Polycomb group proteins are required for maintaining repression of developmental genes, and chromatin compaction is a potential mechanism of such regulation. Here, Kingston and colleagues characterize a highly basic protein domain in mammalian PRC1 that is responsible for compaction. Cross-species analysis of this domain provides further insight into its evolution and function.
Keywords: Polycomb, chromatin, transcriptional repression, intrinsic disorder
Abstract
Polycomb group (PcG) proteins are required for the epigenetic maintenance of developmental genes in a silent state. Proteins in the Polycomb-repressive complex 1 (PRC1) class of the PcG are conserved from flies to humans and inhibit transcription. One hypothesis for PRC1 mechanism is that it compacts chromatin, based in part on electron microscopy experiments demonstrating that Drosophila PRC1 compacts nucleosomal arrays. We show that this function is conserved between Drosophila and mouse PRC1 complexes and requires a region with an overrepresentation of basic amino acids. While the active region is found in the Posterior Sex Combs (PSC) subunit in Drosophila, it is unexpectedly found in a different PRC1 subunit, a Polycomb homolog called M33, in mice. We provide experimental support for the general importance of a charged region by predicting the compacting capability of PcG proteins from species other than Drosophila and mice and by testing several of these proteins using solution assays and microscopy. We infer that the ability of PcG proteins to compact chromatin in vitro can be predicted by the presence of domains of high positive charge and that PRC1 components from a variety of species conserve this highly charged region. This supports the hypothesis that compaction is a key aspect of PcG function.
The coordinated regulation of development requires the faithful maintenance of gene expression programs through multiple cell divisions. Two classes of proteins that maintain the epigenetic inheritance of gene states are the Polycomb group (PcG) and Trithorax group (TrxG) proteins (Kennison 1995; Schuettengruber et al. 2007). These proteins act in complexes that either repress or activate gene transcription, respectively (Lewis 1978; Ingham and Whittle 1980; Kennison and Tamkun 1988; Kennison 1993). PcG protein complexes map to hundreds of genomic loci, the most notable being the Hox clusters (Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006; Schwartz et al. 2006). These proteins form several different complexes, of which the best characterized are Polycomb-repressive complex 1 (PRC1) and PRC2 (Shao et al. 1999; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). In current models of PcG-mediated repression, PRC2 is recruited to target loci, where it methylates Lys 27 of histone H3 (H3K27me3) (Cao and Zhang 2004). This histone modification acts as a binding site for the PRC1 protein Polycomb (PC), although there are indications that other as-yet-uncharacterized mechanisms are also involved in targeting PRC1 action (Muller and Verrijzer 2009; Simon and Kingston 2009; Morey and Helin 2010).
Binding of PRC1 to target loci is believed to be central to the establishment of transcriptional silencing that is stable through cell divisions. The mechanisms via which PRC1 establishes repression are an area of intense study. PRC1 was first defined in Drosophila, where genetic studies initially identified the PcG genes via the phenotype of extra sex combs on the hind legs of male flies (Slifer 1942; Lewis 1947). There are multiple PRC1 family complexes in mammals. Each has some combination of four proteins, encoded by genes homologous to the Drosophila genes Pc, Ph, Psc, and Sce (dRing) (Cao et al. 2005; Schwartz and Pirrotta 2008). Potential mechanisms by which PRC1 family complexes silence genes include ubiquitylating histone H2A to initiate a block to transcription, directly inhibiting the transcriptional machinery, and creating a compacted state in chromatin that is refractory to transcription. Here we explore chromatin compaction by PRC1 and the domain within PRC1 responsible for compaction.
In this study, compaction is defined as the ability to reduce the average distance of nucleosomes from each other as compared with the typical “beads on a string” seen with electron microscopy (EM). In vitro, compaction is thought to be directly related to the ability of PRC1 to inhibit chromatin remodeling, as the ability of PRC1 proteins to perform these two functions is highly correlated. This in vitro work extends to in vivo observations, since the ability of the Drosophila Posterior Sex Combs (PSC) protein to compact nucleosomal arrays correlates with the phenotypes of a set of mutations in PSC (King et al. 2005). In other work, PRC1 is suggested to stabilize nucleosomal turnover rates and create compacted chromatin domains large enough to be detectable by light microscopy in cells (Deal et al. 2010; Eskeland et al. 2010). These studies indicate that compaction is likely to be a biologically relevant mechanism of silencing by PRC1 family complexes.
If compaction is central to PRC1 function, then the ability to compact nucleosomal arrays should be conserved across organisms that contain PRC1. Here we show that the protein that is primarily responsible for compaction in mouse PRC1 is M33 (Cbx2), a homolog of Drosophila PC (Pearce et al. 1992). This was surprising, as PC is not a homolog of PSC or Su(z)2, the proteins responsible for compaction in Drosophila PRC1 (Francis et al. 2004; Lo et al. 2009). We performed a structure/function analysis of M33 and found that it and Drosophila PSC share a region that is highly basic and predicted to have a disordered secondary structure. Using protein charge as a basis, we identified putative PRC1 components in other organisms that are expected to compact nucleosomes and showed that these are functional in both inhibition of remodeling and compaction. These studies define a region in PRC1 proteins that functions similarly to the Drosophila protein PSC. We provide evidence supporting the idea that during evolution this key aspect of PRC1 function diverged onto distinct subunits. That this region appears to be present across evolution is consistent with it playing a key role in PRC1 function.
Results
M33 is a functional homolog of PSC
The Drosophila PcG protein PSC is able to block remodeling and compact nucleosomes in vitro, activities that might directly contribute to PRC1-mediated repression (King et al. 2002; Francis et al. 2004). We hypothesized that if these activities are important to PRC1 function, they would be conserved in mammalian PRC1. To investigate this, we used the mouse PRC1 core complex (mPCC). As in Drosophila, a PRC1 core complex of M33, Ring1A, and Bmi1 retains the majority of activity, so we chose to focus on these three subunits (Francis et al. 2001; Lavigne et al. 2004). We began by expressing mPCC and individual subunits of the core complex (Fig. 1A) to determine which, if any, individual subunits had in vitro activity versus the core.
Figure 1.
We characterized the activity of mPCC subunits and subcomplexes using a solution assay that we previously used to characterize PcG proteins. The restriction enzyme accessibility (REA) assay measures the ability of PcG proteins to antagonize nucleosome remodeling by the ATP-dependent remodeling complex hSWI/SNF (Kwon et al. 1994; Francis et al. 2001). Briefly, we assembled nucleosomes onto a 2.5-kb DNA/chromatin template (G5E4) (Fig. 1B) using salt dialysis (Utley et al. 1998). This DNA fragment contains 10 5S nucleosomal positioning sequences. At the center of this fragment is space for two additional nucleosomes, for a total of 12. With two nucleosomes positioned in the central part of G5E4, one of them occludes a unique HhaI restriction site and prevents cutting. Movement of this nucleosome, which is accomplished efficiently by ATP-dependent nucleosome remodeling, allows HhaI to cut the G5E4 DNA. Preincubation of the arrays with PcG proteins prevents efficient remodeling of nucleosomes by the hSWI/SNF remodeling complex and therefore inhibits cutting by HhaI. By titrating in PRC1 proteins or complexes and measuring the amount of cut and uncut DNA, we are able to quantify their inhibitory activity.
As anticipated from previous studies, when we preincubate nucleosomal arrays with PSC or with a mouse core complex composed of Bmi1, Ring1A, and M33 (mPCC), we see a concentration-dependent inhibition of remodeling (Fig. 1C,D). The inhibition of remodeling activity that we observe with mPCC was lower than what we previously measured, which could reflect differences in the activities of protein preparations (Lavigne et al. 2004).
To determine the subunits of mouse PRC1 responsible for inhibition of remodeling activity, we tested proteins individually using this protocol. Surprisingly, we did not observe any appreciable inhibition of remodeling by Bmi1, the mouse homolog of the biochemically active PSC. Likewise, Ring1A did not exhibit any activity. However, M33, the mouse homolog of Drosophila PC, exhibited activity that was similar in efficiency to the core complex and to PSC (Fig. 1C,D). We conclude that the most active subunit in mouse PCC for inhibition of remodeling is the M33 protein.
Inhibition seems to require the presence of nucleosomes, as preincubation of M33 with naked DNA template prior to adding HhaI results in inhibition of cleavage that is two orders of magnitude less than when the DNA template is assembled into nucleosomes, suggesting that M33 is not directly interfering with the restriction enzyme (Supplemental Fig. S1A,B).
Inhibition of remodeling reactions was performed at ionic concentrations of 60 mM, which is optimal for hSWI/SNF activity (Supplemental Fig. S1C,D). We do not observe significant differences in inhibition of remodeling activity by M33 up to ionic conditions of 175 mM (Supplemental Fig. S1E,F).
These results led us to question the conservation of domain structure between Drosophila PRC1 and mammalian PRC1. The Drosophila PC protein can inhibit remodeling of nucleosomal templates, but is at least fivefold less efficient than PSC, while the mouse homolog of PC (M33) is at least one order of magnitude better at inhibiting remodeling than the mouse homolog of PSC (Bmi1) (Fig. 1C,D) (note that Bmi1 displays a low level of compaction activity when assessed by EM; see below). We previously located the inhibition of remodeling and compaction activities in PSC to its C terminus, a region with no obvious primary sequence homology with any of the PcG proteins in the mammalian complex. We therefore set out to complete a structure/function analysis of M33 to determine what features of M33 were required for activity and how those features compared with PSC.
None of the conserved features of M33 are required for in vitro activity
To identify the domain of M33 required for inhibition of remodeling, we expressed and purified M33 and deletion variants in Escherichia coli. By using a cleavable GST tag on the N terminus and a Flag tag on the C terminus, we were able to obtain M33 and variants that were more homogeneous than the proteins we obtained using the baculovirus system (Supplemental Fig. S2A). We constructed a series of N-terminal and C-terminal truncation mutants, expressed and purified them, and tested their activity in the REA assay (Fig. 2A). In agreement with the above results, full-length M33 purified from E. coli has an inhibitory activity that is similar to M33 purified from Sf9 cells (cf. Figs. 1D and 2B).
Figure 2.
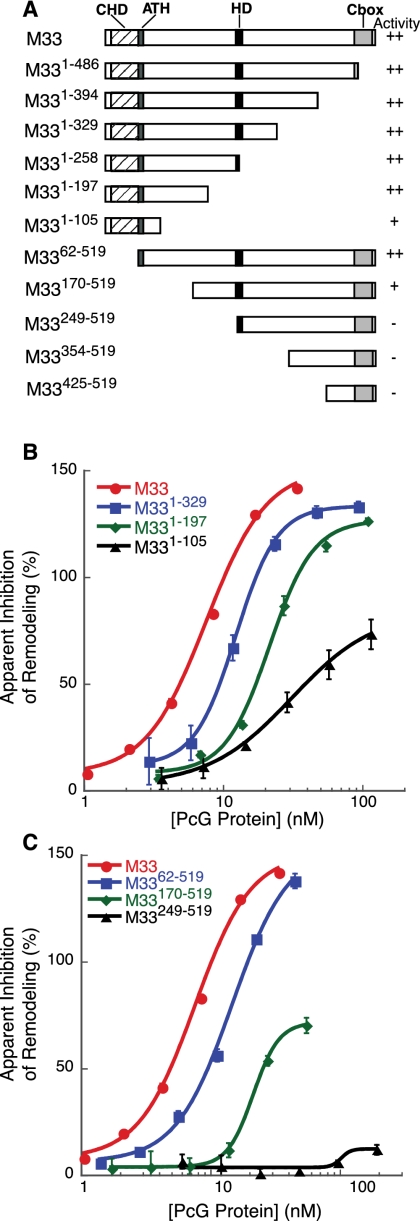
Structure/function analysis of M33-mediated repression. (A) Diagram of M33 truncation mutants tested for biochemical activity. (B) Graph of inhibition activity of selected M33 C-terminal truncation mutants. Data were analyzed as in Figure 1. (C) Graph of inhibition activity of selected M33 N-terminal truncation mutants.
M33 does not share significant sequence alignment with PSC, but contains several domains that are conserved in other chromatin-binding proteins: a chromodomain (CHD), an AT-Hook (ATH), and a PC C-box (CBOX) (Fig. 2A). Another domain, termed here as the homology domain (HD; amino acids 252–266), is a motif that is conserved in M33/Cbx2 homologs but is not found in other Cbx proteins.
We first determined whether any known domains in M33 contributed to its inhibitory activity. Deletion of either the ATH or HD motifs did not result in any reduction in inhibition activity (data not shown). The CHD and CBOX domains interact with nucleosomes, and additionally, the CBOX domain is required for repression activity in Drosophila embryos and in transient transfection assays, as well as for interactions with RING proteins (Muller 1995; Schoorlemmer et al. 1997; Breiling et al. 1999; Satijn and Otte 1999; Fischle et al. 2003; Min et al. 2003; Wang et al. 2008). Surprisingly, deletion of the CHD or CBOX domains did not abolish the inhibition activity (Fig. 2B,C; Supplemental Fig. S2B).
Since none of the conserved features of M33 are required for our in vitro activity, we wondered whether there might be a nonconserved motif that was responsible for the inhibition of remodeling activity. Thus, we decided to create a series of N-terminal and C-terminal deletions.
We cloned and expressed a series of truncation mutants of the M33 protein and tested them in the REA assay for inhibition activity (Fig. 2B,C; Supplemental Fig. S2B). We found that truncations of the C terminus to amino acid 105 (M331–105) still retain repression activity. In contrast, N-terminal deletions to amino acid 248 (M33249–519), completely abolishes activity. Since the M331–197 and M3362–519 constructs retain near wild-type levels of activity, we conclude that an N-terminal region between amino acids 62 and 197 is required for optimal repression activity.
Positively charged residues are required for optimal activity in vitro
We wondered what characteristics are shared between the regions of M33 and PSC that are required for inhibition of remodeling. Examination of the amino acid composition of M33 revealed a bias toward lysines and arginines—the two most positively charged amino acids. The C-terminal region of PSC that is required for in vitro and in vivo activity also has an overrepresentation of arginines and lysines. The predicted overall charge of M33 and PSC is +32.5 and +82.1, respectively. This is in contrast to Bmi1, inactive in the REA assay, which has a predicted overall charge of +10.1.
We wondered whether we could observe any correlation between the predicted overall charge of the M33 variants and inhibition of remodeling activity. Indeed, if we plot the predicted charge of the truncation mutants versus the concentration required for 50% inhibition of remodeling, we obtain a linear relationship with R2 = 0.7 (Fig. 3A). These charged amino acids are spread throughout M33 and PSC, which is reminiscent of another class of repressive proteins: the linker histones (Hansen et al. 2006).
Figure 3.
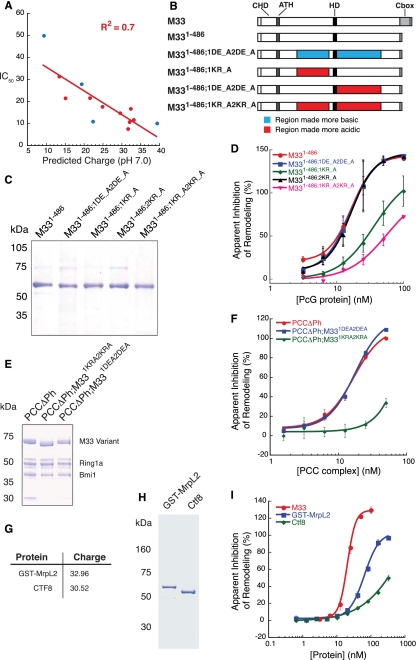
The role of charge in M33-mediated repression activity. (A) Graph of predicted protein charge at pH 7.0 versus IC50 as determined using Kaleidagraph software and performing a linear fit. Red circles represent data from M33 truncation mutants, and blue circles represent data from M33 charge mutants. (B) Schematic representation of the charge mutant proteins that were tested. (C) Coomasie-stained gel of M33 charge mutants expressed and purified from E. coli. (D) Plot of the quantification from the REA done with M33 charge mutants. (E) Coomasie-stained gel of M33 charge mutants in the context of the core PRC1 complex. Proteins were expressed and purified from Sf9 cells. (F) Plot of the quantification from the REA done with the charge mutant complexes. (G) Charge characteristics of the non-PcG basic proteins cloned. The accession numbers for the proteins are MrpL2, NP_079578.1; and CTF8, AAH23107.1. (H) Coomasie-stained gel of the non-PcG basic proteins expressed and purified from E. coli. (I) Plot of the quantification from the REA done with the non-PcG basic proteins.
We hypothesized that these basic residues in M33 are important for binding to chromatin and therefore contribute to PcG-mediated repression. To test this hypothesis, we synthesized mutant variants of the M33 protein that perturb overall protein charge. We systematically mutated every other lysine or arginine in M331–486 to alanine, beginning with K132A and ending with K240A, termed here M331–486;1KR_A, for a total of 13 residues mutated (Fig. 3B; Supplemental Fig. S3B,C). We used the M331–486 construct for this purpose because we found that deletion of the C-terminal 33 amino acids of M33 enhances expression and purification with marginal effect on in vitro activity. Additionally, we generated a construct that mutates every other arginine or lysine from R262A to R415A (M331–486;2KR_A), as well as a combination of the two regions (M331–486;1KR_A2KR_A). These constructs reduce the predicted overall charge of M331–486 at pH 7.0 from +32.5 to +19.3, +22.3, and +9.3, respectively. Finally, we synthesized a construct in which every other aspartic or glutamic acid was mutated to alanine, from E156A to E399A, for a total of seven mutated residues (M331–486;1DE_A2DE_A). This construct is expected to increase the predicted charge to +39.3. If overall positive charge of these domains is important to function, these mutations are expected to reduce and enhance the in vitro activity of M33, respectively.
We expressed and purified these proteins to a level similar to M331–486 (Fig. 3C). When we tested these proteins for inhibition of remodeling activity, we found that M331–486;1DE_A2DE_A has inhibition activity similar to M331–486 (Fig. 3D; Supplemental Fig. S2C). Additionally, M331–486;2KR_A inhibits remodeling to an extent similar to M331–486. However, both M331–486;1KR_A and M331–486;1KR_A2KR_A reduce inhibition activity, approximately threefold in the case of M331–486;1KR_A and fivefold for M331–486;1KR_A2KR_A (data for all of the charge mutants are depicted as blue circles in Fig. 3A). These results are consistent with the idea that the charged residues within the M331–486;1KR_A region are important for optimal repression activity and agree with the deletion analysis above that suggests that the region between amino acids 62 and 197 is required for optimal repression activity. However, since there was no enhancement of activity in the M331–486;1DE_A2DE_A variant and no loss of activity in the M331–486;2KR_A mutant, we conclude that there must be criteria other than simple overall protein charge contributing to inhibition activity of M33.
Since PcG proteins typically function within complexes, we tested whether the results we obtained with M33 alone could be replicated in the context of the core complex. We coexpressed two full-length M33 charge variants with Bmi1 and Ring1A in Sf9 cells and were able to purify both mutant complexes to a level similar to the wild-type complex (Fig. 3E). In agreement with the results obtained with M33 variants alone, PCCΔPh;M331DE_A2DE_A inhibited remodeling to an extent similar to the wild-type core complex, whereas PCCΔPh;M331KR_A2KR_A reduces activity by more than fourfold (Fig. 3F; Supplemental Fig. S2C).
We wondered whether inhibition activity is a general characteristic of any basic protein. We cloned two mouse genes that contain charge characteristics similar to M33 and expressed the proteins MrpL2, a mitochondrial protein with predicted charge of +32.96, and CTF8, a nuclear protein with a predicted charge of +30.52. These non-PcG proteins are sevenfold to 10-fold less active than M33, suggesting that features beyond overall charge are involved in PcG protein activity (Fig. 3G–I; Supplemental Fig. S2E).
To determine whether there are any conserved sequence motifs within the region mutated in the M331–486;1KR_A construct, we performed a sequence alignment with frog, zebrafish, and chicken Cbx2 proteins (Supplemental Fig. S3A). We found that five of the mutated residues are conserved among all four species, raising the possibility that these amino acids are involved in nucleosome interactions. While we cannot rule out that the loss of activity is due to disruption of protein structure, these results are consistent with the idea that the intrinsic charge of PcG proteins is important for repression activity.
Given the high number of hydrophilic amino acids in M33, one possibility is that the charged region is “natively unfolded” or “intrinsically disordered” (Uversky and Dunker 2010). Natively unfolded regions occur in other chromatin architectural proteins, including the PcG proteins RYBP and GAGA factor, and have been proposed to play a role in the function of PSC and Su(z)2 (Agianian et al. 1999; Emmons et al. 2009; Lo et al. 2009; Neira et al. 2009). The linker histone regions required for chromatin compaction are intrinsically disordered in solution. Genome-wide predictions of natively unfolded regions in proteins forecast a high percentage of transcriptional regulators as having some intrinsic disorder (Garza et al. 2009; Sandhu 2009). Consistent with this idea, Metadisorder, a program that uses several different disorder prediction algorithms, predicts that M33 is folded within the CHD and CBOX, while the central portion that contains the 1KR_A region is disordered (Supplemental Fig. S4A; Kurowski and Bujnicki 2003). Additionally, the far-UV circular dichroism (CD) spectra of M331-486 demonstrate that it has the characteristic spectra of a protein that contains intrinsic disorder; specifically, a minima of molar ellipticity at 200 nM (Supplemental Fig. S4B).
Evolution of PcG proteins suggests charge is a predictor for in vitro activity
The above studies collectively suggest that a positively charged domain is responsible for the ability of PRC1 proteins to inhibit nucleosome remodeling. In vitro, M33 appears to inhibit remodeling enzymes in a manner that is dose-responsive to the overall charge of the protein, whereas PSC appears to be more complicated. Two truncation mutants of PSC that disrupt overall charge to variable degrees do not directly correlate with inhibition activity. PSC1–909 contains half as much overall charge as PSC456–1603, yet inhibits remodeling to a similar extent (Lo and Francis 2010).
Since M33 and PSC are not sequence homologs, we hypothesized that these proteins evolved to become functional homologs. This hypothesis predicts that organisms that are more closely related to Drosophila will have PSC homologs that are active. Likewise, it predicts that organisms more related to mice will have M33 homologs that are classified as active. We sought to test these predictions by using a computational approach.
Homologous M33/PC and Bmi1/PSC proteins were identified by searching the UniProt database for conserved CHD (IPR000953) and RING (IPR001841) InterPro signatures (Hunter et al. 2009; The UniProt Consortium 2011). Identified proteins from this search were filtered to PcG-like proteins using additional regions of homology—the CBOX domain for M33/PC and the extended RING HD for Bmi1/PSC—by using hmmsearch (Eddy 2009).
Using PSC and M33 as known active proteins and Bmi1 as a known inactive protein, the PcG proteins were classified as active or nonactive using k-means clustering (Cock et al. 2009). The clustering was based on overall protein charge and regional charge. Overall charge was calculated using Biopython, and regional charge was calculated as the percentage of 75-amino-acid windows with an isoelectric point (pI) >10.2. Proteins of the active class contained a regional charge of at least 10% and an overall charge of at least +15. Additional classification parameters such as protein interaction and domain distribution were considered, but were excluded as noninformative.
The total number of CHD and RING domain proteins identified were 44 and 59, respectively, from 13 species (Supplemental Table S1). Of these proteins, 32 CHD proteins and four RING proteins are predicted to inhibit remodeling. We then used 18S ribosomal RNA (rRNA) sequences from these 13 species to generate a phylogenetic tree to show the evolutionary relationships of these organisms (Fig. 4A). Interestingly, when we did this, we observed an evolutionary point where the predicted activity of RING-containing PcG proteins and CHD-containing proteins diverged. In deuterostomes, the PcG proteins predicted to be active were all CHD-containing. In protostomes, only predicted active RING domain-containing proteins were observed. Notably, despite the expansion of RING proteins in deuterostomes, we could not observe any Bmi1/PSC homologs that the analysis predicted to be active. We used this list to test the accuracy of our predictions.
Figure 4.
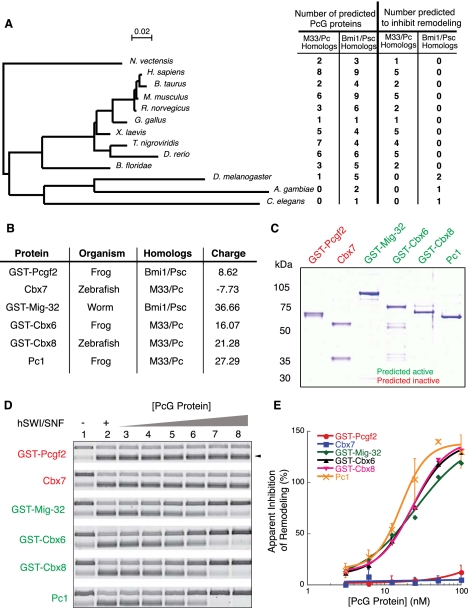
Analysis of evolutionary conservation of PcG function. (A) Phylogenetic tree of species containing RING domain or CHD proteins from UniProtKb protein database. The tree is based on alignments of 18S rRNA from each of the species. Number of predicted PcG proteins represents the number of each class of proteins that was found in the UniProtKb database. Number predicted to inhibit remodeling is the number of proteins from each class that is expected to have inhibition activity based on overall protein charge and regional charge. The bar represents 0.02 substitutions per site. (B) Charge properties of PcG proteins selected for in vitro activity analysis. The accession numbers for the proteins are Pcgf2, NP_001084738.1; Cbx7, NP_001017853.1; Mig-32, NP_502293.2; Cbx6, NP_001088074.1; Cbx8, AAI54356.1; and Pc1, NP_001081900.1. (C) Coomasie-stained gel of PcG proteins expressed and purified from E. coli. (D) Agarose gel of REA assay reaction products. (E) Plot of the quantification of results obtained in D.
To accomplish this, we expressed and purified six evolutionarily divergent CHD- and RING-containing PcG proteins and tested their activity in vitro. These proteins were from the species Xenopus laevis, Danio rerio, and Caenorhabditis elegans (Fig. 4B,C). Overall, we tested one predicted inactive RING protein (frog Pcgf2), one predicted inactive Cbx protein (zebrafish Cbx7), one predicted active RING protein (worm Mig-32), and three predicted active Cbx proteins (zebrafish Cbx8, frog Pc1, and Cbx6) (see Fig. 4B for predicted charge).
Proteins were chosen based on evolutionary divergence from either mice or Drosophila, and the availability of cDNAs. When we tested the activity of these proteins using the REA assay, we found that, as predicted, frog Pcgf2 and zebrafish Cbx7 do not inhibit remodeling activity (Fig. 4D,E). Conversely, preincubation with arrays using worm Mig-32, frog Pc1 or Cbx6, or zebrafish Cbx8 leads to efficient inhibition of remodeling. We did not observe any effect from leaving the GST tag on our proteins or for GST alone (Supplemental Fig. S5). Thus, for these proteins, regional charge was an accurate predictor of in vitro activity.
Compaction of chromatin by divergent PcG proteins
Our results above are consistent with the hypothesis that the domain primarily responsible for inhibition of remodeling in vitro resides on the PSC homolog in flies and worms and resides on the PC homolog in vertebrates. This idea posits that one subunit of PRC1—either an M33/PC homolog or a Bmi1/PSC homolog—will contain a region (or regions) of high positive charge that is important to repression of remodeling and also for compaction. To test whether these proteins are able to compact nucleosomal arrays, we used EM to visualize the interaction of PcG proteins with arrays.
PSC can efficiently compact nucleosome arrays, as observed visually by EM and as quantified by measuring the end-to-end distance of arrays in the EM images (Francis et al. 2004). We used this technique to examine M33 and the evolutionarily conserved PcG proteins to see which of these proteins could compact chromatin in a manner like PSC. EM images of PcG proteins preincubated with G5E4 arrays were acquired, and images of low quality were discarded in a double-blind manner. Nucleosomal arrays alone look like the canonical “beads on a string” conformation (Fig. 5A). Preincubation with Bmi1 tends to reduce the overall array length, but does not appear to promote intranucleosomal interactions. M33, in contrast, promotes the formation of compact particles consisting of multiple nucleosomes.
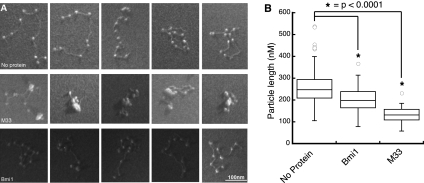
Figure 5.
Compaction of nucleosomal arrays by mouse PcG proteins. (A) Representative EM images of nucleosomal arrays incubated with the indicated PcG protein. (B) Box plot representation of the measured maximal diameter of nucleosomal array particles. Particle length is the diameter of the smallest circle that can entirely surround one nucleosomal array. The box represents the upper and lower quartile, and the line splitting the box represents the mode. The open circles represent outliers, and the asterisks indicate a P-value of <0.0001 using Student's t-test. No protein, n = 72; Bmi1, n = 50; M33, n = 30.
To quantify the ability of these proteins to compact arrays, a single-blind measurement of the end-to-end length of protein/array particles was performed. Full-length M33 creates compacted nucleosome structures as determined by a significant decrease in end-to-end length when compared with arrays incubated with no protein (Students's t-test, P-value < 0.0001) (Fig. 5B). Surprisingly, despite being inactive in the REA, Bmi1 is also able to measurably compact arrays. However, M33 is able to compact chromatin significantly better than Bmi1 (P-value < 0.0001) .
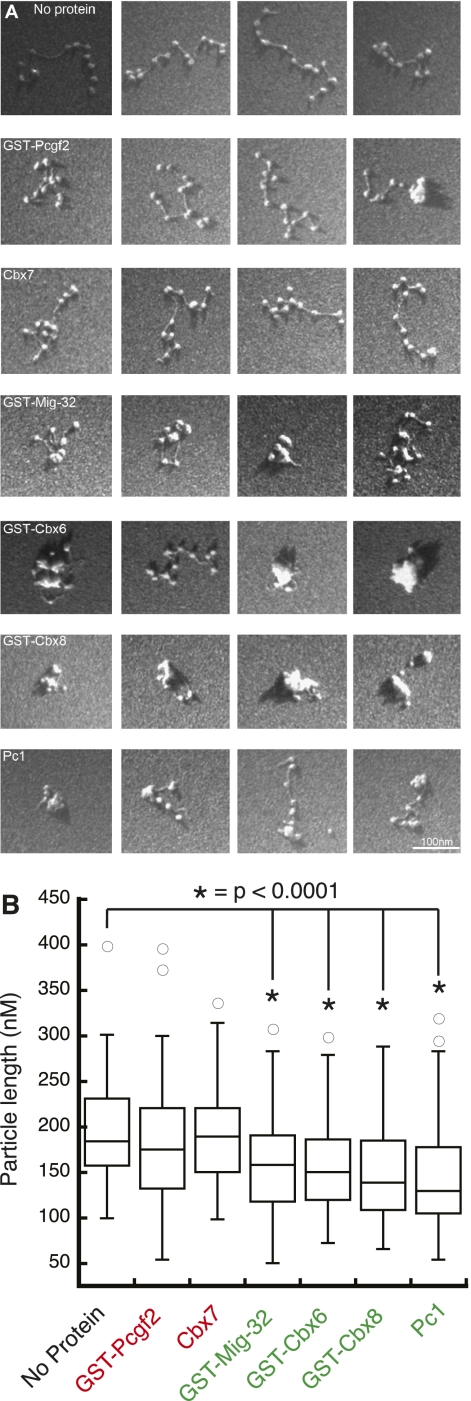
Next we quantified the ability of the PcG proteins from divergent species to compact arrays. In agreement with the previous REA assay, neither GST-Pcgf2 nor Cbx7 (predicted inactive) is able to significantly compact arrays (P-values = 0.1 and 0.8, respectively) (Fig. 6B). These reactions yield arrays that have extended conformations. However, the proteins that inhibit remodeling (GST-Mig32, GST-Cbx6, GST-Cbx8, and Pc1) are able to significantly compact arrays (P-value < 0.0001) (Fig. 6B). As with M33, these proteins promote intranucleosomal interactions.
Figure 6.
Compaction of nucleosomal arrays by diverse PcG proteins. (A) Representative images of nucleosomal particles incubated with various PcG proteins from different species. (B) Box plots of images as described above. No protein, n = 79; GST-Pcgf2, n = 86; Cbx7, n = 113; GST-Mig-32, n = 90; GST-Cbx6, n = 88; GST-Cbx8, n = 87; Pc1, n = 89.
Thus, we predicted PcG protein activity based on regional charge and showed that the predictions appear to hold true. Proteins that we predict to be active both inhibit remodeling and compact nucleosomes in vitro. In contrast, the predicted inactive proteins do not.
Discussion
Here we show that the predicted protein charge of a mouse PcG protein correlates with in vitro activity. We extended this observation by making a computational prediction of PcG activity in a variety of species and demonstrated that we can predict activity based on charge characteristics. These results support the hypothesis that one key function for PRC1 proteins is the ability to compact nucleosomal arrays and repress chromatin remodeling. The conservation of this basic, charged domain suggests that it may be important to silencing by PRC1 family proteins.
Roles of natively unfolded proteins
Natively unfolded or intrinsically disordered proteins were first described in the late 1980s (Sigler 1988). These early descriptions were focused on the proteins that are involved in transcriptional activation. Notably, it was observed that the negatively charged amino acids of proteins required for optimal transcriptional activation did not need to be precisely ordered; Ma and Ptashne (1987) elegantly demonstrated that the critical parameter appeared to be amino acid composition. We found that canonical transcription repressors, the PcG proteins, also appear to have regions of disorder, yet, in contrast to transcriptional activators, contain high concentrations of basic amino acids. It is tempting to speculate that these oppositely charged disordered regions play a “yin-yang” role in transcriptional regulation. It is possible that, in addition to the roles in nucleosome interaction described above, these positively charged transcription repressors could directly interact with and inhibit the negatively charged activation domains of the transcriptional machinery.
There are several proposed reasons why proteins would contain regions of disorder. Disordered regions could potentially adopt different conformations that allow interactions with multiple binding partners. This “hub” function is expected to be beneficial for regulatory proteins; a single protein could potentially regulate many different proteins in a context-specific manner (Gunasekaran et al. 2003). There is also the “fly casting” model, where an extended conformation could allow a protein to “sample” a larger amount of space, forming and breaking low-affinity contacts until conformational change induces tighter binding (Shoemaker et al. 2000). This is expected to promote interactions of low affinity and high specificity. One computational predictor of protein disorder—charge—was found to also be predictive of PcG functional activity, suggesting that charged disordered regions could possibly play a general role in PcG-mediated repression.
Charged domains and PcG function
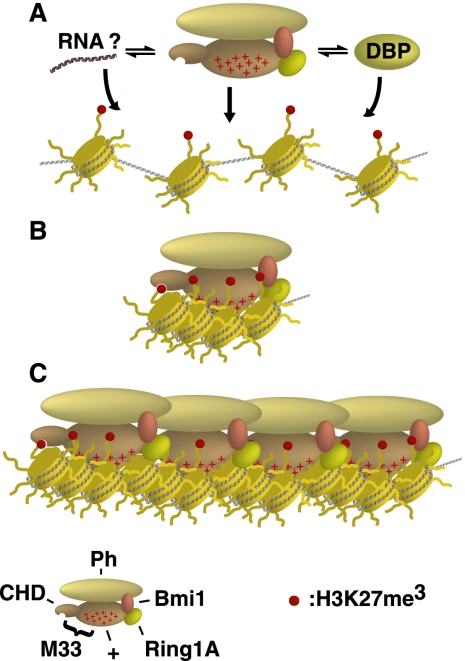
What might be the biological role for PcG charged domains in the repression of transcription? They appear to be predictive for both inhibition of remodeling and compaction of chromatin in vitro. Here we propose a model for how the charged domains of PRC1 function: (1) PRC1 is recruited to target loci and presents the charged domain to linker and/or nucleosomal DNA (Fig. 7A). (2) The charged domain initially interacts with a nucleosome and creates more interactions with other nucleosomes (Fig. 7B). (3) Finally, oligomerization occurs through Ph or other protein–protein interactions to promote spreading or formation of higher-order chromatin fibers (Fig. 7C).
Figure 7.
A model for chromatin compaction by the mPCC. (A) The mPCC is recruited to target loci, potentially through a variety of mechanisms. (DBP) DNA-binding protein. (B) The charged region of M33 (indicated by plus signs) interacts with nucleosomes to compact chromatin. (C) Further protein–protein interactions from other proteins in the core PRC1 complex drive spreading of compacted chromatin.
The CBOX domain of M33 is not required for in vitro repression activities, yet this motif is conserved and required for the repression of template DNA in cell-based assays (Schoorlemmer et al. 1997). This domain is required for interactions with Ring1A/B and Bmi1, which in turn interact with Ph proteins (Alkema et al. 1997; Gunster et al. 1997; Hashimoto et al. 1998; Hemenway et al. 1998; Satijn and Otte 1999). Thus, we imagine that an initially transient nucleosome–nucleosome interaction mediated by charged domains facilitates the further stabilization of a repressed chromatin structure that is mediated by other PRC1 proteins. Studies analyzing the dynamics of Cbx/chromatin interactions in culture cell models observe both transiently and stably associated Cbx proteins, consistent with an initial unstable interaction followed by step(s) that promote stable associations (Ren et al. 2008).
PcG protein evolution
What might be the explanation for how the charged domain evolved to reside on an M33/PC homolog in mammals as opposed to a Bmi1/PSC homolog in flies? We can imagine at least four possible explanations: (1) An early common ancestor had both an M33/PC and a Bmi1/PSC homolog that each contained a charged region; sometime during evolution, the characteristic charge was lost from one or the other homolog. (2) The charged region initially resided on either an M33/PC homolog or a Bmi1/PSC homolog in the common ancestor, but was lost during evolution and subsequently gained on the other PcG homolog. (3) The charged region initially resided on either an M33/PC homolog or a Bmi1/PSC homolog in the common ancestor and was gained on the other before being lost. (4) A convergent evolutionary event occurred: Neither the M33/PC nor the Bmi1/PSC homolog in the common ancestor had a charged domain, but during evolution, one homolog or the other evolved it.
Resolution of the possibilities mentioned above will require a more extensive examination of potential PcG members across evolution than has been performed here. Our phylogenetic tree was not a comprehensive list of all deuterostomes and protostomes, so it is possible that there may be unexpected active homologs in other species. Additionally, protein sequences may have been incorrectly annotated in the database that we queried or missing from organisms with incomplete genomes. One unclear example involves Su(z)2, a homolog of PSC that can inhibit remodeling in vitro. Su(z)2 was found in both the predicted active and inactive lists, although the isoforms in the inactive list appear to be short fragments of the full-length protein that may not exist in vivo. Regardless of a full examination of this issue, the data reported here are consistent with the hypothesis that there was evolutionary pressure to maintain a highly charged domain in a component of PRC1. One possibility is that across multiple species, the core of PRC1 will contain several conserved domains/motifs—two RING fingers, a CHD, a SAM domain, and a disordered/charged domain—defining the basic functional unit of PRC1.
Molecular nature of PcG/chromatin interactions
We do not understand the precise molecular mechanisms behind PcG protein interactions with chromatin. The flexible charged domains might interact with linker DNA, nucleosomal DNA, the histones themselves, or a combination of these chromatin components. We found that two non-PcG proteins with predicted charges similar to M33 do not inhibit remodeling as well as M33. This suggests a mechanism that does not rely solely on the amount of positive charge. It is possible that function involves a specific spacing of the charged residues and/or juxtaposition of the charged surface with other functional domains. For example, the majority of the active proteins that we characterized also contain a CHD, a known histone-binding domain, opening up the possibility that both DNA and histone contacts are required for optimal PcG-repressive activities.
Previous studies have shown that intrinsically disordered regions of proteins can become folded upon interacting with their substrate. This gives us hope that eventually the molecular mechanisms of chromatin condensation by PcG proteins can be unraveled using structural approaches.
Materials and methods
Protein expression
Baculovirus expression was done essentially as described (Francis et al. 2001). Briefly, Sf9 cells were grown in Hyclone CCMIII medium at 27°C with shaking. One liter of cells was either infected with individual viruses for the expression of single subunits or coinfected with multiple viruses for the expression of protein complexes. After 40 h, cells were harvested by centrifugation and washed in PBS prior to making nuclear extracts (NEs) as described (Sif et al. 1998). For the purification of single PcG subunits, NEs were bound to M2 resin (Sigma, catalog no. A2220) for 4 h, then washed extensively with BC buffer (20 mM HEPES at pH 7.9, 0.2 mM EDTA, 20% glycerol, 1 mM DTT, 10 mM PMSF, protease inhibitor cocktail [Roche complete protease inhibitor tablets]) containing 500 mM KCl. The M2 beads were then washed with BC buffer containing higher concentrations of KCl, up to 2 M, before eluting in BC buffer containing 500 mM KCl and 0.4 mg/mL Flag peptide. Purification of PcG complexes was identical, except that the bound protein was washed with BC buffer containing 300 mM KCl.
For expression of GST fusion proteins in E. coli, Rosetta pLysS cells were transformed with pGEX6P1 containing the cDNA of interest. A single colony was used to inoculate 5 mL of LB containing 25 μg/mL chloramphenicol and 50 μg/mL ampicillin and grown overnight. The following morning, 250 μL of the overnight culture was used to inoculate 250 mL of the autoinduction medium ZYP-5052 [12 g/L Bacto tryptone, 24 g/L Bacto yeast extract, 25 mM (NH4)2SO4, 50 mM KH2PO4, 50 mM Na2HPO4, 0.05% glucose, 0.2% α-lactose, 0.5% glycerol, 1 mM MgSO4, 25 μg/mL chloramphenicol, 50 μg/mL ampicillin] (Studier 2005). Cultures were shaken for 5 h at 37°C, then grown overnight at 18°C. The cultures were collected by centrifugation at 4000 rpm in a Beckman J6 MI for 20 min. Cell pellets were resuspended in 40 mL of lysis buffer (50 mM HEPES at pH 7.5, 0.5 mM EDTA, 1.6 M KCl, 20% glycerol, 0.5 mM MgCl2, 0.05% NP40, 1 mg/mL lysozyme, 1 mM DTT, protease inhibitors). The cells were taken through three freeze–thaw cycles, then sonicated to shear DNA before centrifugation at 25,000g for 20 min to remove debris. Five percent polyethelenimine (PEI) in 20 mM HEPES (pH 7.5) was added dropwise to the supernatant while stirring to a final concentration of 0.15%, and stirred an additional 30 min. This step was omitted for GST-MrpL2. The precipitated nucleic acid was removed by centrifugation at 25,000g for 20 min. Extracts were bound to glutathione sepharose beads for 2 h before washing with BC buffer containing increasing salt as described above for M2 purifications. Proteins were either cleaved or eluted off of the resin. For cleavage of proteins, the resin was incubated overnight with 20 U of HRV 3C protease in 5 mL of BC buffer with 500 mM KCl. Proteins were eluted from glutathione sepharose by incubating in BC buffer containing 500 mM KCl and 40 mM reduced glutathione. Eluted proteins were purified over M2 resin as described above. Purified proteins were quantified using the Bradford assay, and then normalized relative to each other by the intensity of protein bands on an SDS-PAGE gel analyzed using ImageJ software.
REA assays
Nucleosome arrays were assembled using HeLa histones as previously described, except that Cy5-labeled G5E4 was used (Sif et al. 2001). Human SWI/SNF was purified from HeLa NEs as described (Sif et al. 1998). Reactions were carried out in a 20-μL volume containing 12 mM HEPES (pH 7.9), 12% glycerol, 60 mM KCl, 0.12 mM EDTA, 0.12 μg/μL BSA, 2 mM ATP, 1.25 mM MgCl2, and 2 nM assembled nucleosomes. Dilutions of PcG proteins were incubated with the arrays for 30 min at 30°C prior to the addition of 100 ng of SWI/SNF and 8 U of HhaI. Reactions were incubated for 1 h at 30°C before the addition of 10 μL of stop buffer (1.5 mg/mL Proteinase K, 70 mM EDTA, 10 mM Tris at pH 7.7, 1% SDS, 0.1% orange G). Reactions were incubated for 30 min at 55°C, then separated on a 1% agarose gel in 1× TAE buffer. DNA was visualized on a Typhoon scanner and quantified using ImageQuant software. Apparent inhibition of remodeling was determined by the equation
and plotted using Kaleidagraph. Curves were generated using an equation for a sigmoidal fit.
Computational classification of PcG proteins
The UniprotKb database was queried for proteins containing CHDs (IPR000953) or RING domains (IPR001841). This list was filtered for PcG proteins using HMMER by keeping only those proteins containing a CBOX for CHD proteins or extended homology for RING proteins. The predicted charge and regional charge of known active and inactive PcG proteins (regional charge is defined as the percentage of sliding windows of 75 amino acids where the average pI is 10.2 or above) was used to cluster the unknown PcG proteins into either active or inactive classes using the k-means method. Proteins of the active class contained at least 10% regional charge and overall charge of 15 or greater. The reproducible Python scripts for this analysis are available (https://github.com/chapmanb/mgh_projects/tree/master/dg_PSC).
The phylogenetic tree was generated by using ClustalW to create an alignment of 18S rRNAs, and then using the maximum likelihood method. The tree was drawn using NJplot.
EM
EM experiments were carried out essentially as described previously (Francis et al. 2004), with the following minor modifications. Binding reactions were carried out in either 30 or 60 mM KCl, and NP40 added to a final concentration of 0.001%. All reactions were set up at molar ratios of eight PcG proteins to one nucleosome, based on total protein concentration. This ratio was selected based on electrophoretic mobility shift assays carried out under the same conditions as EM, which demonstrated binding of M33 to nucleosomal arrays with minimal aggregation at this ratio.
Acknowledgments
We thank Sarah K. Bowman, José M. Antao, Karim Bouazoune, and Leslie Beh for critically reading the manuscript and thoughtful discussions, and Anders Näär and C. Ting Wu for critical discussions and experimental suggestions. This work was supported by grants from the NIH (GM043901 to R.E.K., and GM078456-01 to N.J.F.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17288211.
References
- Agianian B, Leonard K, Bonte E, Van der Zandt H, Becker PB, Tucker PA 1999. The glutamine-rich domain of the Drosophila GAGA factor is necessary for amyloid fibre formation in vitro, but not for chromatin remodelling. J Mol Biol 285: 527–544 [DOI] [PubMed] [Google Scholar]
- Alkema MJ, Bronk M, Verhoeven E, Otte A, van 't Veer LJ, Berns A, van Lohuizen M 1997. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev 11: 226–240 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, Bonte E, Ferrari S, Becker PB, Paro R 1999. The Drosophila polycomb protein interacts with nucleosomal core particles in vitro via its repression domain. Mol Cell Biol 19: 8451–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14: 155–164 [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada YI, Zhang Y 2005. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 20: 845–854 [DOI] [PubMed] [Google Scholar]
- Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25: 1422–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S 2010. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328: 1161–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform 23: 205–211 [PubMed] [Google Scholar]
- Emmons RB, Genetti H, Filandrinos S, Lokere J, Wu CT 2009. Molecular genetic analysis of Suppressor 2 of zeste identifies key functional domains. Genetics 182: 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al. 2010. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38: 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE 2001. Reconstitution of a functional core polycomb repressive complex. Mol Cell 8: 545–556 [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL 2004. Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Garza AS, Ahmad N, Kumar R 2009. Role of intrinsically disordered protein regions/domains in transcriptional regulation. Life Sci 84: 189–193 [DOI] [PubMed] [Google Scholar]
- Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R 2003. Extended disordered proteins: targeting function with less scaffold. Trends Biochem Sci 28: 81–85 [DOI] [PubMed] [Google Scholar]
- Gunster MJ, Satijn DP, Hamer KM, den Blaauwen JL, de Bruijn D, Alkema MJ, van Lohuizen M, van Driel R, Otte AP 1997. Identification and characterization of interactions between the vertebrate polycomb-group protein BMI1 and human homologs of polyhomeotic. Mol Cell Biol 17: 2326–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JC, Lu X, Ross ED, Woody RW 2006. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem 281: 1853–1856 [DOI] [PubMed] [Google Scholar]
- Hashimoto N, Brock HW, Nomura M, Kyba M, Hodgson J, Fujita Y, Takihara Y, Shimada K, Higashinakagawa T 1998. RAE28, BMI1, and M33 are members of heterogeneous multimeric mammalian Polycomb group complexes. Biochem Biophys Res Commun 245: 356–365 [DOI] [PubMed] [Google Scholar]
- Hemenway CS, Halligan BW, Levy LS 1998. The Bmi-1 oncoprotein interacts with dinG and MPh2: the role of RING finger domains. Oncogene 16: 2541–2547 [DOI] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. 2009. InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–D215 doi: 10.1093/nar/gkn785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Whittle JRS 1980. Trithorax: a new homoeotic mutation of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Mol Gen Genet 179: 607–614 [Google Scholar]
- Kennison JA 1993. Transcriptional activation of Drosophila homeotic genes from distant regulatory elements. Trends Genet 9: 75–79 [DOI] [PubMed] [Google Scholar]
- Kennison JA 1995. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet 29: 289–303 [DOI] [PubMed] [Google Scholar]
- Kennison JA, Tamkun JW 1988. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci 85: 8136–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IF, Francis NJ, Kingston RE 2002. Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol Cell Biol 22: 7919–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IF, Emmons RB, Francis NJ, Wild B, Muller J, Kingston RE, Wu CT 2005. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol Cell Biol 25: 6578–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski MA, Bujnicki JM 2003. GeneSilico protein structure prediction meta-server. Nucleic Acids Res 31: 3305–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR 1994. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370: 477–481 [DOI] [PubMed] [Google Scholar]
- Lavigne M, Francis NJ, King IF, Kingston RE 2004. Propagation of silencing; recruitment and repression of naive chromatin in trans by polycomb repressed chromatin. Mol Cell 13: 415–425 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB 1947. New mutants: report of P. Lewis. Drosoph Inf Serv 21: 69 [Google Scholar]
- Lewis EB 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- Lo SM, Francis NJ 2010. Inhibition of chromatin remodeling by polycomb group protein posterior sex combs is mechanistically distinct from nucleosome binding. Biochemistry 49: 9438–9448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SM, Ahuja NK, Francis NJ 2009. Polycomb group protein Suppressor 2 of zeste is a functional homolog of Posterior Sex Combs. Mol Cell Biol 29: 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ptashne M 1987. A new class of yeast transcriptional activators. Cell 51: 113–119 [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Helin K 2010. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 35: 323–332 [DOI] [PubMed] [Google Scholar]
- Muller J 1995. Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J 14: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Verrijzer P 2009. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev 19: 150–158 [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Neira JL, Roman-Trufero M, Contreras LM, Prieto J, Singh G, Barrera FN, Renart ML, Vidal M 2009. The transcriptional repressor RYBP is a natively unfolded protein which folds upon binding to DNA. Biochemistry 48: 1348–1360 [DOI] [PubMed] [Google Scholar]
- Pearce JJ, Singh PB, Gaunt SJ 1992. The mouse has a Polycomb-like chromobox gene. Development 114: 921–929 [DOI] [PubMed] [Google Scholar]
- Ren X, Vincenz C, Kerppola TK 2008. Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol Cell Biol 28: 2884–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS 2009. Intrinsic disorder explains diverse nuclear roles of chromatin remodeling proteins. J Mol Recognit 22: 1–8 [DOI] [PubMed] [Google Scholar]
- Satijn DP, Otte AP 1999. RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol Cell Biol 19: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoorlemmer J, Marcos-Gutierrez C, Were F, Martinez R, Garcia E, Satijn DP, Otte AP, Vidal M 1997. Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J 16: 5930–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G 2007. Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V 2008. Polycomb complexes and epigenetic states. Curr Opin Cell Biol 20: 266–273 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46 [DOI] [PubMed] [Google Scholar]
- Shoemaker BA, Portman JJ, Wolynes PG 2000. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci 97: 8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sif S, Stukenberg PT, Kirschner MW, Kingston RE 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev 12: 2842–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev 15: 603–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler PB 1988. Transcriptional activation. Acid blobs and negative noodles. Nature 333: 210–212 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Slifer EH 1942. A mutant stock of Drosophila with extra sex-combs. J Exp Zool 90: 31–40 [Google Scholar]
- Studier FW 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41: 207–234 [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium 2011. Ongoing and future developments at the universal protein resource. Nucleic Acids Res 39: D214–D219 doi: 10.1093/nar/gkq1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley RT, Ikeda K, Grant PA, Cote J, Steger DJ, Eberharter A, John S, Workman JL 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394: 498–502 [DOI] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK 2010. Understanding protein non-folding. Biochim Biophys Acta 1804: 1231–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Ilangovan U, Robinson AK, Schirf V, Schwarz PM, Lafer EM, Demeler B, Hinck AP, Kim CA 2008. Structural transitions of the RING1B C-terminal region upon binding the polycomb cbox domain. Biochemistry 47: 8007–8015 [DOI] [PMC free article] [PubMed] [Google Scholar]