Summary
During germ cell and preimplantation development, mammalian cells undergo nearly complete reprogramming of DNA methylation patterns. We profiled the methylomes of human and chimp sperm as a basis for comparison to methylation patterns of ES cells. While the majority of promoters escape methylation in both ES cells and sperm, the corresponding hypomethylated regions show substantial structural differences. Repeat elements are heavily methylated in both germ and somatic cells; however, retrotransposons from several subfamilies evade methylation more effectively during male germ cell development, while other subfamilies show the opposite trend. Comparing methylomes of human and chimp sperm revealed a subset of differentially methylated promoters and strikingly divergent methylation in retrotransposon subfamilies, with an evolutionary impact that is apparent in the underlying genomic sequence. Thus, the features that determine DNA methylation patterns differ between male germ cells and somatic cells, and elements of these features have diverged between humans and chimpanzees.
Introduction
In mammals, proper DNA methylation is essential for both fertility and viability of offspring (Bestor, 1998; Bourc’his and Bestor, 2004; Li et al., 1992; Okano et al., 1999; Walsh et al., 1998). DNA methylation in germ cells is required for successful meiosis (Bourc’his and Bestor, 2004), and blastocysts derived from ES cells lacking DNMTs cannot survive past approximately 10 days of development (Li et al., 1992).
Mammalian germ cells are derived from somatic cells, rather than being set-aside during the first zygotic cleavages. During germ cell development, the genome undergoes a wave of nearly complete demethylation and remethylation (Popp et al., 2010; Walsh et al., 1998). This reprogramming event correlates with re-establishment of totipotency and with the creation of sex-specific methylation patterns at imprinted loci (reviewed by (Sasaki and Matsui, 2008)). Germ cell methylation patterns are erased and reset during a second wave of epigenetic reprogramming that occurs during preimplantation development. Post-fertilization, DNA methylation levels reach a nadir around the 8-cell stage, after which methylation is re-written, attaining its somatic level by the blastocyst stage (Mayer et al., 2000). Since this is completed prior to the establishment of the inner cell mass from which cultured embryonic stem (ES) cells are derived, one can view ES cells and mature germ cells as the terminal products of the two landmark epigenetic reprogramming events in mammals.
Mobile genetic elements constitute roughly half of most mammalian genomes (Lander et al., 2001). Repression of transposons relies critically on DNA methylation and is essential for the maintenance of genomic stability in the long term and of germ cell function in the near term (Bestor, 1998; Bourc’his and Bestor, 2004; Okano et al., 1999; Walsh et al., 1998). At least in part, silencing of repeated DNA depends upon an abundant class of PIWI-associated small RNAs, called piRNAs (reviewed in (Aravin and Hannon, 2008)). In the absence of this pathway, methylation is lost on at least some element copies, transposons are de-repressed, and germ cell development is arrested in meiosis.
CpG dinucleotides are underrepresented in mammalian genomes, most likely because a higher rate of spontaneous deamination of methylated cytosines exerts evolutionary pressure for CpG depletion by frequent CpG-to-TpG transitions (Duncan and Miller, 1980; Ehrlich et al., 1990). Mammalian genomes contain areas of relatively high CpG density, called “CpG islands” (CGIs) (Gardiner-Garden and Frommer, 1987), which have avoided CpG depletion over evolutionary time. CGIs are frequently observed at promoters and in some cases have been shown to exert regulatory effects. Thus, selection against CpG depletion may reflect the importance of specific CpG dinucleotides as sequence-based binding sites or simply the requirement for a certain regional density of CpGs. As an alternative, the existence of CGIs may simply be an artifact of longstanding hypomethylation of these regions, and consequent relief from CpG erosion, in mammalian germ cells. Under this hypo-deamination model, selective pressure is independent of CpG density, per se, and CGIs may instead be a secondary consequence of protection from methylation at specific sites combined with prevalent methylation elsewhere in the genome (Cooper and Krawczak, 1989; Duncan and Miller, 1980; Ehrlich et al., 1990).
Studies encompassing evolutionarily distant species have shown that broad features of the epigenome, such as the high methylation levels of gene bodies and repeats, are deeply conserved (Zemach et al., 2010). In closely related species, however, fine-scale analysis of DNA methylation state reveals variation. The chimpanzee and human genomes share more than 95% sequence homology but display regions of differential methylation (Enard et al., 2004). Through focused studies, we have gained glimpses into the characteristics of the methylome and the evolutionary pressures that shape it. We wished to enable genome-wide comparisons of DNA methylation states in closely related species and to examine possible differences between the two major waves of epigenetic remodeling that occur during the mammalian life cycle. We therefore produced full-genome, single-CpG resolution DNA methylation profiles in human and chimp sperm and compared these with methylation maps from human ES cells (Lister et al., 2009).
Results
Methylomes of mature male germ cells in human and chimp
We conducted genome-wide shotgun bisulfite sequencing of individual sperm DNA samples isolated from two human and chimp donors (see supplementary information for details). Basic data analysis was conducted using a custom pipeline. We were able to determine methylation status for 96% of genomic CpGs in the human and chimp samples from a total of 28M and 27M CpGs, respectively (Table 1). Read coverage for CpGs on autosomes averaged 16X in human with an overall methylation level of ~70% for all CpG sites. For chimp we sequenced to an average coverage of nearly 14X and observed an average methylation level of ~67%. We did not observe significant methylation at non-CpG sites in either dataset. For comparison, we applied our analysis pipeline to a whole-genome bisulfite dataset from human ES cells (Lister et al., 2009). This dataset was comparable to our own, with 93% of CpG dinucleotides covered and an average depth of 14X on CpGs genome-wide.
Table 1. Shotgun bisulfite sequencing of human and chimp sperm methylomes.
Reads: total read ends sequenced (before filtering based on quality). Unique mapped: reads mapping optimally to a single location in the reference genome. Distinct reads: number of genomic locations to which a read maps; when multiple reads map to the same position, one with the best mapping score was selected at random, and all others discarded. Mismatches: average number of mismatches for the reads indicated in the distinct fragments column. Bisulfite conversion rate was calculated at non-CpG cytosines. Percent methylation: proportion of Cs in reads mapping over CpG dinucleotides. Parentheses indicate data from individual donors; sperm without parentheses indicates pooled donors.
| Species | Sample | Mapped | Distinct | Mis- matches |
BS conversion |
Methylation | CpG coverage |
CpGs covered |
|---|---|---|---|---|---|---|---|---|
| Human | Sperm (1) | 609,127,589 | 388,835,058 | 1.58 | 0.992 | 0.724 | 8.8 | 0.96 |
| Sperm (2) | 588,920,777 | 316,860,245 | 1.84 | 0.983 | 0.674 | 7.3 | 0.94 | |
|
Sperm
(both) |
1,198,048,366 | 705,695,303 | 1.70 | 0.988 | 0.701 | 16.1 | 0.96 | |
| ESC | 940,731,922 | 366,844,212 | 0.64 | 0.988 | 0.663 | 14.1 | 0.93 | |
| Chimp | Sperm (1) | 459,258,834 | 255,193,493 | 1.87 | 0.985 | 0.665 | 6.2 | 0.95 |
| Sperm (2) | 520,905,232 | 327,796,614 | 1.70 | 0.984 | 0.672 | 7.4 | 0.94 | |
|
Sperm
(both) |
980,164,066 | 582,990,107 | 1.78 | 0.985 | 0.669 | 13.6 | 0.96 |
We identified contiguous domains of low methylation, termed hypomethylated regions or HMRs, in a manner independent of genomic annotations such as CGIs and promoters. Since methylation levels in sperm were generally high, HMRs appeared obvious on browser plots as valleys in which methylation dropped to very low levels. To call HMRs in a statistically principled manner, we designed a novel computational approach, based on a 2-state hidden Markov model with Beta-Binomial emission distributions (see supplementary information). This algorithm identified ~79k HMRs in human sperm and ~70k HMRs in chimp sperm. Only ~44.5k HMRs were identified using the human ES cell dataset, despite similar sequence coverage and overall methylation level ((Lister et al., 2009); see Table 1 and S1A). The size of HMRs also differed between germ and ES cells. In both chimp and human sperm, the mean size of HMRs was ~1.8kb and the median was ~1.3kb. In ES cells HMRs showed a mean size of ~1.2kb with a median of 833bp. HMRs overlapped all classes of genomic annotation (see Table S1B).
Global comparisons among primate sperm methylomes and with human ES cells
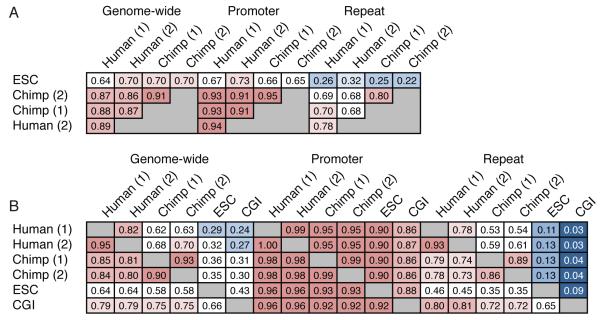
Average methylation levels differed by a small amount among the human donors (donor 1: 72%; donor 2: 67%), but were more similar among chimp donors (donor 1 and 2: 67%). The methylation status of individual CpGs of HMRs correlated very highly between individuals, with divergence being higher in repeats as compared to promoters (Fig. 1A, B). High inter-individual correlations at the CpG and the HMR levels imply that our datasets permit accurate calling of CpG methylation genome-wide.
Figure 1. A global view of sperm and ESC methylomes.
A. Correlations between methylomes with methylation levels measured at individual CpG sites. Correlations are displayed for CpGs genome wide, within promoters and within repeats, and correlation coefficients are colored blue to red to indicate low to high, respectively.
B. Overlap between sets of HMRs from human sperm, chimp sperm and ESC methylomes, along with annotated CGIs. Each cell gives the fraction of HMRs corresponding to the row that overlap HMRs corresponding to the column. Colors are overlaid as in A.
See also Supplementary Tables 1
We also compared methylation between species at an individual nucleotide level (see supplementary methods for details). As expected, the correlations between human and chimp sperm methylation are high, but the correlation remains generally highest within species.
We also directly compared the methylomes from each of the human and chimp donors with the human ES cell (ESC) methylome. The nucleotide-level correlations between sperm methylation of each of the four primate individuals was higher than their correlations with ESC methylation patterns (Fig. 1A). However, the human ESC methylome did show substantially higher correlation with the human germ cell methylomes than with those of chimp donors. Considered together these results indicate that, although waves of reprogramming in developing germ cells and embryos culminate in high genome-wide methylation, these two methylomes bear substantial differences overall.
Comparison of hypomethylated promoters between sperm and ESC methylomes
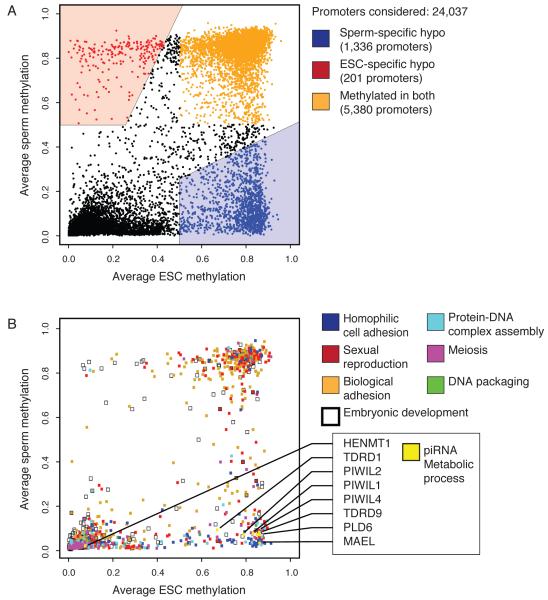
The majority of promoters are associated with HMRs in both sperm and ESC, indicating widespread bookmarking of promoters during both waves of epigenetic reprogramming. A number of promoters did show differential methylation, with 1336 showing sperm-specific HMRs but only 201 showing ESC-specific HMRs (Fig. 2A). Promoters hypomethylated in germ cells were strongly enriched for putative binding sites of transcriptions factors known to function in testis, including NRF1, NF-Y, YY1 and CREB (see Fig S1). A similar analysis of ESC-specific HMRs failed to yield significant results.
Figure 2. Differentially reprogrammed genes and their functions.
A. Average methylation through promoters (−1kbp to +1kbp) in human sperm and ESCs based on RefSeq gene annotations. Promoters that were hypomethylated only in sperm are shown in blue, only in ESCs in red, and promoters methylated in both are shaded orange.
B. Average methylation of promoters associated with GO terms found enriched in the sperm specific hypomethylated fraction (see A), with the addition of genes from the “embryonic development” term. Individual genes involved in the “piRNA metabolic process” are indicated as an example.
See also Supplementary Figure 1 and Supplementary Tables 2
Only the genes with sperm-specific promoter hypomethylation revealed a strong enrichment for functional (GO) categories. These were associated with germ cell functions (Fig. 2B; Tables S2) at distinct stages of gametogenesis (e.g. embryonic germ cell development and spermiogenesis). Thus, genes acting at developmental stages, potentially separated by decades, appear to maintain a permissive epigenetic state. Of the 8 genes analyzed from the piRNA metabolic process category, 7 showed promoter hypomethylation in sperm but not in ES cells and one was hypomethylated in both (Fig. 2B).
Retention of histones in human sperm was reported to be extensive (Hammoud et al., 2009). Our analysis of this data revealed a strong correlation between retained histones marked by H3K4me3 and HMRs at promoters. Among the 25.8k promoters marked by H3K4me3 in sperm, 91% overlapped an identified HMR. In general these results support prior observations that the presence of H3K4me3 at promoters is often accompanied by hypomethylation (Hammoud et al., 2009; Ooi et al., 2007).
It was previously posited that genes involved in early embryonic development had a distinct chromatin status in sperm, being hypomethylated, histone-retained, enriched in H3K4me3 marks, and thus poised for expression (Hammoud et al., 2009). At least with respect to DNA methylation, we do not detect a preferential link between HMRs in sperm and developmental regulators but instead widespread HMRs. One potential explanation for this perceived discrepancy is that our comparisons involve sperm and ES cells, while prior studies used a differentiated cell type to contrast with sperm.
The genes with promoters that lack HMRs in both sperm and ESC (N = 5,380; Fig. 2A) show strong enrichment for G-protein coupled receptors and genes involved in neurological functions (Table S2C and S2D). The reason why many of these genes, associated with highly specialized cell types, seem to lack promoter HMRs in sperm and ESC remains obscure.
Shared HMRs show distinct characteristics in sperm and ES cells
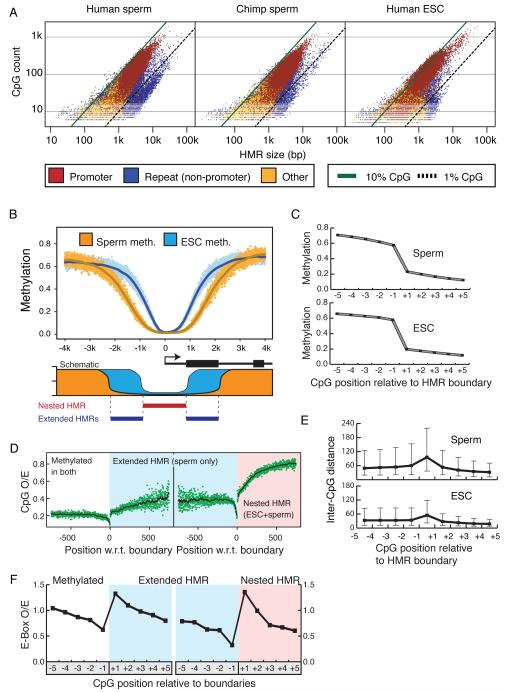
Differences in average size and CpG densities suggest that the HMRs emerging after germ cell reprogramming differ qualitatively from those emerging after zygotic reprogramming (Fig. 3A, Table S1A). The majority of HMRs have CpG density between 1% and 10%, and promoter HMRs fall almost exclusively in this range for the sperm methylomes. Those HMRs falling below 1% CpG density lie almost exclusively in repeats. These are overrepresented in human sperm relative to chimp sperm and human ESCs. Promoter-associated HMRs have sizes concentrated between 1kb and 10kb in human and chimp sperm, with an overall trend to be broader than promoter-associated HMRs in ESCs (Fig. 3A). A notable increase in CpG density accompanies narrowing of HMRs and results in a significant portion of ESC HMRs with a CpG density above 10%.
Figure 3. Characteristics of HMRs emerging from germline and somatic reprogramming.
A. Log-scale plot depicting the sizes (in bases) and number of CpGs for all identified HMRs in human sperm (left), chimp sperm (middle) and human ESCs (right). Diagonal lines indicate 10% CpG density (in green) and 1% CpG density (dashed line). HMRs are colored according to promoter overlap (red), overlap with repeats but not promoters (blue), or overlap with neither (orange).
B. Average methylation around all TSS overlapping HMRs in both sperm (orange) and ESC (blue); solid lines represent data smoothed using a 20 base sliding window. A schematic depicts the concepts of extended and nested HMRs at promoters.
C. Average methylation at the −5 to +5 CpGs around boundaries of extended sperm HMRs and nested ESC HMRs (with the +1 CpG defined as the first inside an HMR on either side).
D. Ratios of observed-to-expected CpG density for each nucleotide position relative to boundaries of extended sperm HMRs (left) and nested ESC HMRs (right). Solid lines indicate values smoothed using a 20 base sliding window.
E. Average inter-CpG distance for −5 to +5 CpGs around HMR boundaries of extended sperm and nested ESC HMRs.
F. Ratio of observed to expected frequencies of the CACGTG pattern at −5 to +5 CpGs for extended sperm and nested ESC HMRs.
See also Supplementary Figure 2 and Supplementary Table 3
To probe structural differences among HMRs in ES cells and sperm, we plotted the average methylation around HMR-associated transcriptional start sites (TSSs), genome-wide (Fig. 3B, upper). This revealed a general principle, that a core HMR in ES cells, referred to as a nested HMR (Fig. 3B, lower), often lies within an extended HMR in sperm. The median size of nested ESC HMRs is 1498, less than half the median size of 3109 for the sperm HMRs in which they reside. This phenomenon was also observed independently in a comparison of somatic and sperm HMRs, where variations in boundaries were additionally correlated with tissue-specific expression (Hodges et al., submitted). Extended HMRs are reminiscent of the concept of CpG shores (Doi et al., 2009), though in comparisons of sperm and ESC, we made no attempt to correlate gene expression with the widespread phenomenon of nesting that we report herein.
The observation of nested HMRs could arise either from a true expansion of the hypomethylated domain in sperm or as an artifact of sperm having less precise HMR boundaries than ESC. Examining degrees of change in methylation states across boundary CpGs in both cell types supports the former conclusion (Fig. 3C). Thus, nesting appears to represent a general phenomenon and likely reflects differences in the underlying mechanisms by which the boundaries of hypomethylated regions are determined during the waves of de novo methylation that lead to sperm and ESC.
As a step toward addressing such mechanisms, we asked whether any features are associated with HMR boundaries in either cell type. Two interesting characteristics emerged. Approaching the boundaries of either the extended sperm HMRs or the nested ES cell HMRs, CpG densities dropped just prior to the start of the HMR and rose dramatically again thereafter, though overall densities were higher in the nested portions (Fig. 3D). This reflects an increase in the average inter-CpG distance at the boundaries of HMRs (Fig. 3E). Because our method of identifying HMRs is agnostic to inter-CpG distance, this is not simply an artifact of our approach. One could imagine increases in inter-CpG distance interrupting a processive activity, preventing the spread of de novo methylation either directly or indirectly.
Though we had no a priori expectation that sequence features would reside at sperm or ESC HMR boundaries, we searched for motifs that might occur at or near boundary CpGs, independent of CpG density. We noted a trend towards enrichment for an ACGT motif at ESC boundary CpGs with a corresponding depletion immediately outside ESC HMRs (Fig. S2). This pattern was not significantly enriched at the boundaries of extended sperm HMRs. Building upon this observation, we also searched for larger motifs, focusing on those containing a central CpG core. Patterns with strong differences across HMR boundaries tended to have the ACGT core (Table S3). The most enriched pattern for sperm was AACGTT. For ESCs we saw a well-known E-box pattern, CACGTG. Plotting observed-to-expected frequencies centered on CpGs around boundaries of extended and nested HMRs (Fig. 3F), there was a clear depletion just outside each boundary followed by a sharp enrichment at the boundary CpG for each pattern in the appropriate cell type (Fig. S2B). These results raise the possibility that one or more DNA binding proteins might localize to HMR boundaries during waves of de novo methylation and help to define transitions in methylation states.
Differential repeat methylation in sperm and ES cells
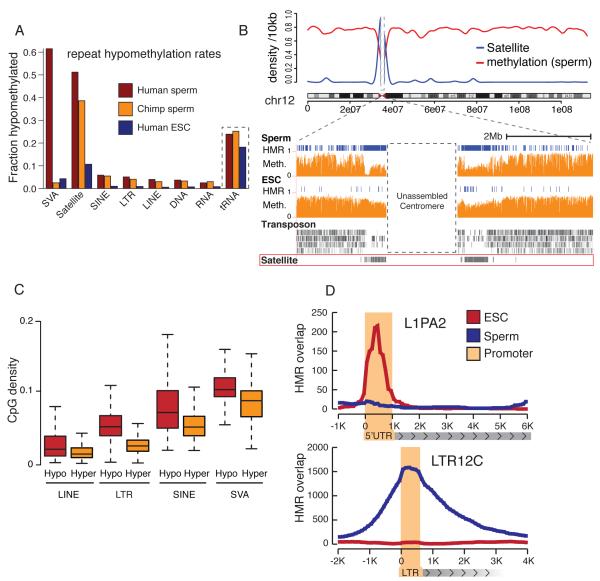
Consistent with prior observations and with the known role of DNA methylation in transposon silencing, most repeat elements were highly methylated in both sperm and ESC. However, a substantial fraction of HMRs overlapped transposons in chimp and human sperm, with all repeat classes represented (Fig 4A, Table S1B). Fewer repeat-associated HMRs appeared in ESCs. In sperm, HMRs collectively contained 4-5% of all bases assigned to repeats, compared to 1.3% in ESC (see Table S1B). Overall, this suggests that different mechanisms, with different stringencies, direct repeat methylation during germ cell and preimplantation development.
Figure 4. Differential repeat methylation during male germ cell and somatic reprogramming.
A. For each repeat class, the proportion of elements that overlap HMRs is shown for human sperm (red), chimp sperm (orange) and ESCs (blue).
B. Upper: Sliding 10kb window average methylation level across chromosome 12 (red) and satellite density (blue). Lower: Chromosome 12 centromeric region with HMRs (blue) and methylation level (orange) for human sperm and ESCs.
C. CpG densities of hypomethylated repeat copies (red) and methylated repeat copies (yellow) for LINEs, LTRs, SINEs and SVAs.
D. HMR overlap distribution around full length L1PA2 and LTR12 ERV9 elements for human sperm (blue) and ESCs (red).
See also Supplementary Figure 3 and Supplementary Tables 4
Sperm-specific satellite hypomethylation is concentrated at centromeres
We noted a strong decrease in methylation of sperm DNA within pericentromeric regions, extending several megabases outward from the unassembled core centromeres (Fig. 4B). This was not seen in ESC or in terminally differentiated cells (Hodges et al., submitted). This striking pattern was attributable to sperm-specific hypomethylation of ~75-80% of the satellite repeats concentrated in pericentromeric regions (Fig. 4A). In ESC, only 16% of pericentromeric satellites were hypomethylated, a figure in accord with the overall hypomethylation rates of non-pericentromeric satellites in ESC and sperm (Table S4A). Prior studies of mouse germ cells using methylation sensitive restriction enzymes had noted selectively low methylation at pericentromeric satellites, suggesting that this is a conserved property (Yamagata et al., 2007).
Retroelement methylation patterns are determined at the subfamily level
Proper methylation of retrotransposons is required for transcriptional silencing of full-length and potentially active copies (Bourc’his and Bestor, 2004; Goodier and Kazazian, 2008; Walsh et al., 1998). However, specific retroelements can be active or unmethylated in male germ cells (e.g., AluY and AluYa5) (Schmid, 1991). Given our read lengths, we were able to address the methylation state of virtually all repeat families and most individual copies (see Table S4B).
Overall, retrotransposon copies that were full length or close to consensus showed a slight bias towards hypomethylation (Fig. S3A, S3B). However, neither of these attributes could explain the variation observed in retrotransposon methylation. Hypomethylated repeat copies did tend to have greater CpG density, especially within the LTR and SVA classes (Fig. 4C). For LINEs, LTR elements, and terminal repeats, HMRs concentrated within regulatory regions, which often show higher CpG density than their coding regions (Figs. 4D, S3C, D, Tables S4D, G). SINE elements displayed a more uniform hypomethylation (Fig. S4E). Thus, similar mechanisms appear to define HMRs in both repeat and non-repeat portions of the genome, since for most repeats, there is a strong association of sperm HMRs with regulatory regions.
Among the LINEs, subfamilies of L1 were often hypomethylated in both sperm and ES cells and these trended strongly towards the active groups (Table S4E and S4H). L1PA subfamilies are considered the most active in the human genome (Khan et al., 2006), and the youngest of these (L1HS and L1PA2) were among the very few subfamilies enriched for hypomethylation in ES cells relative to sperm. Specifically in sperm, we noted hypomethylation of several other L1 families (e.g. L1PA4-16 and L1M3).
Among LTR subfamilies, sperm HMRs were enriched for ERV elements (Table S4C). Hypomethylated copies exist either as part of full-length provirus-like elements or as solo LTRs, with the greatest enrichment for LTRs belonging to “class I” elements (e.g. LTR12; see Table S4D and S4G). The few LTR subfamilies with more hypomethylated copies in ESC than sperm are all recently derived, human-specific ERVs (e.g., LTR5 and 13 and HERVH LTR7).
Sperm hypomethylation has been previously reported for primate Alu elements (Kochanek et al., 1993; Liu et al., 1994), and our data revealed several Alu subfamilies with differential methylation in sperm and ES cells, e,g., the AluY subfamily (Tables S4F and S4I). The more precisely defined AluYa5 (human) and AluYd4 (chimp) showed extreme enrichment for hypomethylation in sperm.
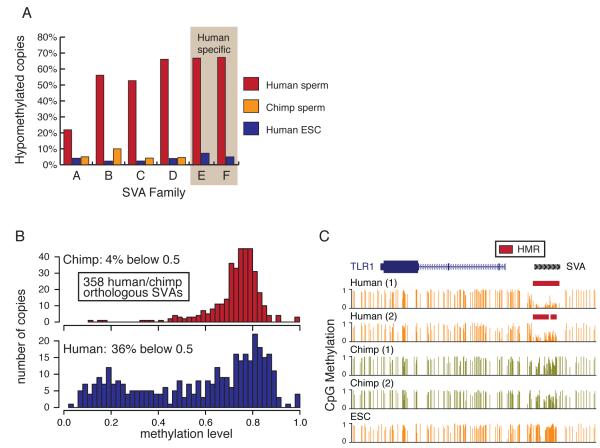
Species-specific methylation of the SVA element
SVA elements showed strong, species-specific differences in methylation in human and chimp sperm (Fig. 4A). SVAs are composite elements consisting of hexameric repeats, an Alu-like region, a VNTR (variable number of tandem repeats) region and a SINE-R (Shen et al., 1994). SVA elements were active in the most recent common ancestor of chimp and human (Mills et al., 2006), and multiple examples of neoinsertions suggest that they still cause genomic rearrangements and disease in human (Ostertag et al., 2003).
Among the SVAs, the youngest subfamilies, D-F (Wang et al., 2005), showed the greatest frequency of hypomethylation in human sperm (Fig. 5A). Notably, these have a higher CpG density than do older subfamilies. 358 SVA insertions can be assigned as high-confidence orthologs between human and chimp, which remain highly similar in sequence (see supplement). Methylation through these element copies was distributed through the full range from very low to very high average methylation, with two modes near 20% and 80% methylation (Fig. 5B). In human sperm, 35% of orthologous SVAs had a methylation level below 50%. In sharp contrast, only 6% of copies fell below 50% methylation in chimp. We also annotated 921 SVA elements that appear to represent new insertions occurring after the human-chimp divergence (Mills et al., 2006). 852 (93%) of these were hypomethylated in sperm compared with only 62 (7%) in ES cells (Fig 5A). Considered together, our data indicate that SVA elements have come under different degrees of epigenetic control in the human and chimp lineages.
Figure 5. Divergent methylation of SVA elements between human and chimp.
A. Proportion of hypomethylated SVA copies hypomethylated according to subfamily (A to F) for human sperm (red), chimp sperm (orange) and ESCs (blue).
B. The distribution of average methylation levels is shown for 358 human (lower) and chimp (upper) SVAs forming high-confidence orthologous pairs.
C. An SVA insertion shared by human and chimp but with differential methylation between species.
Many SVA insertions occur at or around promoters (Lander et al., 2001; Mikkelsen, 2005), and these elements often have a CpG content high enough to fit the traditional definition of a CpG island. Given their properties, SVA elements have the potential to introduce differential, species- and cell type-specific methylation near genes that may be relevant for their regulation. Figure 5C exemplifies such a situation where, in the case of TLR1, no HMR exists near the promoter in chimp sperm or human ES cells, but one is contributed in human sperm by a nearby SVA element. Although sperm are largely transcriptionally silent, similar HMRs are expected to exist in transcriptionally active developing germ cells (data not shown).
Signatures of selection accompany differential methylation between primates
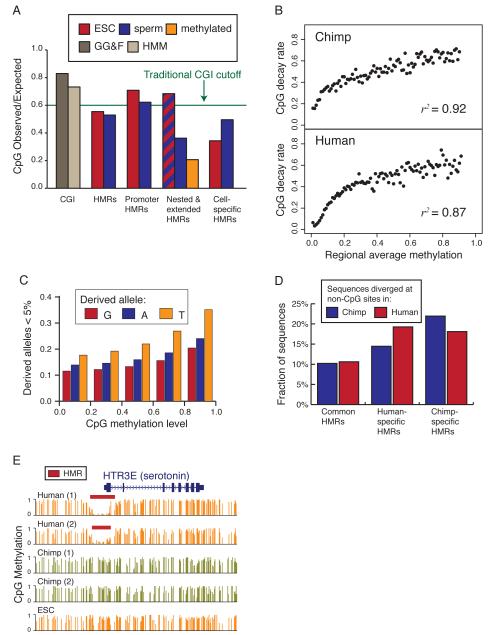
CGIs are the most well known evolutionary signature of vertebrate DNA methylation. Their original definition required a CpG observed-to-expected (o/e) ratio of at least 0.6. Although the full set of HMRs in human sperm and ESCs did not reach this empirical cut off, they did pass the 0.4 benchmark used by Weber and colleagues (Fig. 6A) (Weber et al., 2007). In general promoter-associated HMRs did surpass the 0.6 o/e cut off in both sperm and ESC.
Figure 6. Sequence features associated with methylome divergence.
A. Ratio of observed-to-expected CpG density across all HMRs, those overlapping promoters, those sperm- or ESC-specific, and the extended/nested HMRs. Data for sperm is indicated in blue and for ESC is indicated in red; orange indicates ratio immediately outside extended HMRs.
B. Frequency of regions under CpG decay as a function of methylation for both human and chimp at locations of HMRs in the other species. Decay is presented for chimp in the upper panel and for human in the lower panel.
C. Proportion of sequences displaying over 1% nucleotide divergence relative to the inferred ancestor using gorilla as an out-group and counting only non -CpG sites.
D. The promoter of the human HTR3E (serotonin receptor) gene contains an HMR in both human donors but in neither chimp donor.
See also Supplementary Figure 5 and Supplementary Tables 5
The differences in CpG density in nested and extended HMRs (Fig. 3B) imply distinct CpG depletion pressure in these regions. Average CpG composition genome-wide is ~0.2 o/e, but reaches ~0.35 in extended HMRs and 0.68 in nested HMRs. We analyzed sperm-specific and ESC-specific HMRs in an attempt to decompose the CpG depletion pressure exerted by the two methylomes. The ESC-specific HMRs reached only 0.35 o/e CpG composition, while the sperm-specific HMRs reached a CpG composition of 0.5.
The life cycle of a germ cell can be separated into two components. The first is the time from fertilization to the time that somatically derived, primordial germ cells (PGCs) reach the genital ridge. Second is the time during which the PGC develops into a mature germ cell, which contributes to the zygote. The latter period generally spans from birth to the end of the reproductive life of the animal. Our data suggest a model in which methylation patterns present during both of these intervals shapes genomic CpG distributions but indicate a greater influence of methylation profiles during germ cell maturation (Fig. 6A).
We sought to measure the degree to which differential methylation could lead to CpG decay over the ~6My of divergent evolution separating human and chimp. We focused on regions that qualified as HMRs in either chimp or human. since These regions could have either lost methylation along one lineage or gained methylation along the other. For a given regional methylation level, we measured CpG decay as the proportion of regions having lost more than 5% of inferred ancestral CpGs (using gorilla as outgroup) and plotted the relationship between average methylation and decay rate (Fig. 6B). The correlation between regional methylation level and CpG decay was extremely strong for both human and chimp. These results indicate that CpG decay is appreciable as a function of methylation even over relatively brief evolutionary periods.
This observation predicted that we might see signatures of selective pressure preventing erosion of some CpGs that are maintained despite germline methylation. To address this question, we analyzed segregating sites at CpG dinucleotides using data from the HapMap 3 project (CEU population; (Altshuler et al., 2010)). CpGs were treated symmetrically, so each derived allele at these sites can be classified as A, G or T. As expected, segregating sites with T as the derived allele represent the vast majority.
We generated frequency spectra for each derived allele nucleotide with sites classified according to their methylation level in sperm (Fig. S4). As methylation levels increased, derived allele frequencies shifted towards the low ends of the spectra (Fig. 6C and S4). This shift was observed not only for derived TpG alleles, which could be explained by an extreme bias in mutation rate, but also for ApG and GpG derived alleles. One interpretation of these findings is that selection is on average weaker at individual CpG sites with lower sperm methylation. Such an interpretation is consistent with recent findings of Cohen et al. (Cohen et al., 2011) who used sophisticated evolutionary models to posit that selection for high CpG content is not a significant factor contributing to maintenance of CGIs in the genome.
The strong connection between HMRs and gene promoters suggests that the evolutionary gain or loss of HMRs may be associated with changes in selective pressure on functional regulatory regions. To investigate this possibility we analyzed sequence divergence in HMRs, focusing on those that are human- or chimp-specific. Since these differentially methylated regions will have different rates of C-to-T transitions, we counted changes from the inferred ancestor only at non-CpG sites. Genomic intervals differing by more than 1% relative to the inferred ancestor were counted as having divergent sequences.
Only 10% of HMRs shared between human and chimp showed divergence from the ancestral sequence at non-CpG sites (Fig 6D). At chimp-specific HMRs, 15% of human sequences and 19% of chimp sequences diverged from the inferred ancestor. At human-specific HMRs, 22% of human sequences diverged and 18% of chimp sequences diverged. These results indicated that changes in methylation state between human and chimp are associated with accelerated non-CpG sequence divergence. Interestingly, in both cases the species with lower methylation state had a greater rate of divergence, which is consistent with adaptation at novel regulatory regions as a driver for these changes.
We only identified 104 promoters that are hypomethylated in human but not in chimp sperm and only 52 genes with differential promoter methylation in the opposite orientation. Neither set showed significant enrichment for any ontology category. However, analysis of genes with promoters within 10kb of an identified human-specific sperm HMR revealed a strong enrichment for neuronal functions (see Tables S5). The HTR3E gene, a serotonin receptor subunit, is an example of such a gene, whose promoter is selectively hypomethylated in human sperm (Fig. 6E).
Discussion
Sperm methylation patterns are conserved
Overall, sperm methylation patterns were highly similar in all our samples. However, there were differences, even among individuals. There has been much discussion regarding the role of germline transmission of epigenetic marks in inter-individual variation (Curley et al., 2011). Changes in epigenetic state could allow flexibility in phenotype that could be reverted over short time spans if a trait became disadvantageous. Erosion of CpG content provides a mechanism to allow fixation of a positive trait in the long run. Thus, changes in DNA methylation patterns preceding changes in DNA sequence presents an attractive model for at least one mode of adaptation. While evaluating such hypotheses will require many more datasets, the work presented here builds a firm foundation for such studies.
Most promoters have HMRs in sperm
Global resetting of DNA methylation patterns happens twice during mammalian development: once during germ cell development and once early in embryogenesis. Our data permit a genome-scale analysis of these two events. While high genome-wide levels of methylation are re-established during both waves of epigenetic remodeling, some regions are protected and establish HMR boundaries that appear relevant even in fully differentiated somatic cells (Hodges et al., submitted). A few promoters showed selective hypomethylation in sperm, and these are strongly enriched for annotations related to germ cell processes. Far fewer were selectively hypomethylated in ES cells and these were not enriched in any particular annotation category. Promoters of genes retaining nucleosomes have recently been shown to be hypomethylated in human sperm (Hammoud et al., 2009), and both of these features have been proposed to aid rapid activation during development. We find that gene-associated hypomethylation in sperm can be extended to more than 70% of all annotated genes in both human and chimp. Among these we failed to find any enrichment for regulators of early development. Instead, it seems that promoter regions are instead generally identified and bookmarked in sperm (see (Zaidi et al., 2010)).
Distinct processes of HMR formation shape germ cell and ESC methylomes
Genome-wide, CpG sites seem to adopt a methylated state by default (Edwards et al., 2010). This raises the problem of precisely how regions that become HMRs are identified as such. Regions of hypomethylation at promoters have been correlated with regulatory DNA in various developmental contexts (Illingworth et al., 2008; Lister et al., 2009; Rollins et al., 2006; Straussman et al., 2009). Based upon analysis of histone marks and on the proposed binding properties of Dnmt3s (Dhayalan et al., 2010; Zhang et al., 2010), active transcription and accompanying methylation of K4 on histone H3 are thought to locally inhibit the methylation machinery. This could enable large-scale recognition of promoter regions if widespread transcription occurs during fetal germ cell development as genomic methylation patters are erased and reset. It is also plausible that specific protein/DNA complexes act locally even in the absence of active transcription, to prevent access by de novo methyltransferases. Proteins observed to function as boundary elements, such as CTCF and Sp1 (reviewed in (Gaszner and Felsenfeld, 2006)), provide candidates for such functions.
Despite overall similarity in the sets of promoters they mark, the HMRs observed at promoters in mature male germ cells usually extend beyond the boundaries of HMRs in ES cells when the two overlap. These wider HMRs do not seem to reflect less precision in HMR boundaries, as methylation differences across HMR boundaries are similar between sperm and ES cells. Because this “nested” HMR phenomenon is observed at so many promoters, it does not seem to be associated with the regulation of any specific genes during germ cell development. We have observed a clear increase in CpG content through the extended portion of these HMRs relative to the genome-wide average, suggesting they have to some degree avoided pressure to decay, and hence are more than a transient state. The phenomenon that we observe is similar to the concept of CpG shores (Doi et al., 2009). Perhaps the extended HMRs in germ cells presage the extent of “shores” that correlate with changes in gene expression.
Our data suggest that HMRs emerge from de novo methylation in male germ cells with sizes that differ from those that emerge from somatic reprogramming. Thus, despite involvement of similar methyltransferases and targeting of similar sets of sequences, the determinants of HMR sizes likely differ between the two reprogramming events. We have begun to see hints to the mechanisms determining such differences by comparing boundary-associated motifs in sperm and ESCs.
Transposon hypomethylation in sperm
It is thought that germ cell genomes must be closely guarded from the activity of mobile genetic elements. While repeats were generally heavily methylated, we did find HMRs that overlapped repeats, and these were substantially more prevalent in sperm. We and others have characterized a conserved, small RNA-based silencing pathway, termed the piRNA pathway, that is important for recognizing and silencing mobile elements in germ cells (Aravin and Hannon, 2008). Our data indicate both individual element copies and broader element subfamilies can evade piRNA-based silencing. Yet, both these element copies and element families are often efficiently silenced during preimplantation development. This suggests fundamental differences in the mechanisms that recognize repeats and mark them for repression during the two major waves of epigenetic reprogramming in mammals.
Examining patterns of repeat-associated HMRs is potentially enlightening. HMRs are more prevalent in younger transposon subfamilies and the hypomethylated regions themselves tend to overlap with promoters or regulatory regions, just as they do in genes. Thus, it may be that active elements evade default methylation by being initially recognized as gene-like as a consequence of their binding transcription factors and possibly even being transcribed. In these cases, we imagine that silencing of most elements would be enforced by the piRNA pathway but that some sites, such as those we observe herein, might still escape. A number of examples can be cited in support of this hypothesis. The 5′UTRs of the L1PA subfamilies are known to carry conserved YY1 binding sites, while other recent subfamilies acquired RUNX3 and SRY binding motifs, all of which could promote transcription in developing germ cells (Khan et al., 2006; Lee et al., 2010). Similarly the sperm enriched hypomethylated EVR9 LTR12 elements have been shown to bind NF-Y, MZF1 and GATA-2 in erythroid K562 cells (Yu et al., 2005). In each of these cases, HMRs within these elements tend to encompass such potential transcription factor binding sites.
Similarly, Alu RNAs have been detected in human sperm (Kochanek et al., 1993). This suggests a potential link between Alu HMRs and the transcriptional activity of individual repeats, though previous studies also reported that the binding of SABP across Alu elements in sperm prevents their methylation (Chesnokov and Schmid, 1995). Interestingly, Alu hypomethylation is not seen in female germ cells (Liu et al., 1994) and has been proposed as one mediator of sex-specific imprints.
Centromeric satellite methylation
Satellites resist methylation in sperm when localized in clusters at centromeres, but are generally methylated when located elsewhere even if they are clustered. This is consistent with previous observations made in mouse through the use of methylation sensitive enzymes (Yamagata et al., 2007). Recent reports have shown that the transient transcriptional activation of paternal pericentromeric satellites was essential for centromeric heterochromatin formation in 2-cell zygotes (Probst et al., 2010). This could indicate that hypomethylation of satellite repeats in male germ cell marks paternal centromeres, in a manner similar to imprinting, allowing their rapid transcriptional activation upon fertilization.
In addition to a characteristic location within chromocenters in sperm, centromeres display a distinct chromatin structure differentiating them regionally during meiosis from other chromosomal regions (reviewed by (Dalal, 2009)). This has prompted suggestions that centromeric chromatin states might be critical for proper meiosis, a hypothesis strongly supported by our observation of selective hypomethylation of megabase domains of centromeric satellite clusters. Prior studies have demonstrated that de-repression of satellite repeats in mitotic cells creates segregation defects due to the formation of anaphase bridges (Frescas et al., 2008). Low methylation levels have also been correlated with the ability to bind cohesin complexes (Parelho et al., 2008). Considered as a whole, these observations suggest a model in which selective hypomethylation of centromeric satellites might be critical for accurate chromosome segregation during meiosis.
Differential repeat methylation between species
The most striking example of species-specific methylation to emerge from our analysis involved the SVA elements. These primate-specific composite elements contain a high density of CpGs, remain active in human and chimp and include many copies that are clear orthologs between human and chimp (Bantysh and Buzdin, 2009; Mills et al., 2006). Transduction of SVAs has been implicated in human diseases and gene formation (Damert et al., 2009; Ostertag et al., 2003). Our results indicate that for a subset of SVA elements, the ability to methylate these elements has either been acquired along the chimp lineage or lost in the human lineage during the past six million years, despite very little sequence change in these elements.
Mutual canalization of the genome and the epigenome
It has been thought that CGIs arose as the result of protection from methylation-associated deamination over long evolutionary periods. This is consistent with the observed correlation between the location of CGIs and regions that lack methylation in both germline and somatic cells. However, recent results have pointed to functions for CGIs that may be associated with their high CpG density (Thomson et al., 2010), with the plausible interpretation that selection may be acting to preserve CpG density in CGIs. We find that while most CGIs fall within HMRs of sperm, most HMRs extend well beyond the annotated CGIs, even using weaker CGI definitions. Thus, hypomethylated regions in male germ cells do not appear to require a critical CpG density to avoid methylation. Instead, our results are consistent with CGIs arising as a consequence of different mutational pressures rather than selection for CpG density.
In our datasets, signatures of deamination induced CpG depletion are clear. Yet we also observe CpG depletion from many sperm and ES cell HMRs. Several scenarios could resolve this conundrum. For example, such regions may have been methylated for substantial periods prior to assuming their unmethylated status. Thus, they may have decayed at some time in the past but are now stabilized by their hypomethylated status. Such sites could also actually be methylated during a period of germ cell development to which our current datasets are blind (e.g. in fetal gonocytes or female germ cells). In accord with this explanation, we have observed distinct CpG densities associated with sperm-specific and ESC-specific HMRs. Moreover, at HMRs where the only central, nested portion is hypomethylated in ES cells, we observe greater CpG retention through regions hypomethylated in both ES cells and sperm. Overall, we cannot exclude a model in which selection acts to preserve critical functions requiring specific local CpG densities. However, our results lend additional support to recent conclusions of Cohen et al. (Cohen et al., 2011) whose sophisticated evolutionary modeling showed that CGIs can be explained without invoking selection on CpG sites. Our results suggest a refinement of the hypo-deamination model in which CpG retention is a function of the time spent hypomethylated during each generation in germ cells and their somatic precursors.
The detailed comparative analysis performed here has revealed that, over the ~6M years since the divergence of human and chimp, most patterns of DNA methylation remain conserved in male germ cells. We have directly related evolutionary changes in CpG methylation with loss of CpG dinucleotides and have shown that even small differences in methylation can lead to substantial loss of CpGs over relatively short evolutionary periods. At the same time, there are many genomic regions that are highly conserved in sequence yet show quite different patterns of methylation. This could indicate an ability of the genome and the epigenome to evolve independently. However, we do find that the most drastic changes in methylation between human and chimp, where an HMR in one species shows high levels of methylation in the other, are accompanied by an increased sequence divergence even at non-CpG dinucleotides. One interpretation is that most species-specific HMRs have arisen newly along one lineage with these novel functional elements are showing signs of recent adaptation. On the other hand, if this accelerated sequence change were more a reflection of relaxed selective pressure, we would expect species-specific HMRs to more frequently result from loss of functional elements along the opposite lineage. Resolution of these questions can only come from a broadening of the studies reported herein to many more species.
Experimental procedures
Detailed methods can be found as supplementary information.
Sperm collection
Two anonymous human donors were used and data pooled after sequencing. Two chimp donors were used. Semen was collected at the New Iberia Research Center (New Liberia, LA 70560) or the Southwest National Primate Research Center (San Antonio, TX 78227). Coagulated semen was separated from the liquid phase manually. Both human and chimp samples were diluted (1:1) in HBS buffer (0.01M HEPES ph 7.4; 150mM NaCl) and passed though a silica-based gradient, SpermFilter (Cryobiosystems), by centrifugation (according to manufacturer’s instructions).
Library preparation
DNA from ~100 million cells was extracted and sheared to a size of ~150-200nt by sonication. Double-stranded DNA fragments were end repaired, A-tailed, and ligated to methylated Illumina adaptors. Ligated fragments were bisulfite converted using the EZ-DNA Methylation-Gold Kit (Zymo research). Following PCR enrichment, fragments of 340 to 360bp were size selected and sequenced.
Computational methods
Reads were mapped with RMAPBS (Smith et al., 2009). The accuracy of our mapping method is discussed in Supplementary Information. Mapped reads were used to infer the methylation frequency at each CpG dinucleotide. These frequencies, along with the number of reads contributing to each frequency estimate, were supplied to a segmentation algorithm used to identify HMRs. Ortholog mapping between human and chimp was done with the liftOver tool available through the UCSC Genome Browser. Sequence conservation between human, chimp and rhesus was measured based on MULTIZ 44-way vertebrate alignments, also available through the UCSC Genome Browser. Complete details of all computational methods are provided in Supplementary Information.
Supplementary Material
Acknowledgments
We thank Michelle Rooks, Pramod Thekkat and Colin Malone for help with experimental procedures and Assaf Gordon, Luigi Manna and the CSHL and USC High Performance Computing Centers for computational support. We thank Babette Fontenot (New Iberia Research Center) and Jerilyn Pecotte (Southwest National Primate Center) for help with chimp sperm collection. We thank Sergey Nuzhdin, Ed Green, Peter Calabrese, Maren Friesen, Magnus Norborg, and Marie-Stanislas Remigereau for helpful discussions. This work was supported in part by grants from the NIH (R01HG005238 and 1RC2HD064459) and by a kind gift from Kathryn W. Davis. Data analyzed herein has been deposited in GEO (accession #*).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–290. doi: 10.1101/sqb.2008.73.058. [DOI] [PubMed] [Google Scholar]
- Bantysh OB, Buzdin AA. Novel family of human transposable elements formed due to fusion of the first exon of gene MAST2 with retrotransposon SVA. Biochemistry (Mosc) 2009;74:1393–1399. doi: 10.1134/s0006297909120153. [DOI] [PubMed] [Google Scholar]
- Bestor TH. Cytosine methylation and the unequal developmental potentials of the oocyte and sperm genomes. Am J Hum Genet. 1998;62:1269–1273. doi: 10.1086/301891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Chesnokov IN, Schmid CW. Specific Alu binding protein from human sperm chromatin prevents DNA methylation. J Biol Chem. 1995;270:18539–18542. doi: 10.1074/jbc.270.31.18539. [DOI] [PubMed] [Google Scholar]
- Cohen NM, Kenigsberg E, Tanay A. Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell. 2011;145:773–786. doi: 10.1016/j.cell.2011.04.024. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet. 1989;83:181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- Curley JP, Mashoodh R, Champagne FA. Epigenetics and the origins of paternal effects. Horm Behav. 2011 doi: 10.1016/j.yhbeh.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y. Epigenetic specification of centromeres. Biochem Cell Biol. 2009;87:273–282. doi: 10.1139/O08-135. [DOI] [PubMed] [Google Scholar]
- Damert A, Raiz J, Horn AV, Lower J, Wang H, Xing J, Batzer MA, Lower R, Schumann GG. 5′-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19:1992–2008. doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- Edwards JR, O’Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, Chanrion B, Fu Y, Su T, Hibshoosh H, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome research. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Zhang XY, Inamdar NM. Spontaneous deamination of cytosine and 5-methylcytosine residues in DNA and replacement of 5-methylcytosine residues with cytosine residues. Mutat Res. 1990;238:277–286. doi: 10.1016/0165-1110(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Enard W, Fassbender A, Model F, Adorjan P, Paabo S, Olek A. Differences in DNA methylation patterns between humans and chimpanzees. Curr Biol. 2004;14:R148–149. [PubMed] [Google Scholar]
- Frescas D, Guardavaccaro D, Kuchay SM, Kato H, Poleshko A, Basrur V, Elenitoba-Johnson KS, Katz RA, Pagano M. KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle. 2008;7:3539–3547. doi: 10.4161/cc.7.22.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanek S, Renz D, Doerfler W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 1993;12:1141–1151. doi: 10.1002/j.1460-2075.1993.tb05755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee SH, Cho SY, Shannon MF, Fan J, Rangasamy D. The impact of CpG island on defining transcriptional activation of the mouse L1 retrotransposable elements. PLoS One. 2010;5:e11353. doi: 10.1371/journal.pone.0011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WM, Maraia RJ, Rubin CM, Schmid CW. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994;22:1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Mikkelsen Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, Pittard WS, Devine SE. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet. 2006;78:671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell. 2010;19:625–638. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Schmid CW. Human Alu subfamilies and their methylation revealed by blot hybridization. Nucleic Acids Res. 1991;19:5613–5617. doi: 10.1093/nar/19.20.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem. 1994;269:8466–8476. [PubMed] [Google Scholar]
- Smith AD, Chung WY, Hodges E, Kendall J, Hannon G, Hicks J, Xuan Z, Zhang MQ. Updates to the RMAP short-read mapping software. Bioinformatics. 2009;25:2841–2842. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z, Cedar H. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Yamazaki T, Miki H, Ogonuki N, Inoue K, Ogura A, Baba T. Centromeric DNA hypomethylation as an epigenetic signature discriminates between germ and somatic cell lineages. Dev Biol. 2007;312:419–426. doi: 10.1016/j.ydbio.2007.09.041. [DOI] [PubMed] [Google Scholar]
- Yu X, Zhu X, Pi W, Ling J, Ko L, Takeda Y, Tuan D. The long terminal repeat (LTR) of ERV-9 human endogenous retrovirus binds to NF-Y in the assembly of an active LTR enhancer complex NF-Y/MZF1/GATA-2. J Biol Chem. 2005;280:35184–35194. doi: 10.1074/jbc.M508138200. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.