Abstract
This report investigates the mechanisms by which mammalian cells coordinate DNA replication with transcription and chromatin assembly. In yeast, DNA replication initiates within nucleosome-free regions, but studies in mammalian cells have not revealed a similar relationship. Here, we have used genome-wide massively parallel sequencing to map replication initiation events, thereby creating a database of all replication initiation sites within nonrepetitive DNA in two human cell lines. Mining this database revealed that genomic regions transcribed at moderate levels were generally associated with high replication initiation frequency. In genomic regions with high rates of transcription, very few replication initiation events were detected. High-resolution mapping of replication initiation sites showed that replication initiation events were absent from transcription start sites but were highly enriched in adjacent, downstream sequences. Methylation of CpG sequences strongly affected the location of replication initiation events, whereas histone modifications had minimal effects. These observations suggest that high levels of transcription interfere with formation of pre-replication protein complexes. Data presented here identify replication initiation sites throughout the genome, providing a foundation for further analyses of DNA–replication dynamics and cell-cycle progression.
DNA replication is tightly orchestrated to duplicate the entire genome precisely once each somatic cell cycle. In eukaryotes, replication initiates at multiple sites on each chromosome in a temporally regulated and often tissue-specific manner. Approximately 30 replication initiation sites in the human genome have been identified and characterized in some detail (Aladjem et al. 2006; Hamlin et al. 2008). Additional replication initiation sites have been identified using high-throughput targeted microarray analyses (Cadoret et al. 2008; Karnani et al. 2009; Gilbert 2010). Identified replication initiation sites in human cells do not share clear consensus sequences. In contrast, replication initiation sites exhibit sequence similarities in single-celled eukaryotes such as yeast. In yeast, consensus replication initiation yeast sequences are characterized by a unique, asymmetric AT-rich sequence, leading to a distinct pattern of nucleosome positioning (Eaton et al. 2010). However, even in yeast, replication is limited to a subset of potential initiation sites exhibiting the consensus sequence, and in many instances, potential yeast replication origins do not initiate replication. In metazoans, strong DNA sequence similarity is not observed among replication initiation sites, suggesting that primary DNA sequence is not the sole determinant of replication initiation competence (Aladjem 2007; Mechali 2010). Replication initiation sites in metazoa share several DNA sequence motifs, including AT-rich sequences, matrix attachment sites, and asymmetric purine:pyrimidine sequences (Aladjem 2007). None of these motifs, however, is absolutely essential for replication initiation. Sequence variation within known replication initiation sites suggests that the regulation of site localization and the timing of DNA replication initiation may involve epigenetic mechanisms.
DNA replication may be regulated epigenetically to allow transcription and replication to proceed in a coordinated fashion. In mammalian cells, mapping studies suggest that replication initiation events are enriched in transcription factor binding sites (Cadoret et al. 2008; Karnani et al. 2009; Gilbert 2010; Valenzuela et al. 2011) and CpG islands (Antequera 2004; Gomez and Brockdorff 2004). Consistent with this observation, distal sequences involved in transcription are required for replication initiation at a number of loci, including a region 40 kb upstream of the human beta-globin (HBB) replication origin (Aladjem et al. 1995), the promoter of the Chinese hamster Dhfr locus (Kalejta et al. 1998), and an enhancer of the Th2 locus (Hayashida et al. 2006). In several biological contexts, however, transcription appears to suppress replication initiation. This occurs during differentiation of Xenopus (Hyrien et al. 1995) and Sciara (Lunyak et al. 2002) embryos, at the Chinese hamster Dhfr locus (Saha et al. 2004), and at the murine HoxB locus (Gregoire et al. 2006). Conversely, at the human IGH locus (Zhou et al. 2002) replication initiates in an expanded region after the onset of transcription; and on human (Gray et al. 2007) and murine (Rowntree and Lee 2006) X chromosomes, replication initiates from distinct origins regardless of transcriptional status. Finally, at the human MYC locus, transcription factor binding seems to activate initiation of DNA replication, whereas high transcription rates suppress it (Ghosh et al. 2004).
To investigate the relationship between replication initiation and transcription, we measured replication density and transcriptional activity in nonrepetitive genomes of two human cancer cell lines, which have different patterns of gene expression. Replication initiation events were more frequent and clustered near regions of moderate transcription. In contrast, low levels of replication initiation were found in genomic regions either devoid of transcription, or with very high levels of transcription. High-resolution analyses of initiation event distribution also suggested that replication initiation sites were enriched near promoters, but excluded from transcription start sites (TSSs). Finally, replication tended to initiate in genomic regions containing methylated CpG sequences, which are frequently associated with low rates of transcription
Results
Visualization of replication initiation sites
To map replication initiation at a genome-wide scale, short, RNA-primed nascent DNA was isolated from two human cell lines (myeloid erythroleukemia K562 cells and breast cancer MCF7 cells) (Bielinsky and Gerbi 1998; Wang et al. 2004) and analyzed using massively parallel sequencing. The frequency of initiation events at the individual genomic regions was measured as the ratio between reads obtained from a nascent strand preparation and reads obtained from a corresponding control genomic DNA preparation. Reads were calculated as reads per kilobase per million mapped reads (RPKM). The number of reads and aligned reads from K562 and MCF7 cells are shown in the Supplemental Methods (Supplemental Table 1). Results were visualized using the Integrative Genome Viewer 1.4.1 (Broad Institute), and data validated by analyzing initiation at well-characterized replication initiation sites (Fig. 1; Supplemental Fig. 1).
Figure 1.
Sample replication initiation profiles obtained through massively parallel sequencing of nascent DNA strands. A chromosome map is shown at the top, and the region-of-interest is delineated by a black rectangle. The analyzed region is shown underneath the ideogram, with map coordinates indicated. The experimental tracks (MCF7 or K562 nascent strand [NS]/genome ratios) show the distribution of sequence reads (aligned with the indicated region) obtained from massively parallel sequencing of nascent strands either from MCF7 breast cancer cells or from K562 erythroleukemia cells. All data are shown as the ratio of reads obtained from a nascent strand preparation and reads obtained from a corresponding control genomic DNA preparation. For each track, the y-axis indicates the nascent strand/genomic DNA ratio. Reads were calculated as reads per kilobase per million mapped reads (RPKM); for details, see Supplemental Information. RefSeq genes are aligned under the nascent strand distribution. For RefSeq genes, thick boxes represent exons, whereas thin lines represent introns and untranslated regions. Arrows on RefSeq genes indicate the direction of transcription. Initiation at select sites was verified using real-time PCR, with primers listed in Table 2. Examples of control and nascent strand tracks are shown in Supplemental Figure 1A. (A–C) Mapping replication initiation events at previously characterized replication origins. (A) Data from the MYC locus (human chromosome 8). Replication initiation sites were mapped to the region spanning the promoter to the first exon of the MYC gene. (B) Data from the human beta globin locus (HBB) (human chromosome 11). Replication initiation sites were mapped to the region stretching from the promoter to the first intron of the HBB gene. (C) Data from the HPRT1 gene on the X chromosome. Replication initiation sites were mapped near the HPRT1 promoter. (D) Data from the CTCF locus (chromosome 16 q22.1). This region does not contain a known replication origin. Initiation from gene promoters and from the RLTPR gene region (3′ of the CTCF gene) was verified using real-time PCR on an independent preparation of nascent strands (data not shown).
We first analyzed replication profiles at several, well-characterized genomic loci. At these loci, replication has been mapped several times previously by several independent research groups using methods that rely on nonoverlapping principles. Figure 1A shows the replication initiation ratio (nascent strands vs. genomic RPKM) at the MYC locus on human chromosome 8. At this locus, DNA replication initiates from several sites within and around the promoter and within the first exon of the gene (Ghosh et al. 2004). As expected, our analysis showed that the replication initiation ratio was high in this region, and several replication initiation peaks were observed, consistent with previous observations (e.g., Ghosh et al. 2004). Similarly, replication initiation ratios near the HBB gene (chromosome 11) were highest near the promoter (Fig. 1B), consistent with previous data (Kitsberg et al. 1993; Aladjem et al. 1995). Finally, Figure 1C shows the replication profile at the HPRT1 locus on the X chromosome. Consistent with previous observations (Cohen et al. 2004), replication preferentially initiated near the promoter region.
Although replication initiation sites seemed clustered in some loci, we also identified very large genomic regions that were devoid of replication initiation events. For example, replication initiation sites were entirely absent from the CTCF gene, a region that spans ∼120 kb on chromosome 16 (position 67564943–67704608) (Fig. 1D). Irregular clustering of replication initiation events such as this was evident throughout the genome.
Replication initiation events and transcriptional activity
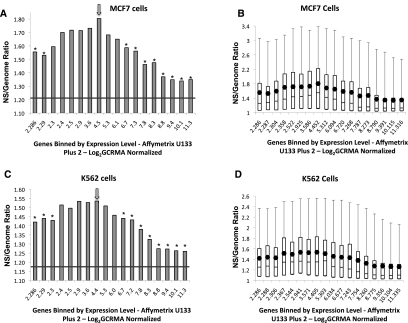
To compare replicative and transcriptional activities, data from Affymetrix Human Genome U133 Plus 2.0 microarrays (Shankavaram et al. 2007) were used to determine gene expression values for K562 and MCF7 cells. Replication initiation data for chromosome regions within 3 kb of known genes were then aligned with the gene expression data. Figure 2, A and B shows that in MCF7 cells, regions with very low transcriptional activity were not enriched in replication initiation events, and that replication density generally increased with gene expression. Peak initiation activity coincided with moderate expression levels. Replication initiation events were again less frequent in genes with high levels of transcriptional activity. Similar results were obtained with K562 cells (Fig. 2C,D). In addition, no replication initiation rate differences were found when 5′ and 3′ flanking gene regions were compared or when chromosomal regions between two genes transcribed either in the same or opposite directions were compared (data not shown). In general, our data suggest that replication initiation events are more frequent in moderately transcribed regions and less frequent in regions exhibiting either high or low levels of transcription.
Figure 2.
Replication and transcription. (A,B) Replication enrichment ratios (nascent strand [NS] versus genomic control RPKM) for all identified genes in MCF7 cells plotted against log2 GCRMA normalized gene expression. Genes on the x-axis were binned according to gene expression, with each bin containing 897 genes. The first column, however, represents a combination of the first five bins, which includes 4485 low-expressing genes that did not show significant differences in log2 GCRMA normalized gene expression. (A) The mean enrichment ratio for each bin is plotted against gene expression. (B) Replication enrichment ratios calculated as in A, showing the distribution of enrichment ratio values as a box plot. (C,D) Replication enrichment ratios calculated as in A and B for K562 cells plotted as a histogram of mean values (C) or as a box plot (D). For mean value histograms (A,C), asterisks represent statistically significant (P < 0.001) divergence from the central bin, which is marked with an arrow. For box plots (B,D), boxes indicate distributions of the second and third quartiles; dots indicate mean values; error bars indicate the fifth and 95th percentiles. The horizontal line in A and C represents the average enrichment ratio of the entire genome.
Replication initiation and TSSs
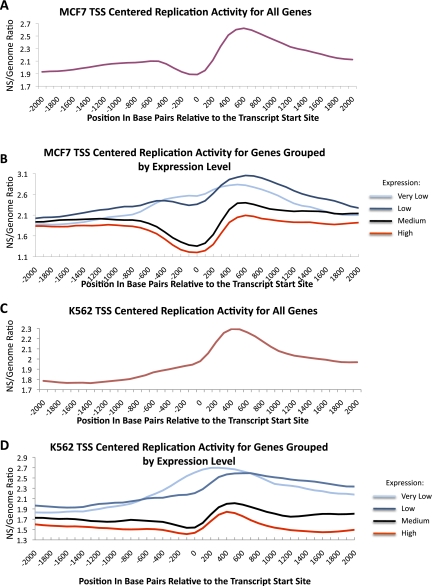
The relationship between replication initiation and distance from a TSS was examined for all predicted genes in K562 and MCF7 cells. Replication initiation events were generally enriched in regions within 10 kb of a TSS (Supplemental Fig. 1B). However, higher-resolution analyses showed that in MCF7 cells, the replication initiation ratio decreased several hundred base pairs upstream of the TSS, and peaked ∼500 bp downstream from the TSS (Fig. 3A). As RPKM values around the TSS were generally elevated in control samples (genomic DNA), the decreased RPKM in nascent strands versus the genomic control was particularly striking (for details, see Supplemental Information). The replication initiation pattern at the TSS indicates that replication initiation events are excluded from regions that nucleate transcriptional complexes. To evaluate whether the depletion of initiation events at the TSS was associated with transcription activity, the analysis was repeated using four groups of genes with different levels of expression. For the purpose of this analysis, log2 GCRMA normalized expression values of <2.3, 2.3–5.3, 5.3–8.5, or >8.5 were classified as very low, low, medium, or high, respectively. For genes with very low and low levels of transcriptional activity, initiation events were not excluded from the TSS, and instead, a preference for initiation near the TSS was detected (Fig. 3B). In contrast, a marked reduction in initiation activity was observed at the TSS in genes with medium and high levels of transcription. Similar results were observed in nascent strands from K562 cells (Fig. 3C,D). These observations suggest that the depletion of replication initiation events from regions adjacent to the TSS are directly associated with transcriptional activity, and possibly the assembly of transcription initiation complexes.
Figure 3.
Replication initiation depletion at the transcription start site (TSS) in transcribed genes. (A) Average replication enrichment ratio in MCF7 cells (calculated as in Fig. 2) plotted against distance from the TSS for all known genes. (B) Distribution of replication enrichment ratios in MCF7 cells for groups of genes that exhibit different levels of expression. Levels include the following: very low (log2 GCRMA <2.3), low (log2 GCRMA 2.3–5.3), medium (log2 GCRMA 5.3–8.5), and high (log2 GCRMA >8.5). (C,D) The same analyses are shown for K562 cells.
Replication initiation activity in discordantly expressed genes
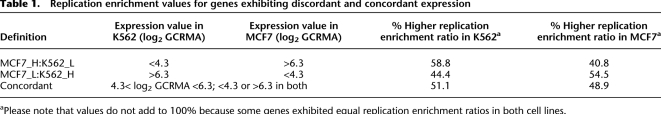
We next tested whether genomic regions that exhibit discordant levels of gene expression between K562 and MCF7 cells exhibit disparate frequencies of replication initiation events. We calculated the replication initiation activity of discordantly expressed genes, defined as genes that exhibit (1) high expression in MCF7 cells and low expression in K562 cells (MCF7_H:K562_L), or (2) low expression in MCF7 cells and high expression in K562 cells (MCF7_L:K562_H). High and low levels of gene expression were defined as >6.3 and <4.3 log2 GCRMA values, respectively. Genes that exhibited similar GCRMA values were defined as concordant genes. As shown in Table 1, a substantial fraction (58.5%) of genes that exhibited high expression in MCF7 cells and low expression in K562 cells had a higher frequency of initiation in K562 cells. Similarly, 54.5% of genes that exhibited high expression in K562 and low expression in MCF7 exhibited a higher frequency of initiation in MCF7 cells. In contrast, in concordantly expressed genes that exhibited similar levels of expression in both cell lines, 51.1% of the genes exhibited a higher frequency of initiation in K562 and 48.9% of the genes exhibited a higher frequency of initiation in MCF7.
Table 1.
Replication enrichment values for genes exhibiting discordant and concordant expression
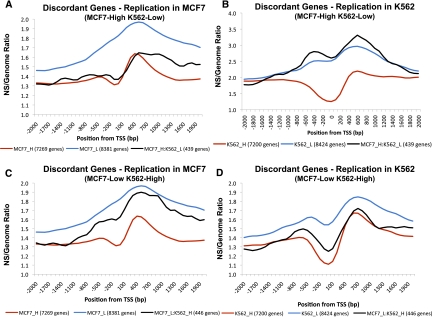
We next tested whether discordantly expressed genes exhibited TSS replication initiation patterns typical of their expression group. Discordantly expressed genes expressed at high levels in MCF7 cells (and therefore low levels in K562 cells) exhibited TSS exclusion patterns in MCF7 cells that resembled those for all MCF7 highly expressed genes (Fig. 4A). In contrast, those same genes did not exhibit TSS exclusion in K562 cells (Fig. 4B). Similarly, genes that exhibited low expression levels in MCF7 cells (and therefore high K562) did not show marked TSS exclusion in MCF7 cells (Fig. 4C) but did show marked TSS exclusion in K562 cells (Fig. 4D).
Figure 4.
Replication initiation in discordantly expressed genes. We calculated replication enrichment ratios (nascent strands RPKM vs. control genomic DNA RPKM) for genes whose level of expression differed significantly between K562 and MCF7 cell types. Genes with log2 GCRMA values <4.3 in MCF7 and >6.3 in K562 were considered MCF7-low and K562-high (MCF7_L:K562_H); genes with log2 GCRMA values <4.3 in K562 and >6.3 in MCF7 were considered MCF7-high and K562-low (MCF7_H:K562_L). (A) Distribution of nascent strand enrichment ratios in MCF7 cells for MCF7_H:K562_L genes. For comparison, enrichment ratio plots for MCF7_H and MCF7_L genes are shown. (B) Distribution of enrichment ratios in K562 cells for MCF7_H:K562_L genes. For comparison, enrichment ratio plots for K562_H and K562_L genes are shown. (C) Distribution of nascent strand enrichment ratios in MCF7 cells for MCF7_L:K562_H genes. For comparison, enrichment ratio plots for MCF7_H and MCF7_L genes are shown. (D) Distribution of enrichment ratios in K562 cells for MCF7_L:K562_H genes. For comparison, enrichment ratio plots for K562_H and K562_L genes are shown.
Replication initiation and chromatin modifications
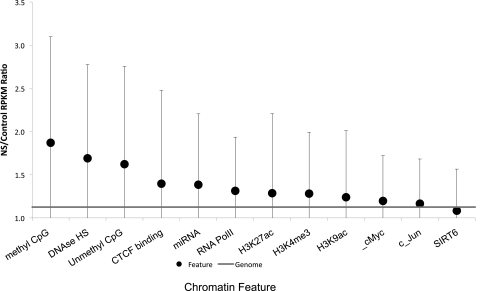
With all replication initiation events precisely mapped for these two cell lines, we next asked whether replication initiation events were associated with particular modifications and properties that affect chromatin conformation. Associations between replication initiation frequency and chromatin modifications in K562 cells were analyzed using the UCSC database of epigenetic modifications. As shown in Figure 5, the highest replication initiation ratios were associated with methylated CpG sequences and DNase hypersensitivity. Other features, such as unmethylated CpGs, CTCF binding sites, regions encoding miRNA transcripts, and RNA Polymerase II binding sites were also enriched, albeit at lower levels. Replication initiation events were more frequent in regions that exhibited both methylated CpGs and DNase hypersensitivity (Supplemental Fig. 2A), but this did not represent a striking synergistic effect. CpG methylation in combination with CTCF binding sites did not enhance the initiation ratio, and CTCF binding sites in combination with other chromatin modifications did not enhance the initiation ratio (Supplemental Fig. 2). Consistent with previous reports, higher replication density was associated with binding sites for the JUN transcription factor (Cadoret et al. 2008), but this enrichment was minimal. Histone modifications such as methylation of H3K4 or acetylation of H3K9 and H3K27 exhibited minor but statistically significant enrichment in replication initiation.
Figure 5.
Chromatin modifications and replication initiation events. The average nascent strand versus genomic DNA ratio (calculated as described for Fig. 2) is shown for genomic regions that contain the indicated chromatin modification features. Modified regions were identified using data from the UCSC Genome Browser (Table 3). Mean NS/control RPKM ratios (dot), and corresponding SDs (error bars), are shown. The horizontal line indicates a matched random control. Chromatin modification features are sorted according to the level of enrichment for replication initiation frequency. Statistically significant deviations from replication ratios of the entire genome (P < 0.001) were observed for all modifications except c-Jun and SIRT6. For intersections between chromatin features (regions that exhibit combinations of chromatin modifications), please see Supplemental Information.
CpG methylation, replication, and gene expression
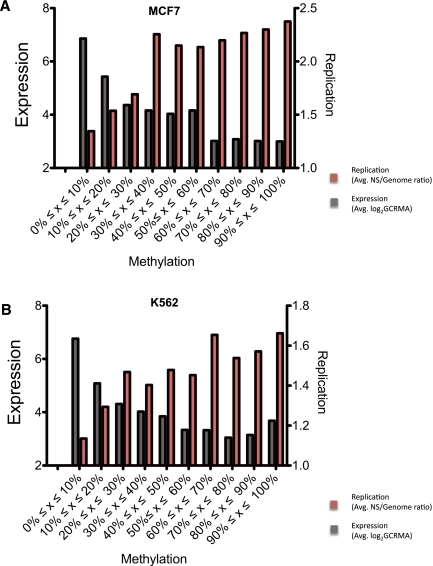
To further explore the effect of CpG methylation on replication initiation and its relationship with gene expression, we plotted replication initiation ratios and gene expression values against the frequency of DNA methylation. As shown in Figure 6, the frequency of replication initiation events generally increased with the extent of methylated CpG sequences, whereas gene expression exhibited an inverse relationship. This was seen for both K562 and MCF7 cell lines.
Figure 6.
Effect of CpG methylation on the frequency of replication initiation events and gene expression. The level of CpG methylation for MCF7 cells (A) and K562 cells (B) is plotted against gene expression levels (log2 GCRMA, gray histograms) and replication enrichment ratio (nascent strands vs. genomic control, red histograms). For box plots of the data , please see Supplemental Information.
Discussion
Results from our study suggest that replication initiation is a dynamic, flexible process that is sensitive to both chromatin conformation and transcriptional activity. We found that replication initiation sites were nonrandomly distributed throughout the genome, with clusters of sites in some regions and other regions completely devoid of sites (replication initiation sites were absent in regions up to several hundred kilobases). Replication initiation events were enriched in open chromatin and moderately transcribed regions. Although replication density was highest immediately adjacent to (and downstream from) TSS, the TSS themselves were characterized by low replication density. Methylated CpG tracks were particularly enriched in replication initiation events. Finally, replication initiation events were excluded from tightly condensed chromatin, as well as from heavily transcribed chromatin.
The replication initiation sites identified in our study are in agreement with previous reports, which examined replication initiation events at individual loci (for reviews, see Aladjem et al. 2006; Hamlin et al. 2008). These previous maps were generated using various methodologies that rely on nonoverlapping principles. Overall, we observed replication initiation enrichment within genic regions, which presumably represent open chromatin. The enrichment of replication initiation sites in open chromatin is concordant with the notion that in budding yeast, initiation events occur at an AT-rich consensus (Newlon and Theis 1993; Aladjem and Fanning 2004) or at several near-consensus sequences (Theis and Newlon 2001). Also, in fission yeast, AT-rich sequences are the primary determinants of replicator activity (Dai et al. 2005). Notably, only a subset of AT-rich consensus sequences bind the origin recognition complex and initiate replication, and selected consensus sequences exhibit a unique asymmetric nucleosome positioning pattern (Eaton et al. 2010). In our current study, human replication initiation sites were remarkably enriched immediately downstream from TSSs, colocalizing with nucleosome-free and variant nucleosome regions (Jin et al. 2009). It should be noted that these observations are in agreement with data from the ENCODE project, which has mapped replication initiation sites in ∼1% of the human genome (ENCODE Project Consortium 2007; Karnani et al. 2009). Our results are similar to those obtained in a microarray-based study reporting enrichment of replication initiation in transcription factor binding sites (Cadoret et al. 2008); although unlike that study, we have not observed a very high enrichment for JUN binding sites. Our studies are also consistent with other microarray-based studies reporting the enrichment of replication initiation events in regions of decondensed chromatin associated with histone modifications such as H3K4Me3 (Karnani et al. 2009; Valenzuela et al. 2011), and RNA Pol II (Karnani et al. 2009; Valenzuela et al. 2011). Here we have shown that while transcription factor binding sites were somewhat enriched near replication initiation events, transcriptional activity and CpG methylation appeared to have the strongest impact on replication density. The effect of transcriptional activity and CpG methylation was more critical than factors such as DNase I hypersensitivity, transcription factor binding, and unmethylated CpGs.
In our analysis, replication initiation events clustered near transcriptionally active regions, as has been reported previously (Cadoret et al. 2008; Karnani et al. 2009; Valenzuela et al. 2011). We also observed, however, that initiation events were less frequent in regions that exhibited very high levels of transcription. Hence, differences in gene expression may explain the low concordance between two previous microarray-based studies that mapped replication initiation events (Cadoret et al. 2008; Karnani et al. 2009). In addition, while those previous studies (Cadoret et al. 2008; Karnani et al. 2009; Valenzuela et al. 2011) noted that replication initiation events are enriched in the vicinity of the TSS, our high-resolution data demonstrated that initiation events were excluded from the TSS but enriched immediately downstream from the TSS. Our data also showed that high levels of transcriptional activity were incompatible with the initiation of DNA replication. The depletion of replication initiation events at the location of transcription initiation complexes is similar to the situation in budding yeast, where replication initiates almost exclusively between genes (Raghuraman et al. 2001). These data imply that a high density of transcription initiation complexes may interfere with the formation of replication initiation complexes.
Previous studies have noted a preference for replication initiation in CpG islands (Delgado et al. 1998), and it has been suggested that CpG islands are associated with promoters that can also function as replication origins (Antequera 2004). Analyses of strand-specific mutational asymmetries in long, intergenic regions also suggest that CpG sequences serve as origins for bidirectional replication (Touchon et al. 2004; Polak and Arndt 2009). In murine embryonic stem cells, 0.4% of all promoters contain TSSs that are also associated with replication initiation (Sequeira-Mendes et al. 2009), further suggesting a preference for replication initiation events in CpG-rich sequences. These suggestions are consistent with our analyses using high-resolution sequencing, as we have detected enrichment of replication initiation events immediately downstream from (but not including) the TSS, and this enrichment was particularly associated with CpG methylated genomic regions. Our combined results are consistent with the hypothesis that promoters and replicator sequences have coevolved to ensure the coordination between replication and transcription (Delgado et al. 1998; Antequera and Bird 1999; Sequeira-Mendes et al. 2009), and suggest that frequent transcription initiation events prevent the assembly of pre-replication complexes on chromatin. Importantly, methylated CpG tracks, which we found associated with a high frequency of initiation events, typically mark transcriptionally inactive regions of the genome (Weber et al. 2007). As such, these regions may be inclined to nucleate pre-replication and pre-initiation complexes.
While the present study does not address the issue of replication timing, the relationship among replication initiation sites, replication timing, and transcription is interesting. For example, replication timing is influenced by and correlated with changes in chromatin condensation, as DNase I hypersensitivity is an effective marker for early replicating regions (Hansen et al. 2009). In our current study, the location of initiation events was affected by transcription, whereas others have shown that replication timing correlates poorly with transcription (in studies involving loci with mono-allelic expression). In agreement, a high-resolution study of replication timing revealed that replication timing correlates with gene density in both pluripotent and differentiating human embryonic stem cells and that changes in replication timing sometimes, but not always, reflect gene expression (Desprat et al. 2009). Cellular differentiation influences replication timing over large genomic regions (400–800 kb) in human and mouse cells, and chromatin domains that replicate concomitantly are often located in distinct nuclear compartments (Ryba et al. 2010). Combined with our studies, current observations are consistent with the notion that chromatin accessibility and nuclear compartmentalization primarily affect DNA replication timing, whereas local metabolic processes (such as transcription) primarily affect the location of replication initiation events.
Methods
Cell culture
Cells were obtained from the Developmental Therapeutics Program at the National Cancer Institute. For replication analyses, K562 erythroleukemia cells and MCF7 breast cancer cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and antibiotics, as needed. K562 cells (2.22 × 108) were harvested after 48 h of growth, and 9.36 × 108 MCF7 cells were grown until 85% confluent and then harvested for the nascent strand abundance assay, or as described previously (Shankavaram et al. 2007). For gene expression analysis, cell growth and harvesting were performed as described previously (Shankavaram et al. 2007). In brief, cells were thawed and placed in RPMI-1640 (Lonza) with 5% fetal calf serum (Atlantic Biologicals Corporation) and 2 mM glutamine (Invitrogen Corporation). Cells were harvested at ∼80% confluence, as assessed by phase microscopy for attached cells, or at ∼0.5 × 106 cells/mL for suspended cells. Total RNA was extracted using the Qiagen RNeasy Midi kit.
Nascent strand abundance assay
Genomic and nascent strand DNA was isolated from K562 and MCF7 cells as previously described (Wang et al. 2004), with some modifications.
Nascent DNA was loaded onto a neutral 5%–30% sucrose gradient in TNE. Gradients were centrifuged for 20 h at 21,000 rpm in an SW40 rotor at 25°C. Fractions containing 400–600 bp DNA fragments were pooled, dialyzed against TE, and precipitated. The sample was divided into two aliquots, and plasmid DNA was added to one aliquot as a carrier or positive control. The sample was extracted with phenol-chloroform, ethanol precipitated, and resuspended in 22 μL DEPC-treated water. DNA was digested overnight at 37°C in 32 μL lambda exonuclease buffer and 2 μL lambda exonuclease. The sample was heated for 10 min at 95°C, ethanol precipitated, and resuspended in 50 μL sterile water. RT-PCR was carried out on an aliquot of the sample, using primers for known replication initiation sites. Nascent strands were random-primed using the DNA polymerase I Klenow fragment and the DNA Prime Labeling System (Invitrogen). Briefly, nascent strand DNA was incubated for 1 h at 37°C, ethanol precipitated, and resuspended in 30 μL sterile water. Double-stranded nascent DNA (1 μg) was sequenced using the Illumnia (Solexa) genome analyzer II. Sheared genomic DNA was sequenced as a control.
Nascent strands and fragmented genomic control strands were sequenced using the Illumina GA II platform. The resultant sequences were aligned to the hg19 (NCBI Build 37) human reference genome using Bowtie (Langmead et al. 2009). Alignments were converted to .bam and .tdf format using SAMtools (Li et al. 2009) and igvtools for visualization in Broad's Integrative Genomics Viewer (http://www.broadinstitute.org/igv) (Robinson et al. 2011).
Reads per kilobase per million aligned reads (RPKM) values were calculated for each sample using 100-base genomic bins (Mortazavi et al. 2008). A Gaussian smoothing algorithm was applied to the bin values. To correct for sequencing biases (Dohm et al. 2008; Harismendy et al. 2009) and copy number variation, an enrichment ratio was defined as the ratio of nascent strand RPKM to control RPKM and was calculated for each 100-base bin.
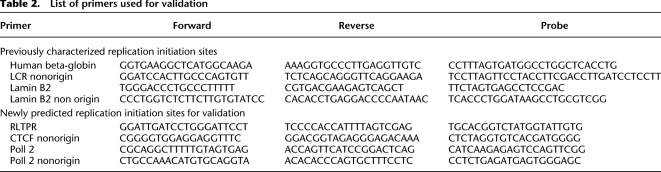
In addition, a .bed format file of the regions with significant replication initiation activity was prepared from the bin-level enrichment ratios. The threshold of enrichment ratios for significant replication initiation activity was selected using a Metropolis Monte Carlo simulation (Metropolis et al. 1953) to find a ratio that identified peaks with a specified target false-discovery rate (FDR) level. The empirical FDR value (Pepke et al. 2009) of a given enrichment ratio threshold was calculated by inverting test and control samples to find the ratio of false (inverted) to true (normal) peaks at the threshold. Initiation at select locations was validated with RT-PCR using primers listed in Table 2.
Table 2.
List of primers used for validation
Transcript expression profiling and data sources
K562 and MCF7 mRNA samples were labeled and processed at Gene Logic using the 54,674 probe-set Human Genome U133 Plus 2.0 Array microarrays (Affymetrix). Each expression profile was done in triplicate. Signal intensities were determined using robust multi-array average (RMA) (Wu et al. 2003), with GC correction (Wu et al. 2003), which is based on perfect match probe intensities that have been background corrected. Intensities were transformed to logarithm base 2. Expression values were GCRMA normalized and log2 transformed. NCBI Gene was used to determine the genomic position of genes, including TSS (NCBI Gene [http://www.ncbi.nlm.nih.gov/gene]). Analysis was restricted to genes with at least one transcript sequence record in RefSeq or GenBank (20,063 genes). GEO record for transcription data is GSE5720.
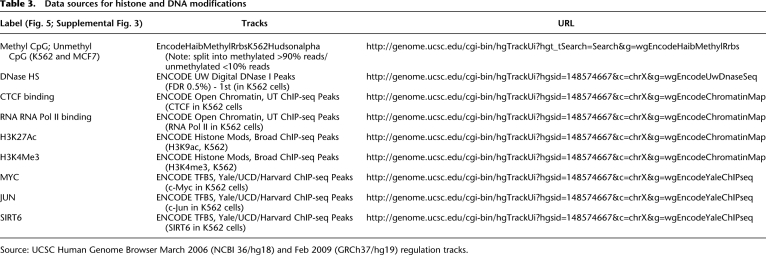
Chromatin modification data were obtained from the UCSC Genome Browser (Rhead et al. 2010). In cases where features were not available in hg19 coordinates, their coordinates were converted to those in hg19 by extracting feature sequences from the original build and realigning them to hg19. Data sources are listed in Table 3. For a more detailed description of the analysis process, please see the Supplemental Data Analysis.
Table 3.
Data sources for histone and DNA modifications
Data access
Replication initiation data reported in this article have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE28911.
Acknowledgments
We thank Bao Tran and Michelle Mehaffey for expert technical assistance in sequencing of nascent strands and Dr. Tobi Guennel for high quality CGH data. This study was supported by the Intramural Research Program of the NIH, Center for Cancer Research, National Cancer Institute; by NIH grant CA138180 to M.S.V.; and by a scholarship from the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.124644.111.
References
- Aladjem MI 2007. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet 8: 588–600 [DOI] [PubMed] [Google Scholar]
- Aladjem MI, Fanning E 2004. The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep 5: 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem MI, Falaschi A, Kowalski D 2006. DNA replication origins. In DNA replication in eukaryotic cells, 2nd ed (ed. DePamphilis ML), pp. 31–61 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Aladjem MI, Groudine M, Brody LL, Dieken ES, Fournier RE, Wahl GM, Epner EM 1995. Participation of the human β-globin locus control region in initiation of DNA replication. Science 270: 815–819 [DOI] [PubMed] [Google Scholar]
- Antequera F 2004. Genomic specification and epigenetic regulation of eukaryotic DNA replication origins. EMBO J 23: 4365–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinsky AK, Gerbi SA 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279: 95–98 [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Hatada S, Brylawski BP, Smithies O, Kaufman DG, Cordeiro-Stone M 2004. Complementation of replication origin function in mouse embryonic stem cells by human DNA sequences. Genomics 84: 475–484 [DOI] [PubMed] [Google Scholar]
- Dai J, Chuang RY, Kelly TJ 2005. DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci 102: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S, Gómez M, Bird A, Antequera F 1998. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J 17: 2426–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprat R, Thierry-Mieg D, Lailler N, Lajugie J, Schildkraut C, Thierry-Mieg J, Bouhassira EE 2009. Predictable dynamic program of timing of DNA replication in human cells. Genome Res 19: 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H 2008. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res 36: e105 doi: 10.1093/nar/gkn425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM 2010. Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Liu G, Randall G, Bevington J, Leffak M 2004. Transcription factor binding and induced transcription alter chromosomal c-myc replicator activity. Mol Cell Biol 24: 10193–10207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM 2010. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M, Brockdorff N 2004. Heterochromatin on the inactive X chromosome delays replication timing without affecting origin usage. Proc Natl Acad Sci 101: 6923–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SJ, Gerhardt J, Doerfler W, Small LE, Fanning E 2007. An origin of DNA replication in the promoter region of the human fragile X mental retardation (FMR1) gene. Mol Cell Biol 27: 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire D, Brodolin K, Mechali M 2006. HoxB domain induction silences DNA replication origins in the locus and specifies a single origin at its boundary. EMBO Rep 7: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin JL, Mesner LD, Lar O, Torres R, Chodaparambil SV, Wang L 2008. A revisionist replicon model for higher eukaryotic genomes. J Cell Biochem 105: 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA 2009. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci 107: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, Schork NJ, Murray SS, Topol EJ, Levy S, et al. 2009. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol 10: R32 doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida T, Oda M, Ohsawa K, Yamaguchi A, Hosozawa T, Locksley RM, Giacca M, Masai H, Miyatake S 2006. Replication initiation from a novel origin identified in the Th2 cytokine cluster locus requires a distant conserved noncoding sequence. J Immunol 176: 5446–5454 [DOI] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270: 994–997 [DOI] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G 2009. H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta RF, Li X, Mesner LD, Dijkwel PA, Lin HB, Hamlin JL 1998. Distal sequences, but not ori-β/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol Cell 2: 797–806 [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A 2009. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell 21: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Keshet I, Cedar H 1993. Replication structure of the human β-globin gene domain. Nature 366: 588–590 [DOI] [PubMed] [Google Scholar]
- Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL 2009. Searching for SNPs with cloud computing. Genome Biol 10: R134 doi: 10.1186/gb-2009-10-11-r134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Ezrokhi M, Smith HS, Gerbi SA 2002. Developmental changes in the Sciara II/9A initiation zone for DNA replication. Mol Cell Biol 22: 8426–8437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M 2010. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738 [DOI] [PubMed] [Google Scholar]
- Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E 1953. Equations of state calculations by fast computing machines. J Chem Phys 21: 1087–1092 [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Newlon CS, Theis JF 1993. The structure and function of yeast ARS elements. Curr Opin Genet Dev 3: 752–758 [DOI] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A 2009. Computation for ChIP-seq and RNA-seq studies. Nat Methods 6: S22–S32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Arndt PF 2009. Long-range bidirectional strand asymmetries originate at CpG islands in the human genome. Genome Biol Evol 1: 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL 2001. Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowntree RK, Lee JT 2006. Mapping of DNA replication origins to noncoding genes of the X-inactivation center. Mol Cell Biol 26: 3707–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Dalton S, Gilbert DM 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Shan Y, Mesner LD, Hamlin JL 2004. The promoter of the Chinese hamster ovary dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries. Genes Dev 18: 397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Díaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gómez M 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5: e1000446. doi: 10.1371/journal.pgen.1000446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankavaram UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK, Reimers MA, Scherf U, Kahn A, Dolginow D, et al. 2007. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther 6: 820–832 [DOI] [PubMed] [Google Scholar]
- Theis JF, Newlon CS 2001. Two compound replication origins in Saccharomyces cerevisiae contain redundant origin recognition complex binding sites. Mol Cell Biol 21: 2790–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Arneodo A, d'Aubenton-Carafa Y, Thermes C 2004. Transcription-coupled and splicing-coupled strand asymmetries in eukaryotic genomes. Nucleic Acids Res 32: 4969–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela MS, Chen Y, Davis S, Fang F, Walker RL, Bilke S, Lueders J, Martin MM, Aladjem MI, Massion PP, et al. 2011. Preferential localization of human origins of DNA replication at the 5′ eds of expressed genes and at evolutionary conserved DNA sequences. PLoS ONE 6: e17308 doi: 10.1371/journal.pone.0017308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lin CM, Brooks S, Cimbora D, Groudine M, Aladjem MI 2004. The human β-globin replication initiation region consists of two modular independent replicators. Mol Cell Biol 24: 3373–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Martinez RG, Murillo F, Spencer F 2003. A model based background adjustement for oligonucleotide expression arrays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Zhou J, Ashouian N, Delepine M, Matsuda F, Chevillard C, Riblet R, Schildkraut CL, Birshtein BK 2002. The origin of a developmentally regulated Igh replicon is located near the border of regulatory domains for Igh replication and expression. Proc Natl Acad Sci 99: 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]