Abstract
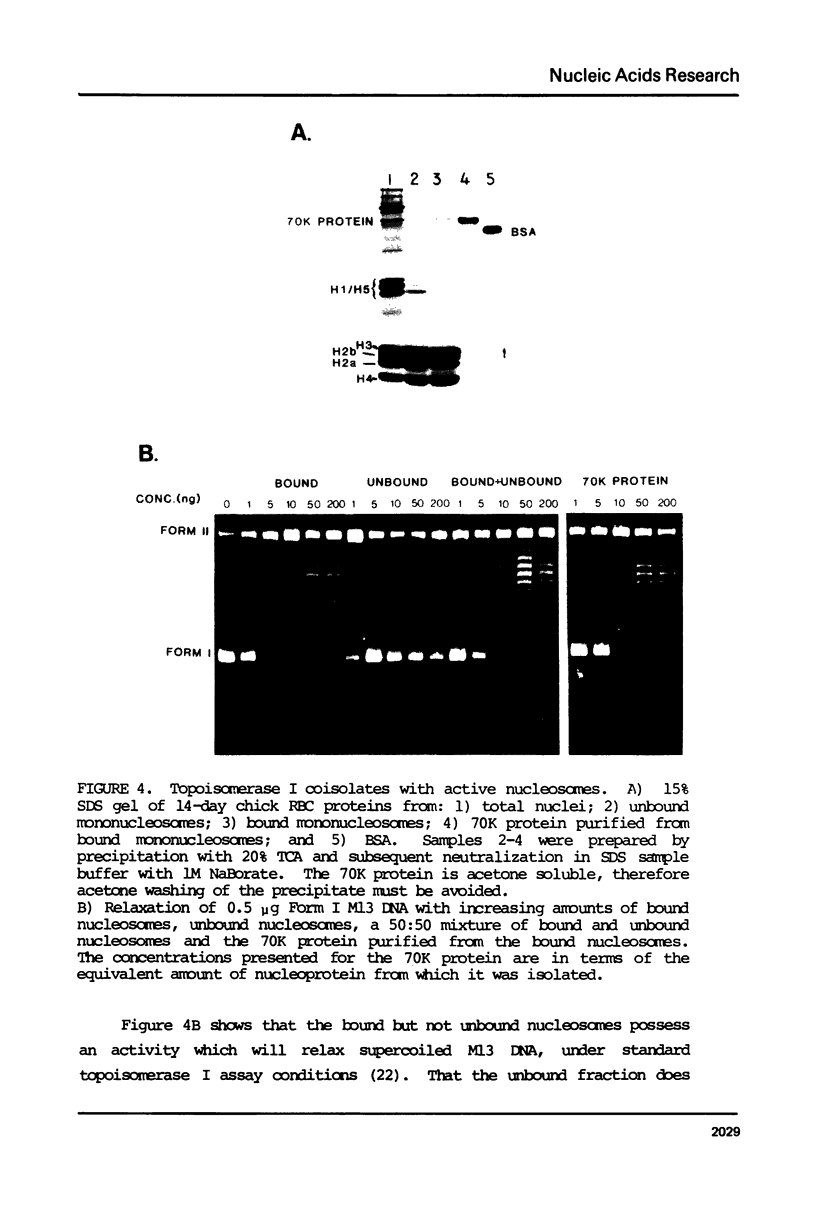
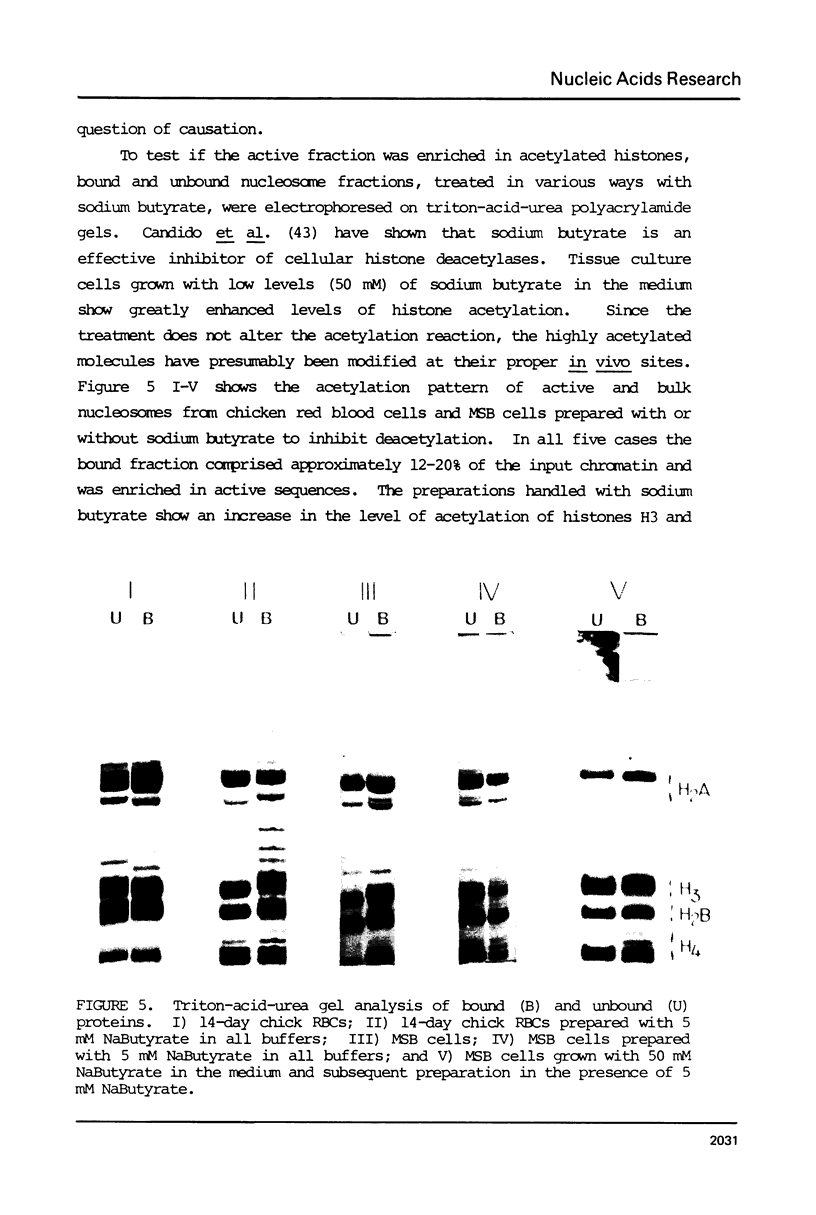
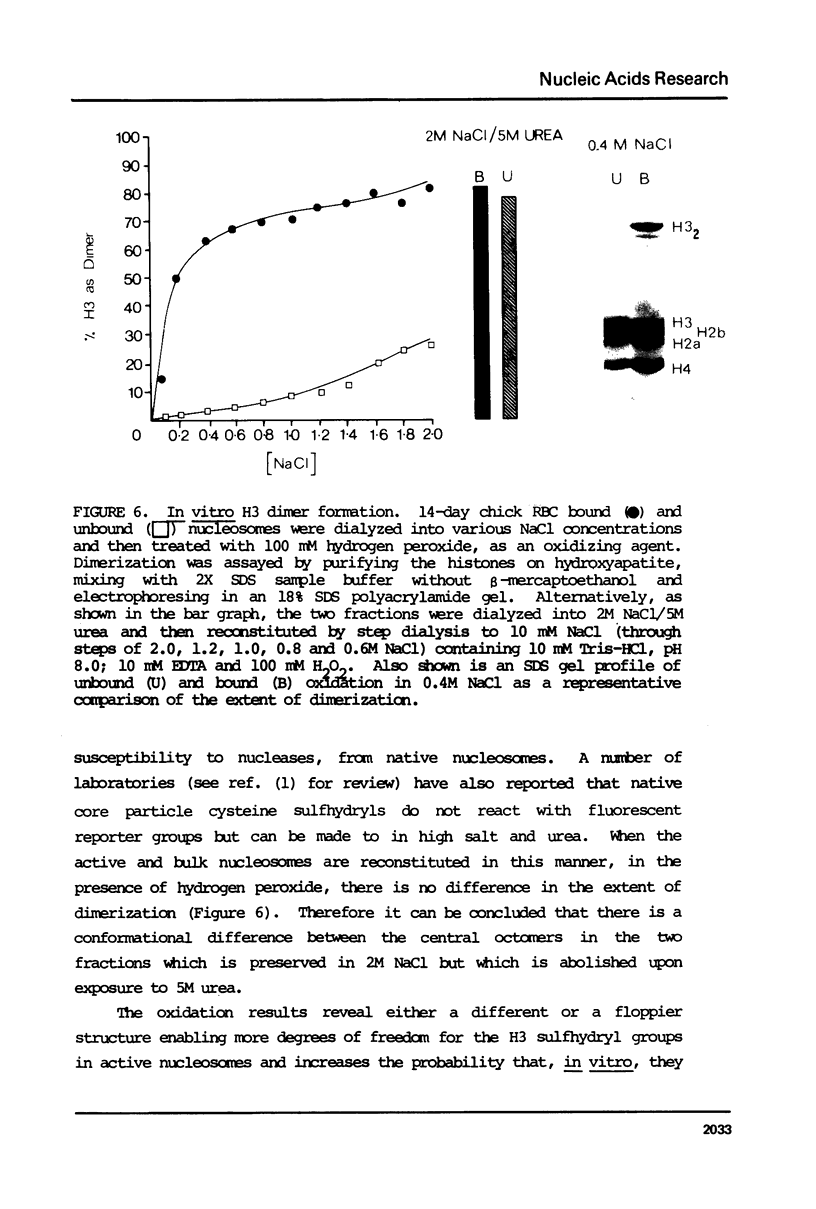
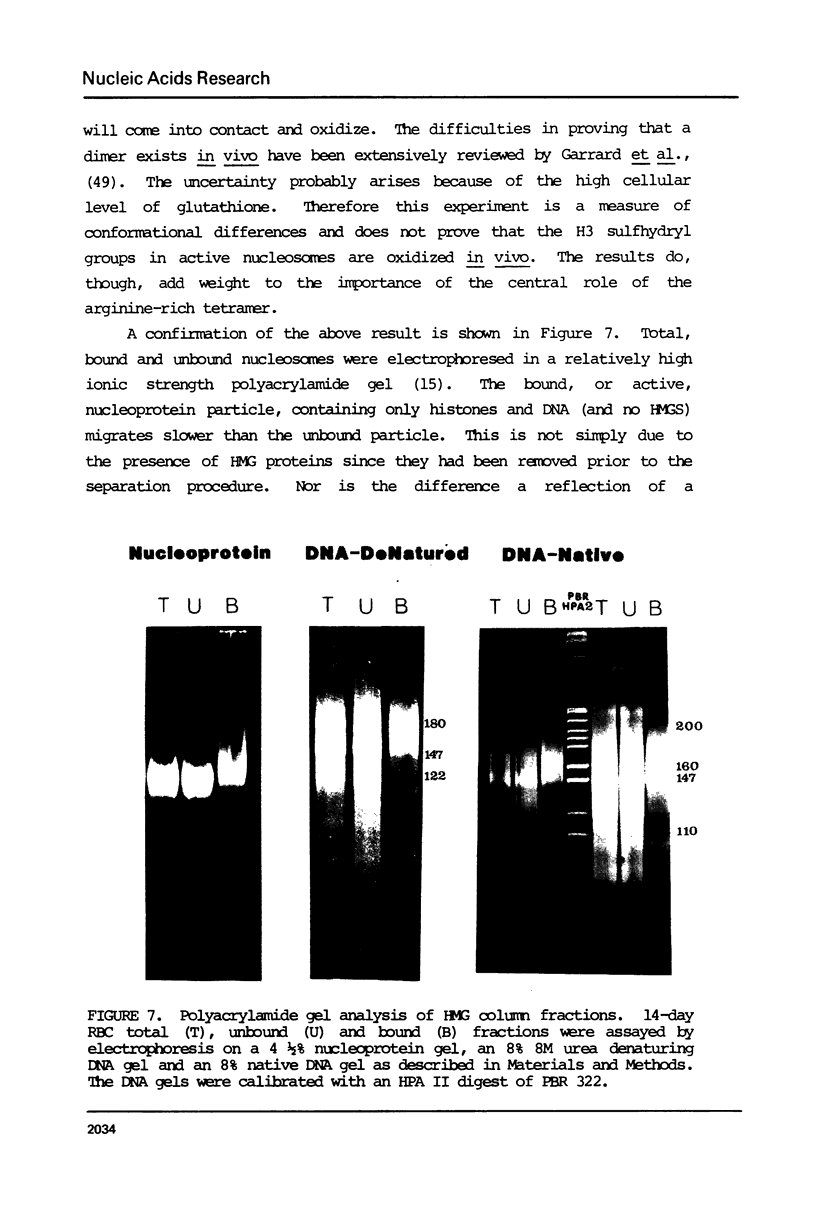
Nucleosomes from actively transcribed genes (active nucleosomes) contain nonhistone proteins HMG 14 and 17 and are preferentially sensitive to digestion by DNAse I. Active nucleosomes isolated by chromatography on an HMG 14 and 17 glass bead affinity column were analyzed with respect to overall structure, accessory nonhistone components and modifications to the DNA and histones. The experiments lead to the following conclusions: the DNA in the active nucleosome is undermethylated compared to bulk DNA; topoisomerase I is a non-stoichiometric component of the active nucleosome fraction; the level of histone acetylation is enriched in active nucleosomes, but the extent of enrichment cannot account for HMG binding; and the two histone H3 molecules in the active nucleosome can dimerize more readily and are, therefore, probably closer together than those in the bulk of the nucleosomes. Additionally it is shown that HMG 14 and 17 prefer to bind to single- vs. double-stranded nucleic acids. The role of HMG 14 and 17 in producing a highly DNAse I sensitive structure and correspondingly helping to facilitate transcription is discussed in terms of these properties.
Full text
PDF



















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albanese I., Weintraub H. Electrophoretic separation of a class of nucleosomes enriched in HMG 14 and 17 and actively transcribed globin genes. Nucleic Acids Res. 1980 Jun 25;8(12):2787–2805. doi: 10.1093/nar/8.12.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albright S. C., Wiseman J. M., Lange R. A., Garrard W. T. Subunit structures of different electrophoretic forms of nucleosomes. J Biol Chem. 1980 Apr 25;255(8):3673–3684. [PubMed] [Google Scholar]
- Alfageme C. R., Zweidler A., Mahowald A., Cohen L. H. Histones of Drosophila embryos. Electrophoretic isolation and structural studies. J Biol Chem. 1974 Jun 25;249(12):3729–3736. [PubMed] [Google Scholar]
- Bakayev V. V., Bakayeva T. G., Varshavsky A. J. Nucleosomes and subnucleosomes: heterogeneity and composition. Cell. 1977 Jul;11(3):619–629. doi: 10.1016/0092-8674(77)90079-4. [DOI] [PubMed] [Google Scholar]
- Bethell G. S., Ayers J. S., Hancock W. S., Hearn M. T. A novel method of activation of cross-linked agaroses with 1,1'-carbonyldiimidazole which gives a matrix for affinity chromatography devoid of additional charged groups. J Biol Chem. 1979 Apr 25;254(8):2572–2574. [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. Supercoiling energy and nucleosome formation: the role of the arginine-rich histone kernel. Nucleic Acids Res. 1977;4(5):1159–1181. doi: 10.1093/nar/4.5.1159-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido E. P., Reeves R., Davie J. R. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978 May;14(1):105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- Diaz M. O., Barsacchi-Pilone G., Mahon K. A., Gall J. G. Transcripts from both strands of a satellite DNA occur on lampbrush chromosome loops of the newt Notophthalmus. Cell. 1981 Jun;24(3):649–659. doi: 10.1016/0092-8674(81)90091-x. [DOI] [PubMed] [Google Scholar]
- Dunn K., Griffith J. D. The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res. 1980 Feb 11;8(3):555–566. doi: 10.1093/nar/8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Goodwin G. H., Mathew C. G., Wright C. A., Venkov C. D., Johns E. W. Analysis of the high mobility group proteins associated with salt-soluble nucleosomes. Nucleic Acids Res. 1979 Dec 11;7(7):1815–1835. doi: 10.1093/nar/7.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M. Methods for fractionation of chromatin into transcriptionally active and inactive segments. Methods Cell Biol. 1977;16:421–436. doi: 10.1016/s0091-679x(08)60117-x. [DOI] [PubMed] [Google Scholar]
- Isackson P. J., Reeck G. R. Nonhistone chromatin proteins HMG-14 and HMG-17 bind preferentially to single-stranded DNA. Nucleic Acids Res. 1981 Aug 11;9(15):3779–3791. doi: 10.1093/nar/9.15.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Klug A., Rhodes D., Smith J., Finch J. T., Thomas J. O. A low resolution structure for the histone core of the nucleosome. Nature. 1980 Oct 9;287(5782):509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- Kuo M. T., Mandel J. L., Chambon P. DNA methylation: correlation with DNase I sensitivity of chicken ovalbumin and conalbumin chromatin. Nucleic Acids Res. 1979 Dec 20;7(8):2105–2113. doi: 10.1093/nar/7.8.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffak I. M., Grainger R., Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977 Nov;12(3):837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- Levinger L., Barsoum J., Varshavsky A. Two-dimensional hybridization mapping of nucleosomes. comparison of DNA and protein patterns. J Mol Biol. 1981 Mar 5;146(3):287–304. doi: 10.1016/0022-2836(81)90389-2. [DOI] [PubMed] [Google Scholar]
- Levy-Wilson B., Watson D. C., Dixon G. H. Multiacetylated forms of H4 are found in a putative transcriptionally competent chromatin fraction from trout testis. Nucleic Acids Res. 1979 Jan;6(1):259–274. doi: 10.1093/nar/6.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. DNA methylation and control of gene expression. Nature. 1981 Apr 2;290(5805):363–364. doi: 10.1038/290363b0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Miller K. G. Eukaryotic DNA topoisomerases: two forms of type I DNA topoisomerases from HeLa cell nuclei. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3487–3491. doi: 10.1073/pnas.78.6.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Bustin M., Miller O. L., Jr Electron microscopic analysis of chromosome metabolism in the Drosophila melanogaster embryo. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):741–754. doi: 10.1101/sqb.1978.042.01.075. [DOI] [PubMed] [Google Scholar]
- Naveh-Many T., Cedar H. Active gene sequences are undermethylated. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4246–4250. doi: 10.1073/pnas.78.7.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ord M. G., Stocken L. A. Metabolic properties of histones from rat liver and thymus gland. Biochem J. 1966 Mar;98(3):888–897. doi: 10.1042/bj0980888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palter K. B., Foe V. E., Alberts B. M. Evidence for the formation of nucleosome-like histone complexes on single-stranded DNA. Cell. 1979 Oct;18(2):451–467. doi: 10.1016/0092-8674(79)90064-3. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Pogo B. G., Allfrey V. G., Mirsky A. E. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc Natl Acad Sci U S A. 1966 Apr;55(4):805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior C. P., Cantor C. R., Johnson E. M., Allfrey V. G. Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: a system for studying changes in nucleosomes assembled in vivo. Cell. 1980 Jul;20(3):597–608. doi: 10.1016/0092-8674(80)90306-2. [DOI] [PubMed] [Google Scholar]
- Quint A., Cedar H. In vitro methylation of DNA with Hpa II methylase. Nucleic Acids Res. 1981 Feb 11;9(3):633–646. doi: 10.1093/nar/9.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman M. M., Levine A. J., Weintraub H. The asymmetric segregation of parental nucleosomes during chrosome replication. Cell. 1979 Oct;18(2):439–449. doi: 10.1016/0092-8674(79)90063-1. [DOI] [PubMed] [Google Scholar]
- Shick V. V., Belyavsky A. V., Bavykin S. G., Mirzabekov A. D. Primary organization of the nucleosome core particles. Sequential arrangement of histones along DNA. J Mol Biol. 1980 May 25;139(3):491–517. doi: 10.1016/0022-2836(80)90143-6. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Stalder J., Groudine M., Dodgson J. B., Engel J. D., Weintraub H. Hb switching in chickens. Cell. 1980 Apr;19(4):973–980. doi: 10.1016/0092-8674(80)90088-4. [DOI] [PubMed] [Google Scholar]
- Stalder J., Seebeck T., Braun R. Degradation of the ribosomal genes by DNAse I in Physarum polycephalum. Eur J Biochem. 1978 Oct;90(2):391–395. doi: 10.1111/j.1432-1033.1978.tb12616.x. [DOI] [PubMed] [Google Scholar]
- Vidali G., Boffa L. C., Bradbury E. M., Allfrey V. G. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc Natl Acad Sci U S A. 1978 May;75(5):2239–2243. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. Assembly of an active chromatin structure during replication. Nucleic Acids Res. 1979 Oct 10;7(3):781–792. doi: 10.1093/nar/7.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Larsen A., Groudine M. Alpha-Globin-gene switching during the development of chicken embryos: expression and chromosome structure. Cell. 1981 May;24(2):333–344. doi: 10.1016/0092-8674(81)90323-8. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Weintraub H. Isolation of actively transcribed nucleosomes using immobilized HMG 14 and 17 and an analysis of alpha-globin chromatin. Cell. 1981 Feb;23(2):391–400. doi: 10.1016/0092-8674(81)90134-3. [DOI] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M. L. Assembly of newly replicated chromatin. Cell. 1978 Nov;15(3):969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]