Abstract
We present a tool to improve quantitative accuracy and precision in mass spectrometry based on shotgun proteomics: protein quantification by peptide quality control, PQPQ. The method is based on the assumption that the quantitative pattern of peptides derived from one protein will correlate over several samples. Dissonant patterns arise either from outlier peptides or because of the presence of different protein species. By correlation analysis, protein quantification by peptide quality control identifies and excludes outliers and detects the existence of different protein species. Alternative protein species are then quantified separately. By validating the algorithm on seven data sets related to different cancer studies we show that data processing by protein quantification by peptide quality control improves the information output from shotgun proteomics. Data from two labeling procedures and three different instrumental platforms was included in the evaluation. With this unique method using both peptide sequence data and quantitative data we can improve the quantitative accuracy and precision on the protein level and detect different protein species.
One reason for the low success of clinical biomarker discovery by proteomics is the difficulty of extracting information that will give answers to clinical biological questions. A major cause of this restraint is that quantitative data analysis methodologies in proteomics are immature (1–9). The large amount of data from mass spectrometry (MS) based protein profiling (10) include a lot of noise and biases arising from biological and chemical variation, sample preparation, instrumental analysis, and data analysis (9). This can lead to over interpretation and misleading conclusions from the data. The present work contributes with a novel algorithm for quantitative analysis of MS/MS proteomics data: Protein Quantification by Peptide Quality control (PQPQ)1. PQPQ improves the quantitative accuracy and precision and hence increases the chances to find clinical biomarkers and reveal biologically relevant information by proteomics research.
Shotgun Proteomics
The identification of the protein components of a biological sample is a complex, multistep procedure. In so called shotgun proteomics, protein samples are digested to peptides by enzymatic cleavage, then typically separated and analyzed in a liquid chromatography-mass spectrometry (LC-MS) system. From the full scan MS spectrum, precursor peptide ions are selected and fragmented for tandem MS analysis (MS/MS). The fragment ion spectra are interpreted to peptide sequences via a database search and the proteins are inferred from the identified peptides. Although today's shotgun proteomics techniques are capable of identifying thousands of proteins from biological samples (11) there are several limitations, especially in terms of protein quantification. The protein data output from shotgun proteomics rely on several assumptions: perfect tryptic cleavage, that a protein can be identified by only a few peptides, that the peptide-matching algorithms works perfect, and that the protein databases are populated with all proteins and their variants (9). However, this is not true for all proteins in a complex sample. Further, the protein inference problem; that a set of peptides may be shared by multiple proteins, puts doubt into the identification and quantification (12). A substantial problem is also that low intensity signals dominates the data set in a typical shotgun proteomics experiment. Furthermore, many protein identifications are based on only a few peptides, limiting the statistical security in the quantitative results (13). All these confounding issues have to be considered when interpreting shotgun proteomics data. The presented algorithm PQPQ is a tool that addresses some of these issues.
Quantitative Accuracy and Precision
Until recently, mass spectrometry based proteomics has mostly focused on assuring the protein identifications; the quantitative accuracy and precision has been less discussed. Nevertheless, the aim of proteomics studies in e.g. biomarker discovery is to find proteins with quantitative differences. Subtle changes in protein levels can have major effect on the underlying biology. Currently, many analyses fail at the point of biological interpretation because of a large quantitative variance and inaccuracy. One part of this problem is that current proteomics data analysis methods are unable to resolve different protein species, such as splice variants or modified subsets of proteins. Because different protein species have different biological functions, it is however essential to be able to detect and quantify those species separately. Today's data analysis output often reports a mean value of different species (14).
Protein Species and Alternative Splicing
Alternative splicing, alternative transcription start sites, post-translational modifications, protein cleavage etc. generate an enormous diversity of protein species. Many of these species are difficult to separate in the protein identification because either the species are not known and mapped, or they are not possible to discriminate because of similar protein sequences or lack of sequence coverage in the experiment. Yet, several diseases have known modifications and predisposition mediated by aberrant splicing. Changes in splicing have for example been shown to contribute in cancer progression (15). Several findings show that different splice variants have different biological functions, and may be useful diagnostic or prognostic tools (15, 16). Methods that detect isoform-specific mRNA changes have been developed for splicing microarrays, but are reported to be incomplete and noisy (14, 17). These methods also miss the protein level generated species (8, 9, 12, 18).
The Scope of This Work
Obtaining an accurate and precise estimate of the protein ratio from peptide intensities can be done in various ways and no standard methodology is yet defined (13, 19–21). Several open source/academic and commercial software for quantitative analysis of proteomics MS/MS data are available supporting different MS instruments and labeling methods (20, 22–25).
This work takes the quantitative protein analysis further. We here present a method for increasing the quantitative accuracy and precision in the protein output from shotgun proteomics data by quality control of the peptide data. The novelty of this algorithm is that it uses quantitative data of each peptide cross multiple samples to find outliers and to detect different protein species. By finding a correlating peptide pattern over several samples, outlier peptides can be detected and excluded. Further, a cluster analysis among the peptides associated to the same protein suggests if there exist several protein species. Different protein species are then accurately quantified separately. The method presented here is unique by using the combination of peptide sequence data and quantitative data to improve the quantitative accuracy and precision of the proteins and detect different protein species in proteomics data.
EXPERIMENTAL PROCEDURES
PQPQ: A New Algorithm for Protein Quantification by Peptide Quality Control
The algorithm PQPQ improves the accuracy in protein ratios calculation by selecting peptides correlating over samples for quantification. The peptides for quantification are selected by analyzing the pattern over several samples to define if the peptides come from the same protein. Pearson's correlation coefficient is used for measuring the similarity of the pattern between peptides:
 |
where pi is an array containing the intensities of peptide i for the different samples, cov denotes covariance, and spi is the standard deviation. The correlation is calculated for all the peptides associated to the same protein as illustrated in Fig. 3. The entire workflow is schematically described in Fig. 1, and in detail below.
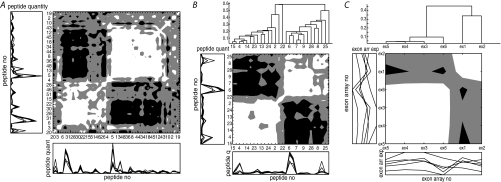
Fig. 3.
Correlation maps of the peptides and exon arrays associated to beta actin. The presented data are from data set I, sample pool 2. White means positive correlation, Pearsons correlation R2 > 0.33. Black is negative correlation, R2 < –0.33. Gray means 0.33 > R2 > –0.33. A–B, The peptides are identified by Protein Pilot from MALDI TOF/TOF MS data of 8-plex iTRAQ labeled lung cancer tumor samples and associated to beta actin by the Paragon algorithm. A, All peptides associated uniquely to the protein beta actin (52 peptides). B, Only the high confident peptides (28 peptides). C, The six exon mRNA arrays analyzed for beta actin and how the quantities are correlating over the samples.
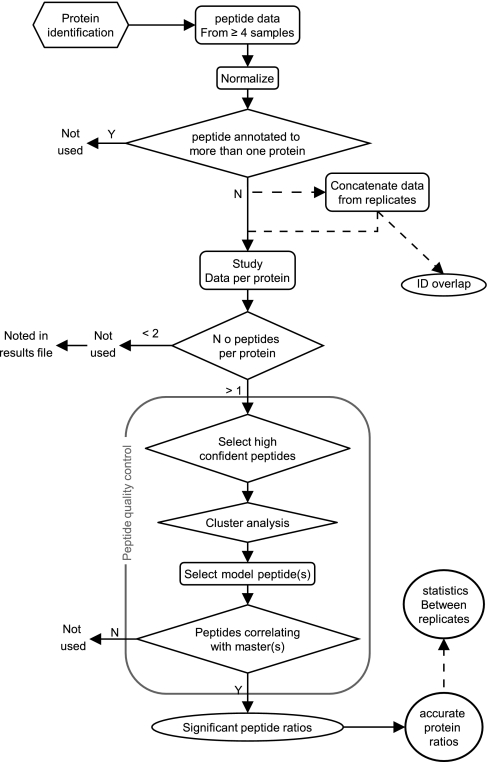
Fig. 1.
The scheme shows the data analysis workflow of the algorithm PQPQ. Data from replicate samples (marked - - -) are concatenated for the quality assessment of the peptides. The detailed peptide quality control scheme is found in Supplementary file S1, Fig. S1.
Experimental Design
To prove the significance and robustness of the developed algorithm PQPQ, complex biological samples, both cell line and clinical sample sets were analyzed, as summarized in Table I. The samples have been analyzed using different experimental setups, sample labeling methods (iTRAQ and TMT), and several MS instruments such as LC-matrix-assisted laser desorption ionization/time of flight (MALDI-TOF)/TOF (4800, Applied Biosystems), High accuracy nanoChipLC-NSI-Q-TOF (Agilent), and LC-NSI-LTQ Orbitrap Velos (high resolution FT instrument, Thermo Scientific). Detailed experimental information is available in Supplementary file S1 online.
Table I. The six cancer related studies that were analysed by PQPQ. Sample pools denotes iTRAQ/TMT sets in this table.
| Study | Samples | Replicates | Sample pools | Labeling | Σ no samples | MS instrument | Peptide data |
|---|---|---|---|---|---|---|---|
| I | Human Lung Cancer samples | 1 | 3 | 8-plex iTRAQ | 24 | MALDIa Q-TOFb Orbitrap | Protein Pilot Spectrum Mill Proteome Discoverer |
| II | Colon cell line | 1 | 1 | 8-plex iTRAQ | 8 | MALDIa | Protein Pilot |
| Fibroplast cell line | 1 | 1 | 8 | ||||
| III | Human Breast Cancer samples | 1 | 4 | 8-plex iTRAQ | 32 | Orbitrap | Proteome Discoverer |
| IV | Cell line: A549 | 1 | 1 | 8-plex iTRAQ | 8 | MALDIa Q-TOFb Orbitrap | Protein Pilot Spectrum Mill Proteome Discoverer |
| V | Cell line: MCF7 Spike in proteins | 1 | 1 | 8-plex iTRAQ 6-plex TMT | 14 | Orbitrap | Proteome Discoverer |
| VI | Cell line: MCF7 (cytosol fraction) | 3 | 1 | 4-plex iTRAQ | 12 | MALDIa Q-TOFb | Protein Pilot Spectrum Mill |
| Cell line: LCC 2 (cytosol fraction) | |||||||
| Cell line: MCF7 (DNA fraction) | 3 | 1 | |||||
| Cell line: LCC 2 (DNA fraction) | |||||||
| VII | Human Vulvar Cancer samples | 1 | 2 | 8-plex iTRAQ | 16 | Orbitrap | Proteome Discoverer |
a Matrix-assisted laser desorption/ionization.
b Quadrupole time-of-flight.
Data Preprocessing
PQPQ is designed to handle output data from ProteinPilot™ (26), Spectrum Mill, Proteome Discoverer, and can also load manually annotated peptide data as .txt, .csv, .xls, or .xlsx files. The data preprocessing is hence done in the software coupled to the instruments. The following information was extracted from the individual software programs: the protein accession number(s) associated with the peptides, a value of the peptide confidence, the peptide sequence, the area (or intensity) of the peptide peak (or reporter ion), one column for each sample, and the corresponding gene name(s) (can be empty). The input data to PQPQ preferably includes quantitative data from all peptides identified in the samples (not filtered). The data can either be from labeled or label-free experiments.
Read and Sort Peptide Data
The extracted peptide data are imported into the PQPQ where the subsequent data processing is done. At first the data is normalized (optional) so that the medians of the peptide intensities are equal across all samples. By this normalization, we are assuming that the samples included are of similar character and the median of peptide content can thus be expected to be equal. It should be noted that in all the experiments, the total protein concentration was measured and adjusted to be equal between samples prior to labeling and the MS runs. In the case of sample replicates, the data were concatenated at this step, and the protein ID overlaps between replicates were calculated. From this step, the data were studied protein by protein. In the case where we had biological replicates, peptides from the same proteins and different replicate samples were collectively treated. To keep information about repeatability, the variation between replicates was calculated after the peptide quality assessment. Further, only proteins supported by at least two peptides were included. Fig. 1 shows the workflow schematically.
Peptide Quality Control
The peptide quality control procedure is described in detail in supplementary Fig. S1. We first select all high confident peptides from the list of peptides defining the actual protein. These have a confidence exceeding the high confidence limit, defined by the user. We identify which of the high confident peptides that correlate positively, with a defined risk of selecting a random correlation, a p value defined by user. From those, a model peptide is chosen. The model peptide is defined as the one with the highest intensity of the ones that correlates with most of the high confident peptides. If the high confident peptides do not correlate at the defined p value, the peptide with the highest intensity of the high confidence ones is selected as the model peptide. If no peptide has a confidence level above the limit, the protein identity and quantity is regarded as unconfident. The quantity is then not calculated and “peptide confidence too low to support protein ID” is noted in the protein output file.
Clusters of High Confident Peptides
To seek for clustering among the high confident peptides a distance between the peptides are calculated. The distance is calculated as 1 – correlation, between all peptides, over samples (Equation [1]). Based on those distances, a hierarchical cluster tree is computed, where the peptides with the shortest distance are linked together. Clusters of peptides are then formed when a node and all of its subnodes have an inconsistent value less than 1. That is, if the node height related to the average height of other nodes at the same level of hierarchy is less than one, those objects are clustered together. If there are several clusters among the high confidence peptides, these probably arise from several protein species. In those cases, a model peptide is defined for each cluster. The clusters are ranked so that the cluster including the most peptides will be noted as protein specie 1, “var 1” in the protein output file.
Include More Peptides
In the next step all peptides associated to the protein (no matter what peptide confidence level they have) that are correlating with the model peptide (at the defined p value for the correlation) are selected as belonging to that protein. Hence, only peptides correlating with a highly confident peptide will be included in the protein quantification. If several clusters were discovered in the previous step, the correlation analysis is done for each model peptide.
Input Parameters
The high confidence limit denotes the peptide confidence above which peptides are defined as high confident. The peptide confidence is the quality measure of the peptide identification, defined by the software used for peptide identification and quantification. In the examples in this paper we have used Spectrum Mill, Protein Pilot, and Proteome Discoverer. A more detailed description of the peptide confidence measures is given in the Supplementary file S1 online. For PQPQ, any definition can be used. The high confidence limit is defined outside the PQPQ algorithm. One way is to define the limit from MAYU (27), where the protein false discovery rate (FDR) is determined. From there the peptide confidence limit can be determined. This was done for data set IV and VII in the present work. Another way of defining the high confidence limit is to estimate the FDR of the peptide identification using a decoy database. This is done by identifying the peptide data from the MS run in a forward and reverse database. Because the reverse hits are known to be false discoveries, the FDR of the database search can be calculated (28). This was done similarly for Spectrum Mill and Protein Pilot for dataset II, III, and VI. In addition, the software connected to the instruments has their own definition of FDR, which was used for the data sets I and III.
The p value defines the probability of getting a correlation as large as the observed value by random chance, when the true correlation is zero. So, if the p value limit is set to 0.4, the risk of defining a correlation although there is none, is 40%. The p values: 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, and 0.7 was tried out to determine a default value for the PQPQ algorithm. The results from this study are presented in the supplementary Fig. S2–S3. Based on this study, the default value for the algorithm was set to 0.4.
Algorithm Availability and Requirements
PQPQ is written in MATLAB (29) and requires also the MATLAB statistics toolbox. A graphical user interface makes it easy to use and a standard PC is powerful enough for the calculations. The algorithm is divided into three processes; Peptide selection, Protein ratio calculation, and Peptide quantity visualization, all or one can be chosen. A detailed manual for the input parameters is found in Supplementary Manual online. The program is free of use and available from the first author at the www.forshed.se/jenny.
PQPQ Data Output
PQPQ creates one Excel file and one MATLAB output file with the cured peptide data. This file includes all the information that was originally in the peptide input file plus information of which peptides to include for protein ratio calculation, noted as valid peptides. Also, the results from the cluster analysis are found in this file. The model peptide of each cluster is also noted. Further, the output file includes a note if the peptide confidence of the ingoing peptides is too low to support the protein quantification. If the protein has support from less than two peptides, this is also noted. If the protein is determined from more than 300 peptides, the 300 most confident peptides are selected, and this is noted in the peptide data output. Those notations are also transferred to the protein data output file.
The protein data output file (Microsoft Excel format) includes the protein ratios; quantitative data and statistics for all proteins with support from assessed peptide data. This file also includes the number of peptides that the quantitative calculations are based on, the standard deviation, and the p value for the probability of the ratio to be equal to 1 (student's t test). In case of replicated sample runs, reproducibility (standard deviation between replicates), and number of replicates are reported. If several variants of one protein were detected these are noted as protein name_var1 etc. The different variants have unique quantitative outputs. Protein name_var1 is regarded as the highest ranked variant/specie because it had most high confident peptides in its cluster. Protein name_var2 had second most high confident peptides etc.
In both the protein and the peptide output files, a sheet with the PQPQ settings is included. This file includes also the normalization factors. Further, one sheet with the following summary of the proteins is included; the number of proteins that were imported, the number of proteins that were left after eliminating redundant peptides, after multiple protein entries were separated, after eliminating proteins with too low intensity (sum of peptide intensity), and finally the number of proteins left after PQPQ selection. See Supplementary file S2 online for further details.
Sample Labeling
To find a correlation pattern between peptides, at least four samples (e.g. patients) are recommended. The samples are preferably from the same study, and must be run with equal instrumental setup to be comparable. An ideal experimental setup to meet those conditions is the iTRAQ or TMT labeling used in this work. Four, six, or eight samples are labeled after digestion, mixed and analyzed together in the mass spectrometer. Technical and laboratory biases after the mixing are consequently “cancelled out.” The samples are then compared by quantification of the different reporter ions (each reporter corresponding to a particular sample) in the MS/MS spectra, where the labels diverge in m/z-position (30, 31). Notably, the PQPQ algorithm can handle other types of labeled data than iTRAQ and TMT, as well as unlabeled data.
Exon Array Analysis
The exon array data we have used for independent detection of splice variants in study I is generated using the 244K array from Agilent, with a custom design of 195,000 probes (60 mers) of exons of around 19,500 genes (hgdownload.cse.ucsc.edu/goldenPath/hg18/database/refGene.txt.gz). The array contains also the probes for the 44 K commercial array 014850 (http://www.chem.agilent.com/cag/bsp/oligoGL/014850_D_GeneList_20070207.txt.zip, and http://www.chem.agilent.com/cag/bsp/oligoGL/014850_D_AA_20070207.txt.zip).
RESULTS
Improved Accuracy and Precision of the Protein Quantification by PQPQ
Precision defines how reproducible measurements can be, and accuracy is used to denote how close measured values are to the true ones (32). The first reason why PQPQ improves the accuracy and precision of protein quantification is the outlier exclusion. Second, peptides with a low confidence in the identification, but still correlating with a high confident peptide (the model peptide) can be included to improve the quantitative precision by increasing the number of included data measurements. Thirdly, different protein species that are not possible to separate based on identified peptides can be identified, as opposed to today's standard methodology. Today's standard methods show the quantitative output as a mean of different species where biological differences between samples can be cancelled out. The possibilities to, by PQPQ detect and quantify different protein species separately will hence increase both the quantitative accuracy and precision.
By reduction of biases (improved accuracy) and variations (improved precision), PQPQ will increase the information output from shotgun proteomics data. This is here proved by processing data from seven proteomics studies (Table I). Below we show examples that provide evidence for the importance of PQPQ.
Improved Precision
Outlier Exclusion Reduces the Variability
To prove the increased quantitative precision by PQPQ, data set IV was generated. Data set IV is an 8-plex iTRAQ mix of human proteome from cell line lysate (A549) in the ratios 2:2:1:1:2:2:1:1. The samples were run on three MS platforms (Table I). We analyzed the precision of the quantitative ratio between the two first samples (ratio 2:2) by the standard deviation of each protein ratio in the sample. PQPQ set with a strict correlation p value was compared with accepting all peptides associated to the protein (if not excluded based on the other control mechanisms in PQPQ: redundancy, to few peptides, etc.). The correlation p value defines the probability of getting a correlation as large as the observed value by random chance, when the true correlation is zero. The standard deviation for the proteins were on average improved by 72%, 39%, and 19% when applying PQPQ with a correlation p = 0.01 for the three instrumental setups MALDI, Q-TOF, and Orbitrap respectively. The improvement for correlation p = 0.1 compared with including all peptides, was 68, 13, and 11% for the respective instruments. This proves the general enhancement by peptide outlier elimination by PQPQ. Graphs of the data distribution is found in supplementary Fig. S4 online.
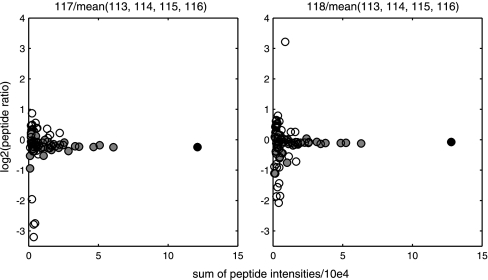
The outlier detection is also illustrated by an example from data set II where isoform A1-B of the heterogeneous nuclear ribonucleoprotein A1 was identified. Ninety-four peptides were uniquely associated to the protein, and were analyzed by PQPQ. Out of the 94 peptides, 52 peptides were selected by PQPQ to be included in the quantitative calculations because of a good correlation with the model peptide. This is illustrated by Fig. 2. Removing the outliers in this case improved the quantitative precision (standard deviation) of the protein ratio by 88% (from 0.87 to 0.097) and 84% (0.90 to 0.14) for the respective ratios shown in Fig. 2.
Fig. 2.
Outlier detection for quantification of isoform A1-B of the heterogeneous nuclear ribonucleoprotein A1. Each spot represents one peptide of the protein from data set II (colon cell line samples) quantified by 8-plex iTRAQ. Each graph shows one sample related to the mean of the control samples represented by iTRAQ reporter ions 113, 114, 115, and 116. The ratios (x axis) are plotted against the sum of the ingoing peptide intensities (y axis) from MALDI MS data, extracted from Protein Pilot. The filled circles are the ratios accepted as quantitative accurate by PQPQ. The outlier peptides are marked as empty circles. The “model peptide” is black. In this specific case the precision (standard deviation) of the protein ratio were improved from 0.87 to 0.097 and 0.90 to 0.14 for the respective ratios by outlier exclusion.
Inclusion Based on Correlation Improves the Precision
Pearson's correlation coefficient calculated between all peptides, over samples, is illustrated by a correlation map in Fig. 3. In study I, sample pool 2, 118 peptides detected by MALDI TOF/TOF were associated to the protein beta actin. Of those, 52 were uniquely associated to beta actin and showed no overlap with any other proteins, Fig. 3A. These 52 peptides were further analyzed by PQPQ. Twenty-eight of the 52 peptides had a peptide confidence exceeding the determined limit for a high confident peptide, Fig. 3B. The peptides in Fig. 3A and 3B are sorted on similarity (hierarchical clustering) based on Pearson correlation. It can be seen from the figures that several peptides are not correlating with the others (gray and black fields). However, groups of peptides correlating with each other are forming two clusters of peptides (Fig. 3A and 3B). Also, a few outlier peptides which do not correlate with any of the clusters of peptides are seen in Fig. 3A, these were discarded as outliers in the PQPQ analysis. Further evidence on the correct, biologically relevant clustering, is shown on this example by comparison with exon mRNA array data on same samples (Fig. 3C). This is discussed later in section “Splice variants verified by exon array data.”
Fig. 3A and 3B, show that several of the low confident peptides (not included in Fig 3B, but seen in Fig 3A) correlate with the high confident ones. The quantitative accuracy and precision of the protein quantity will improve by including these in the calculations as it is done in PQPQ. This is shown by comparing the standard deviations from the current example. As indicated in Fig. 3, beta actin was separated into two variants by PQPQ. For each variant we have compared the standard deviation from calculating ratios by using only high confident peptides (as is the standard procedure) with using the peptides selected by PQPQ, where also low confident peptides can be included. For variant 1, the standard deviation decreased by 13–50% for different sample ratios, for variant 2 the standard deviation decreased by 0–22%. This is illustrated in supplementary Fig. S5 online.
Improved Accuracy
Detection of the Existence of Different Protein Species
The quantitative clustering of peptides by the algorithm presented here, PQPQ, enables detection of protein species, e.g. spliced variants or different post-translation modified species. The existence of different protein species is indicated by peptide clusters in the peptide quality control in PQPQ. The ability to detect different protein species makes it possible to quantify those separately. This will improve the protein quantification accuracy as is illustrated by the following examples. The putative protein species are flagged in the PQPQ output file. This allows further analyses of subsets of proteins, for example splice variant detection.
Spike in Samples
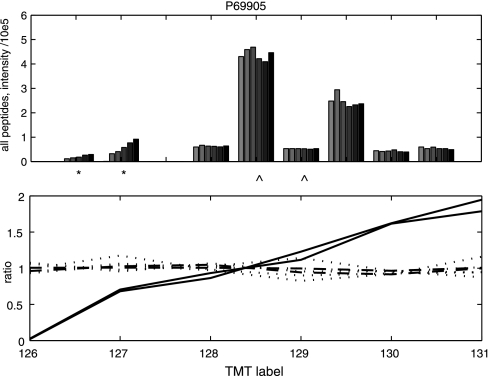
The spike in data set V consisted of a complex proteome (human cell line MCF7) in each sample as a background sample matrix and different spiked in proteins in dilution series. To illustrate the possibility of detecting different protein species by PQPQ we have chosen the protein hemoglobin subunit alpha. PQPQ detected two variants of the protein, of which one followed the dilution pattern of the added protein. Although the second variant was identified based on high confident peptides, it did not follow the spike in pattern, see Fig. 4. The peptides from variant 2 are likely detected remnants of cell culture media hemoglobin. PQPQ will quantify the two species separately. This demonstrates further the accurate quantification of proteins species and clustering of data for downstream protein species analysis.
Fig. 4.
Peptide intensities and ratios from hemoglobin subunit alpha showing two different patterns. The upper graph shows the nine peptides that were detected from hemoglobin subunit alpha in the Orbitrap-MS analysis of the spike in samples in data set V. One color for each sample and one group of bars for each peptide. The * and ⋀ denotes which peptides that belong to the different variants that PQPQ detected. Two distinct patterns are seen. The bottom graph shows the same data related to the mean of all sample intensities (the ratio). The solid lines (—) represents variant 1 (*) which is the spiked in variant. The dashed lines (- - -) represents variant 2 (⋀), interpreted as coming from the background cell line. The dotted lines (. . . .) represent peptides excluded as outliers in the present PQPQ data analysis. The p value for detecting a correlation was set to 0.1 in this calculation.
Splice Variants Detection
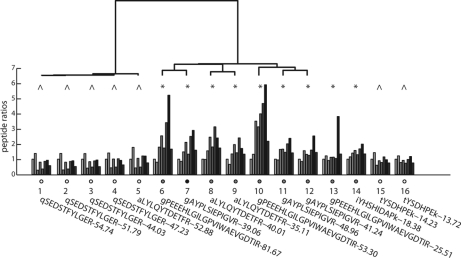
Both Fig. 3 and 4 shows examples of how peptides associated to the same protein can be divided into different clusters. Fig. 5 shows an additional example of this phenomenon from a clinical study analyzing human breast cancer samples (data set III). It is seen from Fig. 5 that the peptides associated to Ceruloplasmin showed two clusters. Peptides 1–12 had a peptide confidence above the high confidence limit. These peptides were analyzed for clusters, represented in Fig. 5 as a dendogram. Peptides 13–16 represent low confident peptides in this example. Peptides 13–14 correlate with the left cluster (*) and peptides 15–16 with the right (⋀). As the low confident peptides are not outliers and clearly correlates with high confident peptides cross samples, including these peptides in the calculations will improve the protein quantitative statistics and hence the accuracy and precision.
Fig. 5.
Illustration of clustering of peptides associated to Ceruloplasmin. The 16 peptides detected by LC-LTQ Orbitrap MS and associated to Ceruloplasmin in human breast cancer samples, data set III, sample pool 4. The peak areas are in relation to the internal standard peak iTRAQ 113. One color for each sample/iTRAQ label and one group of bars for each peptide. Above is a dendogram showing the clustering of peptides 1–12, which had a peptide confidence above the high confidence limit at 30. Peptides 13–16 had confidence score below the limit. The two clusters with the symbols * and ⋀ includes peptides correlating with each other over samples. Below the peptide ratio bars are the sequences and the peptide confidences noted.
The peptides associated to Ceruloplasmin were also analyzed by SpliceCenter (33). SpliceCenter is a tool identifying possible splice variants that could potentially be detected by the given peptides. The SpliceCenter analysis confirmed that two different splice variants of Ceruloplasmin were possible to distinguish based on the detected peptides. The results can be found in supplementary Fig. S6 online.
Improved Quantitative Accuracy by Detecting Protein Species
An additional example, which illustrates improved protein quantification accuracy by sorting out different protein species is taken from the MALDI data set I (human lung cancer samples). The Paragon algorithm (34) (Protein Pilot) identified 11 peptides associated to peroxiredoxin 1. It was not possible to distinguish the isoform CRA_b of the same protein based on the identified peptide sequences. The relative quantification was hence reported as a mean quantity from all of the associated peptides from Protein Pilot, as is the standard methodology. However, by PQPQ we were able to separate two species of the protein and quantify them separately. All but one of the peptides had a high peptide confidence. From the PQPQ protein data we identified 4 and 5 patients for the two protein species respectively that were shown to differ from the patient mean. The Protein Pilot protein data showed only two patient samples that were significantly different from the patient mean. This protein species quantification illustrates the obvious benefit of analyzing the data by means of PQPQ when aiming for quantitative analysis of clinical samples, as in biomarker discovery. The result is shown in supplementary Table S1 online.
That several protein identities cannot be distinguished from each other by the detected peptides is common in shotgun proteomics data. A common way of solving this problem is to discard all of the proteins that do not have one unique protein identity connected to the peptides. However, the risk is then to miss several “true protein hits.” In the above example with MALDI data, 443 out of 661 proteins would then have been discarded, i.e. 67% (!). Looking at high resolution data (Orbitrap) of the same sample, 554 out of 2926 proteins (19%) would have been discarded.
Splice Variants Verified by Exon Array Data
To verify that PQPQ can detect splice variants on protein level, the samples from data set I (human lung cancer samples) were analyzed further by exon array data on mRNA level. Beta actin here figures as an example. The PQPQ analysis of the proteomics data showed two possible species of the protein (Fig. 3B). The exon array data for the corresponding protein (six probes) also shows two clusters (Fig 3C). This indicates that the sample contains two splice variants of beta actin. To confirm this hypothesis, we studied which spliced variants that are known for beta actin. Based on the here detected peptides, five different splice variants of the protein beta actin were registered by SpliceCenter. The analysis shows that only protein variant 1 (defined by PQPQ) includes peptides translated from exon 2. This agrees with the cluster analysis of the exon array data, where exon 2 forms one cluster together with exon 1 (noncoding). This strongly indicates that we detect the same two splice variants in both the proteomics and the exon array data. Supplementary Fig. S7 online shows screenshots from the SpliceCenter analysis of the peptides forming the two variants detected by PQPQ.
DISCUSSION
The incomplete tryptic digestion, the few peptides per protein, not perfect peptide matching algorithms, incomplete databases, the protein inference problem (9), and low intensity signals are all confounding issues that have to be handled when quantifying proteomics data. The presented algorithm PQPQ is a tool for that. By PQPQ the proteomics data is analyzed protein by protein, and by combining peptide sequencing data and quantitative data from several samples we are able to improve the quantitative accuracy and precision of the proteins and detect different protein species. By means of PQPQ we can discover if the detected peptides show different patterns over samples. In that case, PQPQ can distinguish different proteins species and quantify them separately. This has so far been impossible by current state of the art methodology.
One of the most crucial steps when generating, using and interpreting proteome specific information is to be able to analyze information on protein species. We show that PQPQ can decipher and accurately quantify protein subspecies. This is illustrated by the examples beta-actin, hemoglobin, ceruloplasmin, and peroxiredoxin 1 where PQPQ quantified the protein species from these proteins separately. The protein lists from PQPQ include separate protein entries for the different protein variants if any. In a subsequent statistical analysis for e.g. biomarker discovery, the found protein species will be treated as separate entries. The outcome of the statistical analysis will show if any of the species have an effect in the clinical and biological question. These protein species can then be focus of further analyses by plotting the selected protein and its peptides in PQPQ. The PQPQ algorithm hence serves as a tool to direct biomarker analysis and biological interpretation of proteomics data to include protein species.
There might be several reasons for detecting different protein species from the same protein identification. The protein species might for example originate from different post translational modifications or different splice variants of the protein. If splice variants for a specific protein is included in the protein database used for the protein identification, and the variants cannot be separated based on the detected peptides, it will be noted as multiple identities. However, if the different variants have different patterns over samples, PQPQ will be able to separate them. As shown in supplementary Fig. S6–S7 such a case can be investigated further by analyzing output data from PQPQ by for example SpliceCenter.
It should be noted that peptides shared between different protein species will have a quantitative pattern over samples that is a mix of the two proteins. A mixed quantitative pattern can also be seen if the MS precursor includes several peptides (35). These mixed peptide peaks will be discarded as outliers if they are extreme and single occurrences. If several peptides are quantified from the same protein species mix, these peptides will be distinguished as a separate protein species. If one of the protein species are dominant, these peptides will be associated with the most dominating pattern. This phenomenon is the reason for the very tolerant default correlation p value of 0.4 (40% risk of having no correlation although a correlation is detected).
We have also shown the improved precision of protein quantification by outlier elimination. A majority of the outliers has a low intensity, illustrated by Fig. 2. Peptides with high intensity can become outliers if they are erroneously connected to the protein because of a low quality peptide MS/MS spectrum leading to false peptide sequence determination. And, as stated above, if peptides from other species of the same protein are present these may be erroneously associated to the same protein. By the data set IV, constructed from cell line samples in known concentration ratios, we were able to prove the outlier elimination effect of PQPQ. The largest improvement was on the MALDI data, followed by the Q-TOF data and the Orbitrap data. This is in accordance to our expectations because it follows the number of identified proteins, assuming that the number of protein identities correlates with the data quality. Further, we have shown that the peptide quality control in PQPQ allows us to use low intensity, and low confident peptides in the quantification to improve the statistics of the protein quantification.
PQPQ has been validated on two constructed human cell line proteome samples with proteins of known ratios as well as five different cancer related proteomics studies on three different mass spectrometry platforms. Further, two different sample labeling methods have been included in the validation. Notably, PQPQ can also handle data from other labeling methods as well as label free proteomics data. This shows the broad use of this algorithm. By using PQPQ for peptide selection, we are able to improve the quantitative precision as well as the quantitative accuracy of the protein output from shotgun proteomics. We are also able to detect some of the shortcomings in the proteomics analysis workflow, and correct for them to some extent. PQPQ can hence enrich the information of the output from mass spectrometry based proteomics. This will facilitate to find actual changes on protein level, which is a well needed development in clinical biomarker discovery research.
Footnotes
* This work is supported by the Cancer Society in Stockholm, The Swedish Research Council, the Swedish Cancer Society, Stockholm County Council, Agilent Foundation, EU FP 7th project GlycoHit and FP 6th project Chemores.
 This article contains supplemental Manual, Figs. S1 to S8 and Table S1.
This article contains supplemental Manual, Figs. S1 to S8 and Table S1.
1 The abbreviations used are:
- PQPQ
- Protein Quantification by Peptide Quality control
- FDR
- False Discovery Rate.
REFERENCES
- 1. Mischak H., Apweiler R., Banks R. E., Conaway M., Coon J., Dominiczak A., Ehrich J. H. H., Fliser D., Girolami M., Hermjakob H., Hochstrasser D., Jankowski J., Julian B. A., Kolch W., Massy Z. A., Neusuess C., Novak J., Peter K., Rossing K., Schanstra J., Semmes O. J., Theodorescu D., Thongboonkerd V., Weissinger E. M., Van Eyk J. E., Yamamoto T. (2007) Clinical proteomics: A need to define the field and to begin to set adequate standards. Proteomics Clin. Appl. 1, 148–156 [DOI] [PubMed] [Google Scholar]
- 2. Aebersold R. (2009) A stress test for mass spectrometry-based proteomics. Nat. Methods 6, 411–412 [DOI] [PubMed] [Google Scholar]
- 3. Reymond M. A., Schlegel W. (2007) Proteomics in cancer. Adv. Clin. Chem. 44, 103–142 [DOI] [PubMed] [Google Scholar]
- 4. Service R. F. (2008) Proteomics. Proteomics ponders prime time. Science 321, 1758–1761 [DOI] [PubMed] [Google Scholar]
- 5. Service R. F. (2008) Proteomics. Will biomarkers take off at last? Science 321, 1760. [DOI] [PubMed] [Google Scholar]
- 6. Sung H. J., Cho J. Y. (2008) Biomarkers for the lung cancer diagnosis and their advances in proteomics. Bmb Reports 41, 615–625 [DOI] [PubMed] [Google Scholar]
- 7. Lescuyer P., Hochstrasser D., Rabilloud T. (2007) How shall we use the proteomics toolbox for biomarker discovery? J. Proteome Res. 6, 3371–3376 [DOI] [PubMed] [Google Scholar]
- 8. Meyer H. E., Stuhler K. (2007) High-performance proteomics as a tool in biomarker discovery. Proteomics 7 Supple 1, 18–26 [DOI] [PubMed] [Google Scholar]
- 9. Duncan M. W., Aebersold R., Caprioli R. M. (2010) The pros and cons of peptide-centric proteomics. Nat. Biotechnol. 28, 659–664 [DOI] [PubMed] [Google Scholar]
- 10. Kumar C., Mann M. (2009) Bioinformatics analysis of mass spectrometry-based proteomics data sets. FEBS Lett. 583, 1703–1712 [DOI] [PubMed] [Google Scholar]
- 11. Eriksson H., Lengqvist J., Hedlund J., Uhlén K., Orre L. M., Bjellqvist B., Persson B., Lehtiö J., Jakobsson P. J. (2008) Quantitative membrane proteomics applying narrow range peptide isoelectric focusing for studies of small cell lung cancer resistance mechanisms. Proteomics 8, 3008–3018 [DOI] [PubMed] [Google Scholar]
- 12. Nesvizhskii A. I., Aebersold R. (2005) Interpretation of shotgun proteomic data - The protein inference problem. Mol. Cell. Proteomics 4, 1419–1440 [DOI] [PubMed] [Google Scholar]
- 13. Karp N. A., Huber W., Sadowski P. G., Charles P. D., Hester S. V., Lilley K. S. (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell. Proteomics 9, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robinson T. J., Dinan M. A., Dewhirst M., Garcia-Blanco M. A., Pearson J. L. (2010) SplicerAV: a tool for mining microarray expression data for changes in RNA processing. BMC Bioinformatics 11:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang G. S., Cooper T. A. (2007) Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Gen. 8, 749–761 [DOI] [PubMed] [Google Scholar]
- 16. Nilsen T. W., Graveley B. R. (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Laajala E., Aittokallio T., Lahesmaa R., Elo L. L. (2009) Probe-level estimation improves the detection of differential splicing in Affymetrix exon array studies. Gen. Biol. 10, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Malmstrom J., Beck M., Schmidt A., Lange V., Deutsch E. W., Aebersold R. (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–U112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boehm A. M., Putz S., Altenhofer D., Sickmann A., Falk M. (2007) Precise protein quantification based on peptide quantification using iTRAQ (TM). BMC Bioinformatics 8:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lau K. W., Jones A. R., Swainston N., Siepen J. A., Hubbard S. J. (2007) Capture and analysis of quantitative proteomic data. Proteomics 7, 2787–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park S. K., Venable J. D., Xu T., Yates J. R. (2008) A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mueller L. N., Brusniak M. Y., Mani D. R., Aebersold R. (2008) An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J.f Proteome Res. 7, 51–61 [DOI] [PubMed] [Google Scholar]
- 23. Panchaud A., Affolter M., Moreillon P., Kussmann M. (2008) Experimental and computational approaches to quantitative proteomics: Status quo and outlook. J. Proteomics 71, 19–33 [DOI] [PubMed] [Google Scholar]
- 24. D'Ascenzo M., Choe L., Lee K. H. (2008) iTRAQPak: an R based analysis and visualization package for 8-plex isobaric protein expression data. Briefings in Functional Genomics and Proteomics 7, 127–135 [DOI] [PubMed] [Google Scholar]
- 25. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 26. (2003–2007) Protein Pilot Software 2.0.1. p., Applied Biosystems/MDS Sciex [Google Scholar]
- 27. Reiter L., Claassen M., Schrimpf S. P., Jovanovic M., Schmidt A., Buhmann J. M., Hengartner M. O., Aebersold R. (2009) Protein Identification False Discovery Rates for Very Large Proteomics Data Sets Generated by Tandem Mass Spectrometry. Mol. Cell. Proteomics 8, 2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 29. (2010) MATLAB. 7.11, R2010b Ed., The MathWorks Inc., Natick, MA [Google Scholar]
- 30. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 31. Song X., Bandow J., Sherman J., Baker J. D., Brown P. W., McDowell M. T., Molloy M. P. (2008) iTRAQ experimental design for plasma biomarker discovery. J. Proteome Res. 7, 2952–2958 [DOI] [PubMed] [Google Scholar]
- 32. Miller J. C., Miller J. N. (1993) Statistics for Analytical Chemistry, 3:rd Ed., Ellis Horwood Limited [Google Scholar]
- 33. Ryan M. C., Zeeberg B. R., Caplen N. J., Cleland J. A., Kahn A. B., Liu H., Weinstein J. N. (2008) SpliceCenter: A suite of web-based bioinformatic applications for evaluating the impact of alternative splicing on RT-PCR, RNAi, microarray, and peptide-based studies. Bmc Bioinformatics 9:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 35. Ow S. Y., Salim M., Noirel J., Evans C., Rehman I., Wright P. C. (2009) iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly.” J. Proteome Res. 8, 5347–5355 [DOI] [PubMed] [Google Scholar]