Abstract
Transmissible spongiform encephalopathies are fatal neurodegenerative diseases attributed to misfolding of the cellular prion protein, PrPC, into a β-sheet-rich, aggregated isoform, PrPSc. We previously found that expression of mouse PrP with the two amino acid substitutions S170N and N174T, which result in high structural order of the β2–α2 loop in the NMR structure at pH 4.5 and 20°C, caused transmissible de novo prion disease in transgenic mice. Here we report that expression of mouse PrP with the single-residue substitution D167S, which also results in a structurally well ordered β2–α2 loop at 20°C, elicits spontaneous PrP aggregation in vivo. Transgenic mice expressing PrPD167S developed a progressive encephalopathy characterized by abundant PrP plaque formation, spongiform change, and gliosis. These results add to the evidence that the β2–α2 loop has an important role in intermolecular interactions, including that it may be a key determinant of prion protein aggregation.
Introduction
Prion diseases are infectious and invariably fatal neurodegenerative disorders caused by the conversion of cellular prion protein, PrPC, into a misfolded and aggregated isoform known as PrPSc (Prusiner, 1982, 1998; Collinge, 2001; Deleault et al., 2007; Wang et al., 2010). In humans, prion diseases occur in rare cases spontaneously, can be initiated by infection, or are related to inherited mutations in the prion gene, PRNP, which encodes for 208 residues in the mature polypeptide chain (Aguzzi and Calella, 2009). PrPC consists of a flexible, unstructured N-terminal tail and a globular C-terminal domain comprising three α-helices and a short β-sheet, and is highly conserved among mammals (Riek et al., 1996; Zahn et al., 2000; Gossert et al., 2005; Lysek et al., 2005). Pathogenic PRNP mutations can affect all regions of the protein but display some clustering toward the C terminus (Riek et al., 1998). Particular mutations are destabilizing, such as T183A, which eliminates two hydrogen bonds linking helix α2 and the β-sheet, yet mutations in the flexibly extended N-terminal domain do not affect stability (Riek et al., 1998). Certain Prnp mutants in transgenic mice can reproduce PrP aggregation, clinical neurologic signs, and PrP plaques in the brain as seen in the familial diseases (Hsiao et al., 1990; Chiesa et al., 1998; Dossena et al., 2008; Jackson et al., 2009; Sigurdson et al., 2009).
NMR spectroscopy has shown that in the solution structure at 20°C, the β2–α2 loop (aa 166–172) can be either structurally well ordered (“rigid loop,” RL), or disordered (Riek et al., 1996; Gossert et al., 2005). We previously obtained a well defined β2–α2 loop structure by two amino acid exchanges in mouse PrP, S170N and N174T, and expressed the mutated Prnp gene in transgenic mice. The resulting RL mice developed a spontaneous prion disease and also showed altered susceptibility to infection by prions derived from other species (Sigurdson et al., 2009, 2010). Thus, the β2–α2 loop emerges as a critical region in the PrPC structure that influences prion self-association and cross-species infections, yet the underlying molecular mechanism is incompletely understood.
To further investigate how the loop topology impacts PrP aggregation in vivo, we have developed a second rigid loop transgenic mouse that expresses mouse PrP containing the single-amino acid residue substitution D167S. The horse Prnp sequence normally encodes a serine at position 167, and the NMR structures of horse PrPC and of mouse PrPC with the D167S substitution (MoPrP167) both show a structurally well defined β2–α2 loop in solution at ∼20°C (Pérez et al., 2010). We now find that overexpression of MoPrP167 leads to widespread PrP aggregation in the brain of transgenic mice, similar to that seen in the previously studied RL mice (MoPrP170,174).
Materials and Methods
Generation of transgenic mice expressing MoPrP167.
Single-point mutations (GAT→AGT) that alter the amino acid sequence to 167S were created within a pMECA subclone, based on pHGPrP (Fischer et al., 1996), using the Stratagene point mutagenesis kit (primers: forward, 5′-GGCCA GTG AGT CAG TAC AGC AAC CAG AAC AAC TTC GTG CAC GAC-3′ and reverse, 5′-GTC GTG CAC GAA GTT GTT CTG GTT GCT GTA CTG ACT CAC TGGCC-3′). The PmeI/NheI pMECA subclone was then cloned into the PmeI/NheI sites of the pHGPrP plasmid, and the entire ORF was sequenced (Rosenberg et al., 1977). The pHGPrP plasmid containing the mutated Prnp was propagated in Escherichia coli Top10 cells (Invitrogen), and the PrP mini-gene sequence was excised with NotI/SalI. Constructs were microinjected into the pronucleus of fertilized B6;129S5-Prnp+/o oocytes (Prnp-KO Zurich I) using conventional methods (Rülicke, 2004). Founder lines were identified by PCR for the transgene using the exon-2 primer pE2+ (5′-CAA CCG AGC TGA AGC ATT CTG CCT-3′) and the exon-3 primer Mut217 (5′-CCT GGG ACT CCT TCT GGT ACC GGG TGA CGC-3′), and founders were bred to Prnpo/o mice. Six transgene-carrying founder mice were identified; Tg(PrnpD167S)77–82Biat. Two lines were maintained by crossing with Prnpo/o mice. To identify the KO neo-cassette, PCR analysis was performed using the primers P10 (Prnp exon 3, 5′-GTA CCC ATA ATC AGT GGA ACA AGC CCA GC-3′, 3′NC (non-coding region at 3′of exon 3, 5′-CCC TCC CCC AGC CTA GAC CAC GA-3′), and P3 (neoR gene, 5′-ATT CGC AGC GCA TCG CCT TCT ATC GCC-3′); P10 and 3′NC gave a 560-bp signal for the Prnp+ allele, and P3 and 3′NC gave a 362-bp product for the Prnp− allele. As an alternative test for the presence or absence of the endogenous Prnp+ allele, an additional PCR was performed using the primers P2 (Prnp int2, 5′-ATA CTG GGC ACT GAT ACC TTG TTC CTC AT-3′) and P10rev (reverse complementary of P10 5′-GCT GGG CTT GTT CCA CTG ATT ATG GGT AC-3′), generating a 352-bp amplicon for the Prnp+ allele.
Tga20 transgenic mice, which overexpress murine PrP (Fischer et al., 1996), were used as controls for many of these experiments. Male and female mice were used in these studies and were maintained under specific pathogen-free conditions. This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The study protocol was approved by the University of California San Diego Institutional Animal Care and Use Committee as well as the Zürich Cantonal Veterinary Office in Switzerland.
Western blots.
Ten percent tissue homogenates were prepared in PBS using a Beadbeater tissue homogenizer. Extracts of 50–90 μg of protein were diluted in a Tris HCl-based buffer (10 mm Tris, 10 mm EDTA, 100 mm NaCl, 0.5% NP40, and 0.5% DOC) and digested with 5–50 μg/ml proteinase K (PK) for 30 min at 37°C. A lithium dodecyl sulfate (LDS)-based buffer was then added, and the samples were heated to 95°C for 5 min before electrophoresis through a 12% Bis-Tris precast gel (Invitrogen), followed by transfer to a nitrocellulose membrane by wet blotting. Proteins were detected with anti-PrP antibody POM1 (epitope in the globular domain, aa 121–230) (Polymenidou et al., 2008). For secondary detection we used an HRP-conjugated anti-mouse IgG antibody (Jackson Immunolabs). Signals were visualized with the ECL detection kit (Pierce).
Flotation assay.
Brain homogenates were lysed in cold TNE buffer (25 mm Tris HCl, pH7.5, 150 mm NaCl, 5 mm EDTA, and 1% Triton X-100). Extracts were mixed with two volumes of 60% Optiprep (Nycomed) to reach a final concentration of 40%. The mixture was carefully overlaid with a 5 and 30% Optiprep step-gradient and ultracentrifuged at 100,000 × g for 20 h at 4°C. Thirteen fractions were collected and used for SDS-PAGE and immunoblot analysis using anti-PrP (Pom1) and anti-flotillin (Abcam) antibodies.
Detection of insoluble PrP.
Brain homogenate samples were lysed in a Tris HCl-based buffer (10 mm Tris HCl, pH 8.0, 10 mm EDTA with 2% sarcosyl) and incubated at 37°C for 1 h before ultracentrifugation at 150,000 × g for 1 h. Supernatant and pellet fractions were collected and treated with a nuclease (Benzonase). Supernatant proteins were concentrated by methanol precipitation before Western blotting.
Velocity sedimentation.
Brain homogenates were lysed in a Tris HCl-based buffer containing 1% sarcosyl and incubated at 37°C for 10 min before layering over a 10–54% iodixanol gradient. Dextran blue (2000 kDa) was layered over a separate gradient as a reference standard. The samples were ultracentrifuged at 200,000 × g for 1 h and 175 μl fractions were collected and examined by Western blotting.
PrP peptide ELISA.
The peptide ELISA was performed as described by Lau et al. (2007) with minor modifications. Brain homogenate was mixed with an equal volume of lysis buffer (100 mm Tris HCl, pH 7.5, 150 mm NaCl) containing 2% sarcosyl, incubated for 10 min at 37°C, and centrifuged at 500 × g to remove cell debris. The supernatant was incubated with peptide-coated magnetic beads (M-280, Invitrogen) for 1 h at 37°C with constant shaking. The beads were washed with TBS containing 0.1% Tween 20, before denaturation with 0.1 m NaOH and neutralization with 0.3 m NaH2PO4. PrP was then measured by standard sandwich ELISA using a 96-well plate precoated with 2.5 μg/ml POM-2 antibody, and using a biotinylated POM-1 antibody (50 ng/ml), followed by streptavidin HRP and an Ultra TMB-ELISA substrate (Thermo Scientific) for detection. RML-infected and uninfected control brain samples were included in every experiment. Samples were run in triplicate.
Immunoprecipitation
15B3 immunoprecipitation.
Samples were homogenized in Prionics buffer (Prionics, Switzerland) supplemented with protease inhibitors (1 mm PMSF and Complete TM; Roche). IgM-Dynabeads (Invitrogen) were used for preclearing the samples for 2 h at 25°C. For the immunoprecipitation (IP), the sample was added to 15B3-conjugated IgM Dynabeads and shaken at 25°C for ∼16 h. Beads were washed and bound sample was eluted with an LDS-based sample buffer.
PrP 136–158 IP.
Brain homogenate was lysed in PBS buffer containing 1% Triton X-100 and protease inhibitors, and centrifuged at 500 × g for 15 min. The supernatant was incubated with 3 μg of 136–158 antibody in 450 μl of lysis buffer and shaken for 2 h at 25°C. A total of 25 μl of goat anti-human (Fab′)2-conjugated Dynal beads were added to each tube followed by a second round of incubation at 25°C for 16 h at 1000 rpm. The beads were washed in lysis buffer and eluted as described above. The eluted material was analyzed by SDS-PAGE, and immunoblotting was performed using the anti-PrP Pom1 antibody.
Histopathology and immunohistochemical stains.
Two-μm-thick sections were cut onto positively charged silanized glass slides and stained with hematoxylin and eosin, or immunostained using antibodies for PrP (SAF84), for astrocytes (GFAP), or microglia (Iba1). For PrP staining, sections were deparaffinized and incubated for 6 min in 98% formic acid. Sections were heated to 100°C in a pressure cooker or autoclaved in citrate buffer, pH 6.0, and then cooled for 3 min. Immunohistochemical stains were performed on an automated Nexus staining apparatus (Ventana Medical Systems) using an IVIEW DAB Detection Kit (Ventana). After incubation with protease 1 (Ventana) for 16 min, sections were incubated with anti-PrP SAF-84 (SPI bio; 1:200) for 32 min. Sections were counterstained with hematoxylin. GFAP immunohistochemistry for astrocytes (1:1000 for 24 min; DAKO) and Iba1 (1:2500 for 32 min; Wako Chemicals) for microglia was similarly performed, however with antigen retrieval by heating to 100°C in EDTA buffer (pH = 8.0).
Luminescent conjugated polymer staining of tissue sections.
The synthesis of polythiophene acetic acid (PTAA) (mean molecular weight, MW = 3 kDa) has been reported (Ding et al., 2000; Ho et al., 2002). Frozen mouse brain sections were dried for 1 h and fixed in 100% ethanol for 10 min. After washing with deionized water, the sections were equilibrated in the incubation buffer, consisting of 100 mm sodium carbonate at pH = 10.2. Luminescent conjugated polymers (LCPs) were diluted in the sodium carbonate buffer (0.01 μg/μl), then added to the brain sections and incubated for 30 min at room temperature, and finally washed with the sodium carbonate buffer.
Fluorescence microscopy.
Spectra were collected with a Zeiss Axioplan 2 microscope, fitted with a Spectraview 4.0 (Applied Spectral Imaging, Migdal, Israel) and a Spectra-Cube (interferometrical optical head SD 300) module with cooled CCD-camera, through a 405/30 nm (LP 450) or a 470/40 nm (LP 515) bandpass filter. The data were processed with SpectraView 3.0 EXPO. Spectra were collected from 10 individual spots within 3–5 plaques and from unstained regions and mock-inoculated negative control Tga20 mice. Fluorescent spectral unmixing was performed using the function in the software.
Statistical methods.
For spectral collection of PTAA bound to prion aggregates, brain sections were analyzed as follows: 10 individual spots within each of 3–5 plaques from each case were examined, yielding 30–50 measurements per mouse. The fluorescent intensity ratios were calculated (intensity at 532 nm/emission maximum, and 532/641) and mean and SD were recorded for each spectral ratio for each individual. An unpaired, two-tailed Student's t test was performed using mean values of single animals as observations. Differences were considered statistically significant at p < 0.05 (2-tailed).
Results
Generation and characterization of MoPrP167 transgenic mice
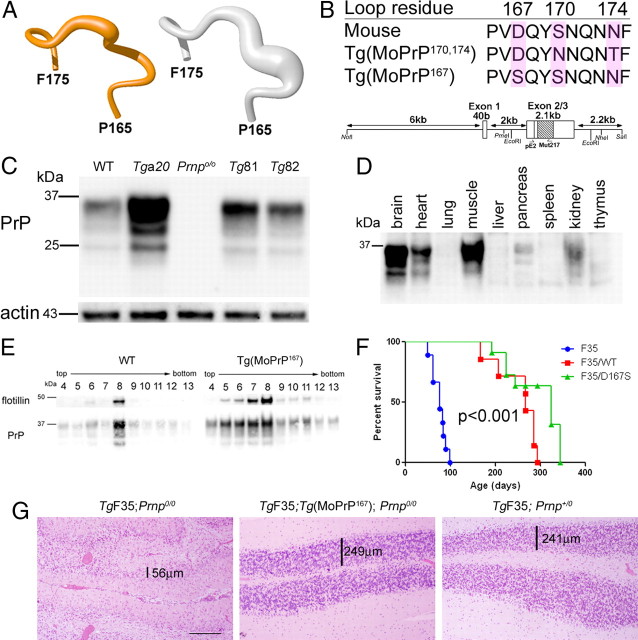
We generated 6 founder lines of transgenic mice that express mouse PrP with a D167S substitution, which imparts a structurally well ordered β2–α2 loop in the NMR solution structure determined at 20°C (Fig. 1A), similar to that in variant mouse PrP with S170N and N174T substitutions (MoPrP170,174, line Tg1020) (Sigurdson et al., 2009). To do this, we cloned the transgene into the “half-genomic” mouse Prnp minigene (pHGPrP) containing the prion promoter and the Prnp open reading frame (Fischer et al., 1996) (Fig. 1B), and introduced the construct into Prnp+/− zygote pronuclei by microinjection. We bred the Tg(MoPrP167) founders to B6J;129S5 Prnpo/o mice, and quantified the PrPC expression in brain from each mouse line by Western blot and ELISA. We chose two lines, 81 and 82, which express ∼3- to 5-fold and 1- to 2-fold PrP levels in brain when compared with WT mice, respectively (Fig. 1C). Heart, lung, liver, stomach, spleen, and kidney were histologically normal. The highest PrP-expressing tissues in the Tg(MoPrP167) mice were the CNS, skeletal muscle, and heart muscle (Fig. 1D).
Figure 1.
Tg(MoPrP167) mice express mouse PrP with a D167S substitution. A, Backbone fold of the β2–α2 loop (shown are residues 165–175) in the NMR structures at 20°C of MoPrP167 (orange; left) and MoPrP (white; right). The drawings are based on the atomic coordinates of the PDB deposits 2KU5 and 1XYX, respectively, as previously published. The thickness of the spline function through the Cα-atoms represents the precision of the structure determination. B, Amino acid sequence comparison of MoPrP, the previously studied MoPrP170,174, and the newly generated MoPrP167. Below the sequences is the half-genomic mouse Prnp gene construct with the enzyme restriction sites and PCR primer sites used for cloning. C, Brain PrP expression levels in wild-type, Tga20, Prnpo/o, and two lines of Tg(MoPrP167) mice, denoted Tg81 and Tg82. D, PrPC expression in peripheral organs of the Tg(MoPrP167) mouse shows that the highest-expressing organs were heart, muscle and kidney. E, Flotation assay of PrPC after density gradient centrifugation of brain shows a concurrence of PrP and flotillin in WT and Tg(MoPrP167) mice, indicating residency of PrP within detergent-resistant membranes. F, Survival curves of F35 mice expressing amino-proximally truncated MoPrP (Δ32–134) show rescue of the F35 phenotype through coexpression of MoPrP (WT) or MoPrP167. A log-rank test indicated further that the F35; Prnpo/o and the F35; Tg(MoPrP167) curves are significantly different. G, Cerebellum from TgF35 mice that co-express either MoPrP167 (middle) or MoPrP (right) show a rescue of the severe CGC layer degeneration that occurs in the TgF35; Prnpo/o mice (left). The width of the CGC layer is noted. Scale bar, 250 μm.
MoPrP167 in young mice is biochemically and functionally indistinguishable from WT-PrP in vivo
PrPC from the Tg(MoPrP167) mice was glycosylated and appeared to be processed normally (Fig. 1C). Similar to WT PrP, MoPrP167 was buoyant in density gradients and colocalized with flotillin, indicating residency within detergent-resistant membranes (Fig. 1E). Mutant PrPC functionality was assessed indirectly by crossing the Tg(MoPrP167) mice with hemizygous TgF35 transgenic mice expressing amino-proximally truncated PrP(Δ32–134) (Shmerling et al., 1998; Barmada and Harris, 2005; Radovanovic et al., 2005). TgF35 mice develop cerebellar granular cell (CGC) degeneration followed by death at 3–4 months of age, which is rescued by coexpression of wild-type MoPrP. To assess whether the MoPrP167 would also rescue the F35 phenotype, we crossed the Tg(MoPrP167) (line 82) and TgF35 mice. Similar to the TgF35 mice that coexpress wild-type MoPrP, the TgF35 mice that coexpressed MoPrP167 survived beyond 300 d of age (Fig. 1F) (p-value <0.001, log rank test) and did not develop CGC degeneration (Fig. 1G), indicating that the MoPrP167 was functioning similarly to MoPrP.
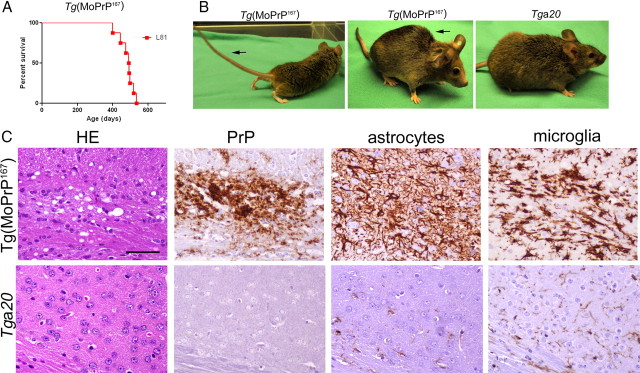
Aged Tg(MoPrP167) mice develop spontaneous neurologic disease, spongiform degeneration, and PrPSc deposits in brain
Aging Tg(MoPrP167) mice from line 81 developed a slowly progressive neurologic disease characterized by ataxia, tremors, weight loss, kyphosis, and in some cases priapism, which was associated with striking degenerative lesions in the brain (Fig. 2A–C). Both diffuse and scattered PrP-immunoreactive plaques and moderate neuropil vacuolation occurred in the basal ganglia, thalamus, and cerebral cortex (Fig. 2C). We detected intense astrogliosis and severe microglial activation in the vacuolated brain regions (Fig. 2C), as typically seen in transmissible spongiform encephalopathies.
Figure 2.
Aged Tg(MoPrP167) mice develop spontaneous neurologic disease with spongiform encephalopathy and prion plaques in brain. A, Survival curves of a line (L) of transgenic mice (L81) that expresses high levels of MoPrP167. B, Aged Tg(MoPrP167) mice (L81) with clinical neurologic signs including a stiff tail (left, arrow) and kyphosis (middle, arrow) are compared with a healthy Tga20 mouse (right). C, Brain at the level of the frontal cortex has extensive spongiform change (HE), PrP aggregates, and activated astrocytes and microglia in Tg(MoPrP167) mice but not in Tga20 mice. Scale bar, 100 μm.
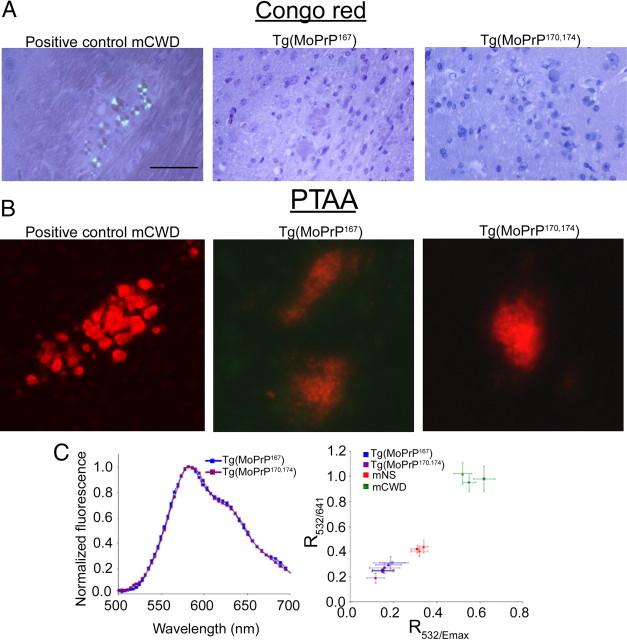
MoPrP167 aggregates in the brain are labeled by LCPs
The PrP aggregates in the Tg(MoPrP167) mice were not stained by Congo red (Fig. 3A), yet were stained by PTAA, which we have previously described as a new tool to distinguish prion strains (Fig. 3B) (Ding et al., 2000). PTAA molecules interact with amyloid (Nilsson et al., 2005, 2010) and emit light with a distinct pattern of spectral intensities depending on the amyloid bound (Nilsson et al., 2007; Sigurdson et al., 2007), and may therefore serve as a surrogate marker of conformation. To compare the conformation of PTAA bound to de novo MoPrP167 and MoPrP170,174 aggregates, we measured the emission spectrum from PTAA labeled brain sections. We found no differences in the PTAA emission spectra between the MoPrP167 and MoPrP170,174 PrP aggregates (Fig. 3C), suggesting that aggregates from mice expressing RL-PrP based on different residue substitutions have a common underlying structural feature.
Figure 3.
Congo red and PTAA binding properties of de novo MoPrP167 aggregates. A, PrP aggregates in Tg(MoPrP167) or Tg1020 (MoPrP170,174) mice do not stain with Congo red. B, PrP aggregates in Tg(MoPrP167) and Tg1020 (MoPrP170,174) mice bind PTAA. C, Emission spectra of PTAA bound to MoPrP167 and MoPrP170,174 plaques (left). Correlation diagram of the ratio of light intensity emitted at 532/emission maximum (R532/Emax) and 532/641 (R532/641) from PTAA bound to MoPrP167 and MoPrP170,174 plaques compared with mouse-adapted strains of chronic wasting disease (mCWD) or sheep scrapie (mNS) in individual mice (right). Scale bar, 100 μm.
Aggregated PrP in the Tg(MoPrP167) mice from line 81 is insoluble and PK sensitive
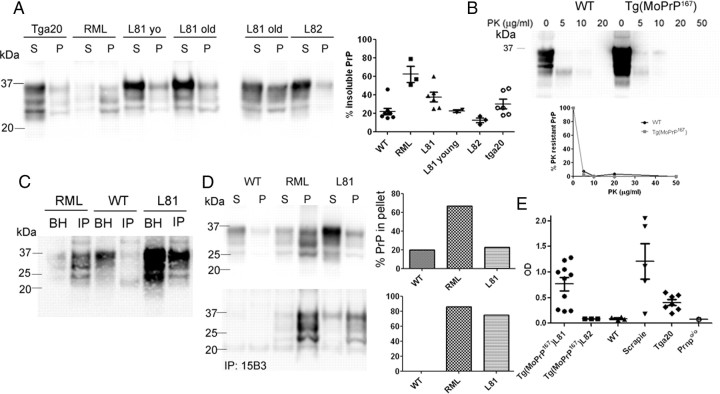
To assess whether the sarcosyl-insoluble PrP occurred in brains of the Tg(MoPrP167) mice, we performed ultracentrifugation on brain homogenates from young and old Tg(MoPrP167) mice, WT mice, mice infected with mouse-adapted scrapie (RML strain), and Tga20 mice, which overexpress WT mouse PrP (4- to 6-fold WT). We found that ∼40% of the PrP was sarcosyl-insoluble in aged line 81 mice, which was significantly more than in WT (22%), lower expressing line 82 (13%), and young line 81 (23%) mice, but less than in mice infected with RML prions (62%) (Fig. 4A). Also, aged untreated Tga20 mice, 370–500 d old, spontaneously showed moderate amounts of insoluble PrP (30%) (Fig. 4A).
Figure 4.
Detection of aggregated PrP in the brain of Tg(MoPrP167) mice, line Tg81. A, Soluble (S) and insoluble (P) PrP from brain homogenate were separated by ultracentrifugation and immunoblotting. Percentage of PrP in the pellet fractions was quantified and graphed, with each point representing an individual mouse. Samples labeled L81 yo were from 40-d-old mice, where as all other uninoculated mice were older than 300 d. B, PK digest of WT and Tg(MoPrP)167 brain homogenate using a gradient of PK concentrations at 37°C for 30 min reveals that the modified moPrP167 is sensitive to PK at >10 μg/ml PK. C, IP of PrP aggregates using a peptide-grafted antibody, 136–158, reveals abundant PrPSc in the immunoprecipitated fractions of RML-infected and line Tg81 (L81) mice, but not WT mice. D, Soluble and insoluble fractions were each divided for direct immunoblotting or for IP using PrPSc-specific antibody 15B3 followed by immunoblotting using anti-PrP antibody POM1. Quantification of the percentage of pellet to total PrPSc shows that PrPSc is enriched in the pellet fractions after 15B3 IP (bottom), indicating that the PrPSc is primarily insoluble. E, PrPSc-peptide ELISA of Tg81 mice of various ages was positive in all Tg81, Tga20 and scrapie-infected mice, and negative in Tg82 mice. Prnpo/o and WT mice were used to set the upper threshold for the negative samples.
Since PrPSc in prion-infected brain is typically PK resistant, we next measured the extent to which PK-resistant PrP could be detected in the brain of Tg(MoPrP167) mice. Brain homogenate samples from Tg(MoPrP167) and WT mice were incubated with increasing concentrations of PK. The PrP in the brain of aged Tg(MoPrP167) mice was primarily PK sensitive (Fig. 4B), as often seen in mice expressing mutant PrP (Hsiao et al., 1990; Jackson et al., 2009; Sigurdson et al., 2009) and in humans with certain sporadic and familial prion diseases, including fatal familial insomnia and Gerstmann-Sträussler-Scheinker disease (Medori et al., 1992; Colucci et al., 2006; Gambetti et al., 2008).
We next determined whether the aggregates were captured by PrPSc-specific antibodies, which may indicate conformational similarities with infectious PrPSc. We performed immunoprecipitation using the PrP peptide 136–158 grafted to an antibody scaffold that has been shown to bind PrPSc (Moroncini et al., 2004). Antibody 136–158 immunoprecipitated abundant PrPSc in brain from RML-infected and Tg(MoPrP167) mice, but not from WT mice (Fig. 4C). To determine whether the Tg(MoPrP167) PrPSc was soluble or insoluble, we performed ultracentrifugation and immunoprecipitated PrPSc from the supernatant and pellet fractions using the PrPSc-specific antibody 15B3 (Korth et al., 1997) (conformational epitope formed by PrP aa 142–148, 162–170, and 214–226). Here we found that the PrPSc detected in the line 81 mice was primarily in the insoluble fraction, similar to RML scrapie (Fig. 4D, bottom).
To further detect and quantify PrPSc aggregates, we assessed whether a PrPSc-specific peptide (aa 23–30) binds MoPrP167 aggregates in a prion assay that excludes PK or other enzyme treatment (Lau et al., 2007). We precipitated the PrPSc aggregates using PrP peptide bound to magnetic beads, then denatured the bound aggregates and detected PrP by antibody ELISA. We could detect PrP aggregates in all 13 of the line 81 and scrapie-infected mice, but in none of the WT, line 82, or Prnpo/o mice. Interestingly, aggregates were also detected in the brains of all aged Tga20 mice (Fig. 4E), yet signals were significantly lower than in the line 81 mice exceeding 180 d of age (line Tg81 average 0.77, range 0.25–1.3; Tga20: average 0.38, range 0.20–0.60; p = 0.04, Student's t test).
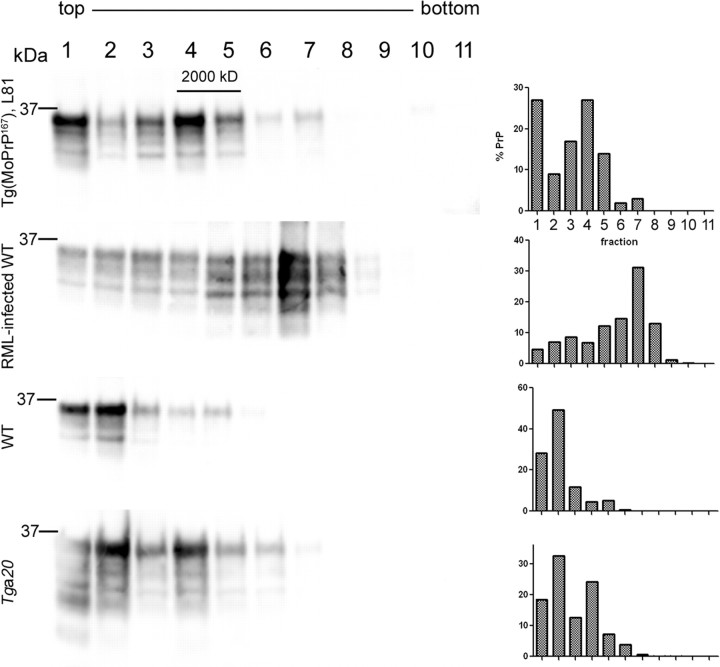
PK-sensitive PrPSc has been shown to correspond to lower density fractions after density gradient centrifugation (Pastrana et al., 2006). To determine whether the aggregates from the Tg(MoPrP167) mice were also in the lower density fractions, we measured the sedimentation properties of PrP from Tg(MoPrP167), uninfected WT, Tga20 and RML-infected mice in a 10–54% iodixanol gradient. WT PrPC was present primarily in the low density fractions 1 and 2, whereas RML was primarily in fraction 7. By comparison, the aged Tg(MoPrP167) showed abundant PrP in low density fraction 4 (27%), but with a very small amount (0.5%) detected in the most dense fractions 10 and 11, which was also seen with RML. These findings indicated that most MoPrP167 aggregates were smaller and more buoyant than RML, yet a rare dense aggregate population existed in common with RML (Fig. 5).
Figure 5.
Sedimentation properties of the MoPrP167, RML PrPSc, WT PrPC, and Tga20 PrPC from brain homogenates. The MoPrP167 migrated into fractions 4 and 5, which are more dense than WT PrPC, but less dense than RML-containing fractions.
Discussion
Here we show that a single-residue substitution in the β2–α2 loop leads to neurodegeneration and prion plaque formation, as seen in humans with various PRNP mutations (Masters et al., 1981). The Tg(MoPrP167) and the previously described Tg(MoPrP170,174) mice are based on rationally designed modifications of mouse PrP that mimic the well defined β2–α2 loop in the NMR structures at 20°C of the elk and horse prion proteins (Gossert et al., 2005; Pérez et al., 2010). We have previously found that the mutations in the β2–α2 loop of Tg(MoPrP170,174) led to de novo generation of PrP plaques, spongiform degeneration, gliosis, and infectious prions in vivo (Sigurdson et al., 2009). Here we showed that except for its PK sensitivity, the PrPSc in the Tg(MoPrP167) mice has biochemical properties in common with mouse scrapie PrPSc, including insolubility, recognition by conformational PrPSc-specific antibodies and peptides, and binding by PTAA.
Several of the familial prion diseases in humans and related mouse models also develop PrPSc with biochemical properties common to our Tg(moPrP167) mice, such as PK sensitivity and insolubility (Hsiao et al., 1990; Medori et al., 1992; Colucci et al., 2006; Gambetti et al., 2008; Jackson et al., 2009; Sigurdson et al., 2009). Together, these data suggest that aggregates precipitated by a Prnp mutation might form different multimeric structures than those acquired by infection. In support of this hypothesis, we found that PrP aggregates from our Tg(MoPrP167) mice were found primarily in lower density fractions than RML mouse PrPSc, indicating that the aggregates may be smaller or have a different shape. Our observations are consistent with previous work showing PK-sensitive PrP in lower density fractions (Pastrana et al., 2006). It will be interesting to determine in future studies the specific aggregate size and whether this low sedimentation property occurs with additional PK-sensitive prion strains in animals and humans.
In conclusion, we now have two lines of transgenic mice expressing MoPrP with different β2–α2 loop substitutions that confer RL-loop characteristics to the NMR structures at 20°C, which both develop PrP aggregates and spongiform degeneration (Sigurdson et al., 2009). The S170N, N174T substitutions also had a major impact on species barriers (Sigurdson et al., 2010). Whether the increased aggregation tendency and altered species barriers are due to the altered spatial loop secondary structure, the primary sequence, or a combination of both is not yet clear. Future studies of the β2–α2 loop substitutions should provide insights into the relative contribution of primary versus secondary structure of the PrPC substrate in prion assembly. The implicated key role of the β2–α2 loop region in aggregation and species barriers may thus provide a starting platform for novel approaches to unravel details of the roles of wild-type prion proteins in health and disease, with possible leads into the rational design of therapies to block PrP aggregation.
Footnotes
This study was supported by the National Institutes of Health (Grants NS055116, NS069566, and U54AI065359 to C.J.S.), the U.S. National Prion Research Program (to C.J.S. and A.A.), the European Union (TSEUR to A.A. and UPMAN to K.W.), the Swiss National Science Foundation, the Novartis Research Foundation (to A.A.), the National Competence Centers for Research on Neural Plasticity and Repair (to A.A.) and on Structural Biology (to K.W.), and ETH Zürich (to K.W.). K.W. is the Cecil H. and Ida M. Green Professor of Structural Biology at The Scripps Research Institute. A.A. is the recipient of an Advanced Investigator Award of the European Research Council. The confocal microscopy experiments were performed at the Human Embryonic Stem Cell Core facility at UC San Diego. We thank Dr. Fred Damberger for illustrating the β2–α2 loop in the NMR structures of the MoPrP167 and MoPrP, Dr. Michael Oldstone for the 136-158 antibody, and Marianne König, Donna Harclerode, Mona Farahi, Melanie Lucero, and the animal caretakers for technical assistance. The confocal microscopy experiments were performed at the Stem Cell Core Facility at University of California, San Diego.
The authors declare no competing financial interests.
References
- Aguzzi A, Calella AM. Prions: protein aggregation and infectious diseases. Physiol Rev. 2009;89:1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- Barmada SJ, Harris DA. Visualization of prion infection in transgenic mice expressing green fluorescent protein-tagged prion protein. J Neurosci. 2005;25:5824–5832. doi: 10.1523/JNEUROSCI.1192-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa R, Piccardo P, Ghetti B, Harris DA. Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron. 1998;21:1339–1351. doi: 10.1016/s0896-6273(00)80653-4. [DOI] [PubMed] [Google Scholar]
- Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- Colucci M, Moleres FJ, Xie ZL, Ray-Chaudhury A, Gutti S, Butefisch CM, Cervenakova L, Wang W, Goldfarb LG, Kong Q, Ghetti B, Chen SG, Gambetti P. Gerstmann-Sträussler-Scheinker: a new phenotype with ‘curly’ PrP deposits. J Neuropathol Exp Neurol. 2006;65:642–651. doi: 10.1097/01.jnen.0000228198.81797.4d. [DOI] [PubMed] [Google Scholar]
- Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci U S A. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Jonforsen M, Roman LS, Andersson MR, Inganas O. Photovoltaic cells with a conjugated polyelectrolyte. Synth Metals. 2000;110:133–140. [Google Scholar]
- Dossena S, Imeri L, Mangieri M, Garofoli A, Ferrari L, Senatore A, Restelli E, Balducci C, Fiordaliso F, Salio M, Bianchi S, Fioriti L, Morbin M, Pincherle A, Marcon G, Villani F, Carli M, Tagliavini F, Forloni G, Chiesa R. Mutant prion protein expression causes motor and memory deficits and abnormal sleep patterns in a transgenic mouse model. Neuron. 2008;60:598–609. doi: 10.1016/j.neuron.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- Gambetti P, Dong Z, Yuan J, Xiao X, Zheng M, Alshekhlee A, Castellani R, Cohen M, Barria MA, Gonzalez-Romero D, Belay ED, Schonberger LB, Marder K, Harris C, Burke JR, Montine T, Wisniewski T, Dickson DW, Soto C, Hulette CM, et al. A novel human disease with abnormal prion protein sensitive to protease. Ann Neurol. 2008;63:697–708. doi: 10.1002/ana.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossert AD, Bonjour S, Lysek DA, Fiorito F, Wüthrich K. Prion protein NMR structures of elk and of mouse/elk hybrids. Proc Natl Acad Sci U S A. 2005;102:646–650. doi: 10.1073/pnas.0409008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HA, Boissinot M, Bergeron MG, Corbeil G, Dore K, Boudreau D, Leclerc M. Colorimetric and fluorometric detection of nucleic acids using cationic polythiophene derivatives. Angew Chem Int Ed Engl. 2002;41:1548–1551. doi: 10.1002/1521-3773(20020503)41:9<1548::aid-anie1548>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Hsiao KK, Scott M, Foster D, Groth DF, DeArmond SJ, Prusiner SB. Spontaneous neurodegeneration in transgenic mice with mutant prion protein. Science. 1990;250:1587–1590. doi: 10.1126/science.1980379. [DOI] [PubMed] [Google Scholar]
- Jackson WS, Borkowski AW, Faas H, Steele AD, King OD, Watson N, Jasanoff A, Lindquist S. Spontaneous generation of prion infectivity in fatal familial insomnia knockin mice. Neuron. 2009;63:438–450. doi: 10.1016/j.neuron.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth C, Stierli B, Streit P, Moser M, Schaller O, Fischer R, Schulz-Schaeffer W, Kretzschmar H, Raeber A, Braun U, Ehrensperger F, Hornemann S, Glockshuber R, Riek R, Billeter M, Wüthrich K, Oesch B. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature. 1997;390:74–77. doi: 10.1038/36337. [DOI] [PubMed] [Google Scholar]
- Lau AL, Yam AY, Michelitsch MM, Wang X, Gao C, Goodson RJ, Shimizu R, Timoteo G, Hall J, Medina-Selby A, Coit D, McCoin C, Phelps B, Wu P, Hu C, Chien D, Peretz D. Characterization of prion protein (PrP)-derived peptides that discriminate full-length PrPSc from PrPC. Proc Natl Acad Sci U S A. 2007;104:11551–11556. doi: 10.1073/pnas.0704260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysek DA, Schorn C, Nivon LG, Esteve-Moya V, Christen B, Calzolai L, von Schroetter C, Fiorito F, Herrmann T, Güntert P, Wüthrich K. Prion protein NMR structures of cats, dogs, pigs, and sheep. Proc Natl Acad Sci U S A. 2005;102:640–645. doi: 10.1073/pnas.0408937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters CL, Gajdusek DC, Gibbs CJ., Jr Creutzfeldt-Jakob disease virus isolations from the Gerstmann-Straussler syndrome with an analysis of the various forms of amyloid plaque deposition in the virus-induced spongiform encephalopathies. Brain. 1981;104:559–588. doi: 10.1093/brain/104.3.559. [DOI] [PubMed] [Google Scholar]
- Medori R, Tritschler HJ, LeBlanc A, Villare F, Manetto V, Chen HY, Xue R, Leal S, Montagna P, Cortelli P, Tinuper P, Avoni P, Mochi M, Baruzzi A, Hauw JJ, Ott J, Lugaresi E, Autilio-Gambetti L, Gambetti P. Fatal familial insomnia, a prion disease with a mutation at codon 178 of the prion protein gene. N Engl J Med. 1992;326:444–449. doi: 10.1056/NEJM199202133260704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroncini G, Kanu N, Solforosi L, Abalos G, Telling GC, Head M, Ironside J, Brockes JP, Burton DR, Williamson RA. Motif-grafted antibodies containing the replicative interface of cellular PrP are specific for PrPSc. Proc Natl Acad Sci U S A. 2004;101:10404–10409. doi: 10.1073/pnas.0403522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson KP, Herland A, Hammarström P, Inganäs O. Conjugated polyelectrolytes: conformation-sensitive optical probes for detection of amyloid fibril formation. Biochemistry. 2005;44:3718–3724. doi: 10.1021/bi047402u. [DOI] [PubMed] [Google Scholar]
- Nilsson KP, Aslund A, Berg I, Nyström S, Konradsson P, Herland A, Inganäs O, Stabo-Eeg F, Lindgren M, Westermark GT, Lannfelt L, Nilsson LN, Hammarström P. Imaging distinct conformational states of amyloid-beta fibrils in Alzheimer's disease using novel luminescent probes. ACS Chem Biol. 2007;2:553–560. doi: 10.1021/cb700116u. [DOI] [PubMed] [Google Scholar]
- Nilsson KP, Ikenberg K, Aslund A, Fransson S, Konradsson P, Röcken C, Moch H, Aguzzi A. Structural typing of systemic amyloidoses by luminescent-conjugated polymer spectroscopy. Am J Pathol. 2010;176:563–574. doi: 10.2353/ajpath.2010.080797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana MA, Sajnani G, Onisko B, Castilla J, Morales R, Soto C, Requena JR. Isolation and characterization of a proteinase K-sensitive PrP(Sc) fraction. Biochemistry. 2006;45:15710–15717. doi: 10.1021/bi0615442. [DOI] [PubMed] [Google Scholar]
- Pérez DR, Damberger FF, Wüthrich K. Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein. J Mol Biol. 2010;400:121–128. doi: 10.1016/j.jmb.2010.04.066. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Moos R, Scott M, Sigurdson C, Shi YZ, Yajima B, Hafner-Bratkovic I, Jerala R, Hornemann S, Wuthrich K, Bellon A, Vey M, Garen G, James MN, Kav N, Aguzzi A. The POM monoclonals: a comprehensive set of antibodies to non-overlapping prion protein epitopes. PLoS One. 2008;3:e3872. doi: 10.1371/journal.pone.0003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovanovic I, Braun N, Giger OT, Mertz K, Miele G, Prinz M, Navarro B, Aguzzi A. Truncated prion protein and Doppel are myelinotoxic in the absence of oligodendrocytic PrPC. J Neurosci. 2005;25:4879–4888. doi: 10.1523/JNEUROSCI.0328-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. NMR structure of the mouse prion protein domain Prp(121–231) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- Riek R, Wider G, Billeter M, Hornemann S, Glockshuber R, Wüthrich K. Prion protein NMR structure and familial human spongiform encephalopathies. Proc Natl Acad Sci U S A. 1998;95:11667–11672. doi: 10.1073/pnas.95.20.11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M, Segal S, Kuff EL, Singer MF. The nucleotide sequence of repetitive monkey DNA found in defective simian virus 40. Cell. 1977;11:845–857. doi: 10.1016/0092-8674(77)90296-3. [DOI] [PubMed] [Google Scholar]
- Rülicke T. Pronuclear microinjection of mouse zygotes. Methods Mol Biol. 2004;254:165–194. doi: 10.1385/1-59259-741-6:165. [DOI] [PubMed] [Google Scholar]
- Shmerling D, Hegyi I, Fischer M, Blättler T, Brandner S, Götz J, Rülicke T, Flechsig E, Cozzio A, von Mering C, Hangartner C, Aguzzi A, Weissmann C. Expression of amino-terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell. 1998;93:203–214. doi: 10.1016/s0092-8674(00)81572-x. [DOI] [PubMed] [Google Scholar]
- Sigurdson CJ, Nilsson KP, Hornemann S, Manco G, Polymenidou M, Schwarz P, Leclerc M, Hammarström P, Wüthrich K, Aguzzi A. Prion strain discrimination using luminescent conjugated polymers. Nat Methods. 2007;4:1023–1030. doi: 10.1038/nmeth1131. [DOI] [PubMed] [Google Scholar]
- Sigurdson CJ, Nilsson KP, Hornemann S, Heikenwalder M, Manco G, Schwarz P, Ott D, Rülicke T, Liberski PP, Julius C, Falsig J, Stitz L, Wüthrich K, Aguzzi A. De novo generation of a transmissible spongiform encephalopathy by mouse transgenesis. Proc Natl Acad Sci U S A. 2009;106:304–309. doi: 10.1073/pnas.0810680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson CJ, Nilsson KP, Hornemann S, Manco G, Fernández-Borges N, Schwarz P, Castilla J, Wüthrich K, Aguzzi A. A molecular switch controls interspecies prion disease transmission in mice. J Clin Invest. 2010;120:2590–2599. doi: 10.1172/JCI42051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn R, Liu A, Lührs T, Riek R, von Schroetter C, López García F, Billeter M, Calzolai L, Wider G, Wüthrich K. NMR solution structure of the human prion protein. Proc Natl Acad Sci U S A. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]