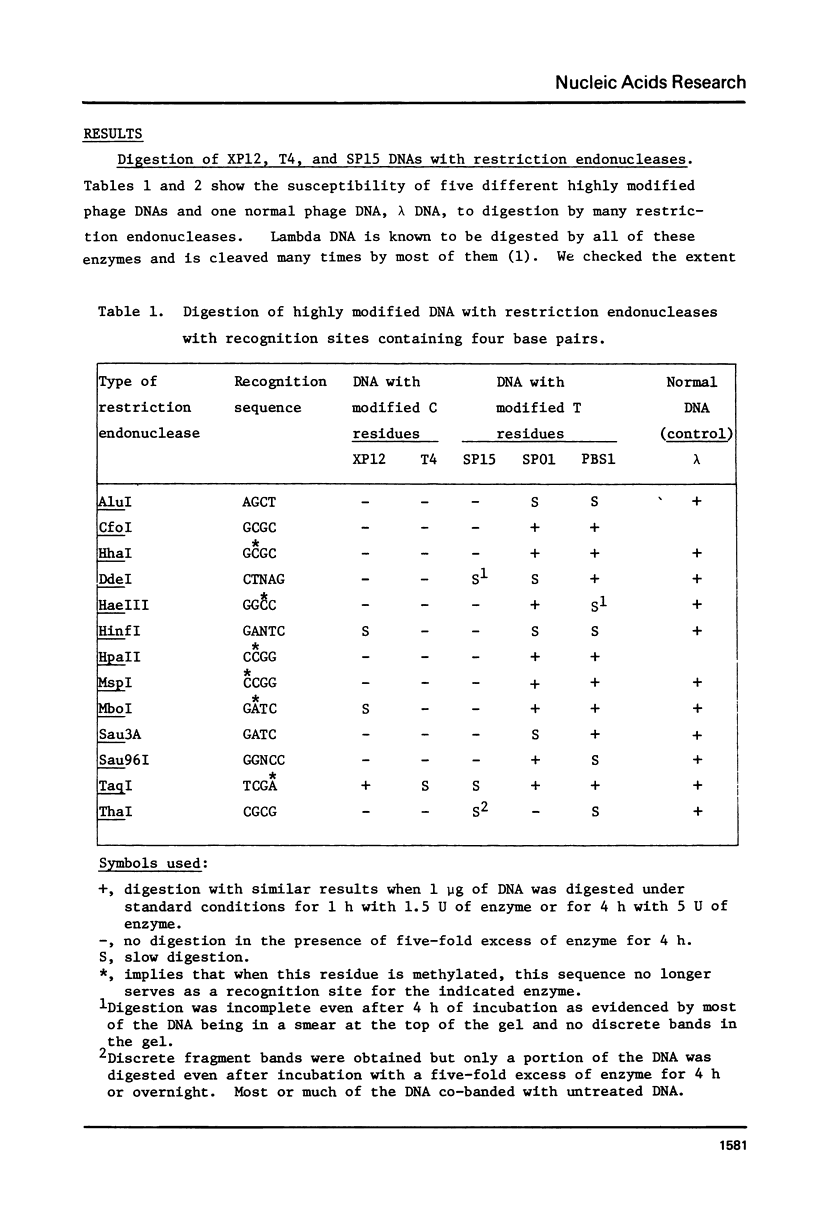
The ability of thirty Type II restriction endonucleases to cleave five different types of highly modified DNA has been examined. The DNA substrates were derived from relatively large bacteriophage genomes which contain all or most of the cytosine or thymine residues substituted at the 5-position. These substituents were a proton (PBS1 DNA), a hydroxymethyl group (SP01 DNA), a methyl group (XP12 DNA), a glucosylated hydroxymethyl group (T4 DNA), or a phosphoglucuronated, glucosylated 4,5-dihydroxypentyl group (SP15 DNA). Although PBS1 DNA and SP01 DNA were digested by most of the enzymes, they were cleaved much more slowly than was normal DNA by many of them. 5-Methylcytosine-rich XP12 DNA and the multiply modified T4 and SP15 DNAs were resistant to most of these endonucleases. The only enzyme that cleaved all five of these DNAs was TaqI, which fragmented them extensively.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alden C. J., Kim S. H. Solvent-accessible surfaces of nucleic acids. J Mol Biol. 1979 Aug 15;132(3):411–434. doi: 10.1016/0022-2836(79)90268-7. [DOI] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. The effects of substituted pyrimidines in DNAs on cleavage by sequence-specific endonucleases. J Biol Chem. 1979 Apr 10;254(7):2551–2560. [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J. M., Stewart C. R. EcoRI cleavage of DNA from Bacillus subtilis phage SPO1. Virology. 1978 Apr;85(2):601–605. doi: 10.1016/0042-6822(78)90464-6. [DOI] [PubMed] [Google Scholar]
- Crosa J. H., Luttropp L. K., Falkow S. Nature of R-factor replication in the presence of chloramphenicol. Proc Natl Acad Sci U S A. 1975 Feb;72(2):654–658. doi: 10.1073/pnas.72.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Ehrlich K. C. A novel, highly modified, bacteriophage DNA in which thymine is partly replaced by a phosphoglucuronate moiety covalently bound to 5-(4',5'-dihydroxypentyl)uracil. J Biol Chem. 1981 Oct 10;256(19):9966–9972. [PubMed] [Google Scholar]
- Ehrlich M., Ehrlich K., Mayo J. A. Unusual properties of the DNA from Xanthomonas phage XP-12 in which 5-methylcytosine completely replaces cytosine. Biochim Biophys Acta. 1975 Jun 16;395(2):109–119. doi: 10.1016/0005-2787(75)90149-5. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res. 1981 Jun 11;9(11):2509–2515. doi: 10.1093/nar/9.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Freund M., Trautner T. A. Restriction and modification in Bacillus subtilis: two DNA methyltransferases with BsuRI specificity. I. Purification and physical properties. J Biol Chem. 1981 Sep 10;256(17):9340–9345. [PubMed] [Google Scholar]
- Hattman S., Gribbin C., Hutchison C. A., 3rd In vivo methylation of bacteriophage phi X174 DNA. J Virol. 1979 Dec;32(3):845–851. doi: 10.1128/jvi.32.3.845-851.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., van Ormondt H., de Waard A. Sequence specificity of the wild-type dam+) and mutant (damh) forms of bacteriophage T2 DNA adenine methylase. J Mol Biol. 1978 Mar 5;119(3):361–376. doi: 10.1016/0022-2836(78)90219-x. [DOI] [PubMed] [Google Scholar]
- Hayashi H., Nakanishi K., Brandon C., Marmur J. Structure and synthesis of dihydroxypentyluracil from bacteriophage SP-15 deoxyribonucleic acid. J Am Chem Soc. 1973 Dec 26;95(26):8749–8757. doi: 10.1021/ja00807a041. [DOI] [PubMed] [Google Scholar]
- Hemphill H. E., Whiteley H. R. Bacteriophages of Bacillus subtilis. Bacteriol Rev. 1975 Sep;39(3):257–315. doi: 10.1128/br.39.3.257-315.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer B., Köster H. Enzymatic synthesis, ligation, and restriction of DNA containing deoxy-4-thiothymidine. Nucleic Acids Res. 1981 Feb 25;9(4):753–767. doi: 10.1093/nar/9.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hughes S. G. The sensitivity of bacteriophage lambda DNA to restriction endonuclease RII. J Mol Biol. 1975 Nov 5;98(3):645–647. doi: 10.1016/s0022-2836(75)80093-3. [DOI] [PubMed] [Google Scholar]
- Hänggi U. J., Zachau H. G. Isolation and characterization of DNA fragments containing the dihydrofolate-reductase gene of coliphage T4. Gene. 1980 May;9(3-4):271–285. doi: 10.1016/0378-1119(90)90327-n. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Günthert U., Trautner T. A. DNA methyltransferases affecting the sequence 5'CCGG. Nucleic Acids Res. 1981 Jun 25;9(12):2753–2759. doi: 10.1093/nar/9.12.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KALLEN R. G., SIMON M., MARMUR J. The new occurrence of a new pyrimidine base replacing thymine in a bacteriophage DNA:5-hydroxymethyl uracil. J Mol Biol. 1962 Aug;5:248–250. doi: 10.1016/s0022-2836(62)80087-4. [DOI] [PubMed] [Google Scholar]
- Kaplan D. A., Nierlich D. P. Cleavage of Nonglucosylated Bacteriophage T4 deoxyribonucleic acid by Restriction Endonuclease Eco RI. J Biol Chem. 1975 Mar 25;250(6):2395–2397. [PubMed] [Google Scholar]
- Kuo K. C., McCune R. A., Gehrke C. W., Midgett R., Ehrlich M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 1980 Oct 24;8(20):4763–4776. doi: 10.1093/nar/8.20.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T. T., Huang T. C., Teng M. H. 5-Methylcytosine replacing cytosine in the deoxyribonucleic acid of a bacteriophage for Xanthomonas oryzae. J Mol Biol. 1968 Jul 14;34(2):373–375. doi: 10.1016/0022-2836(68)90263-5. [DOI] [PubMed] [Google Scholar]
- LEHMAN I. R., PRATT E. A. On the structure of the glucosylated hydroxymethylcytosine nucleotides of coliphages T2, T4, and T6. J Biol Chem. 1960 Nov;235:3254–3259. [PubMed] [Google Scholar]
- Leslie A. G., Arnott S., Chandrasekaran R., Ratliff R. L. Polymorphism of DNA double helices. J Mol Biol. 1980 Oct 15;143(1):49–72. doi: 10.1016/0022-2836(80)90124-2. [DOI] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh R. C., Hepburn M. L. Map of restriction sites on bacteriophage T4 cytosine-containing DNA for endonucleases bamHI, BglII, KpnI, PvuI, SalI, and XbaI. J Virol. 1981 Apr;38(1):104–114. doi: 10.1128/jvi.38.1.104-114.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. Structures and mechanisms of DNA restriction and modification enzymes. Q Rev Biophys. 1979 Aug;12(3):315–369. doi: 10.1017/s0033583500005461. [DOI] [PubMed] [Google Scholar]
- Pero J., Hannett N. M., Talkington C. Restriction cleavage map of SP01 DNA: general location of early, middle, and late genes. J Virol. 1979 Jul;31(1):156–171. doi: 10.1128/jvi.31.1.156-171.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribnow D., Sigurdson D. C., Gold L., Singer B. S., Napoli C., Brosius J., Dull T. J., Noller H. F. rII cistrons of bacteriophage T4. DNA sequence around the intercistronic divide and positions of genetic landmarks. J Mol Biol. 1981 Jul 5;149(3):337–376. doi: 10.1016/0022-2836(81)90477-0. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1981 Jan 10;9(1):r75–r96. doi: 10.1093/nar/9.1.213-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O. Nucleotide sequence specificity of restriction endonucleases. Science. 1979 Aug 3;205(4405):455–462. doi: 10.1126/science.377492. [DOI] [PubMed] [Google Scholar]
- Streeck R. E. Single-strand and double-strand cleavage at half-modified and fully modified recognition sites for the restriction nucleases Sau3a and Taqi. Gene. 1980 Dec;12(3-4):267–275. doi: 10.1016/0378-1119(80)90109-2. [DOI] [PubMed] [Google Scholar]
- TAKAHASHI I., MARMUR J. Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature. 1963 Feb 23;197:794–795. doi: 10.1038/197794a0. [DOI] [PubMed] [Google Scholar]
- Truffaut N., Revet B., Soulie M. O. Etude comparative des DNA de phages 2C, SP8*, SP82, phi e, SP01 et SP50. Eur J Biochem. 1970 Aug;15(2):391–400. doi: 10.1111/j.1432-1033.1970.tb01020.x. [DOI] [PubMed] [Google Scholar]
- Tyeryar F. J., Jr, Taylor M. J., Lawton W. D., Goldberg I. D. Cotransduction and cotransformation of genetic markers in Bacillus subtilis and Bacillus licheniformis. J Bacteriol. 1969 Nov;100(2):1027–1036. doi: 10.1128/jb.100.2.1027-1036.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. Y., Shedlarski J. G., Farber M. B., Kuebbing D., Ehrlich M. Two sequence-specific endonucleases from Xanthomonas oryzae. Characterization and unusual properties. Biochim Biophys Acta. 1980 Feb 29;606(2):371–385. doi: 10.1016/0005-2787(80)90047-7. [DOI] [PubMed] [Google Scholar]
- Warren R. A. Modified bases in bacteriophage DNAs. Annu Rev Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- Woodhead J. L., Malcolm A. D. Non-specific binding of restriction endonuclease EcoR1 to DNA. Nucleic Acids Res. 1980 Jan 25;8(2):389–402. doi: 10.1093/nar/8.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]