PKA and CaM kinase II both target the histone deacetylase HDAC4 such that the former antagonizes MEF2 activity and the latter promotes it.
Abstract
Histone deacetylase 4 (HDAC4) regulates numerous gene expression programs through its signal-dependent repression of myocyte enhancer factor 2 (MEF2) and serum response factor (SRF) transcription factors. In cardiomyocytes, calcium/calmodulin-dependent protein kinase II (CaMKII) signaling promotes hypertrophy and pathological remodeling, at least in part by phosphorylating HDAC4, with consequent stimulation of MEF2 activity. In this paper, we describe a novel mechanism whereby protein kinase A (PKA) overcomes CaMKII-mediated activation of MEF2 by regulated proteolysis of HDAC4. PKA induces the generation of an N-terminal HDAC4 cleavage product (HDAC4-NT). HDAC4-NT selectively inhibits activity of MEF2 but not SRF, thereby antagonizing the prohypertrophic actions of CaMKII signaling without affecting cardiomyocyte survival. Thus, HDAC4 functions as a molecular nexus for the antagonistic actions of the CaMKII and PKA pathways. These findings have implications for understanding the molecular basis of cardioprotection and other cellular processes in which CaMKII and PKA exert opposing effects.
Introduction
It is has long been known that sustained catecholaminergic stress promotes heart failure (Cohn et al., 1984; MERIT-HF Study Group, 1999), whereas short repetitive catecholaminergic stimulation as occurs during physical exercise training exerts cardioprotective effects (Keteyian et al., 2010). Thus, a deeper understanding of the downstream mediators of catecholamine signaling may unveil maladaptive versus adaptive molecular pathways, leading to new strategies for therapeutically manipulating the catecholaminergic pathway.
The G protein GS couples β-adrenergic receptors (β-ARs) to adenylyl cyclase, generating cAMP, which activates PKA (Wettschureck and Offermanns, 2005). PKA, in turn, phosphorylates numerous proteins involved in excitation-contraction coupling (Bers, 2002). However, sustained catecholaminergic signaling, a hallmark of heart failure, results in β-AR down-regulation, uncoupling of adenylyl cyclase from β-ARs, and reduced PKA activity (Fowler et al., 1986; Hausdorff et al., 1990; Rockman et al., 1996; Osadchii, 2007; Rajagopal et al., 2010). In contrast to PKA, calcium/CaM-dependent protein kinase II (CaMKII) remains activated under sustained β-AR stimulation (Wang et al., 2004). Originally, it was thought that CaMKII activation resulted from PKA-mediated increases in cytosolic calcium levels (Grimm and Brown, 2010), but recent evidence suggests the existence of PKA-independent mechanisms of CaMKII activation (Zhu et al., 2003; Erickson et al., 2008; Timmins et al., 2009; Mangmool et al., 2010; Métrich et al., 2010; Wagner et al., 2011).
Class II histone deacetylases (HDACs; HDACs 4, 5, 7, and 9) function as nodal regulators of striated muscle stress responses by linking upstream calcium-dependent protein kinases to downstream gene regulatory programs involved in myocyte hypertrophy, fibrosis, and metabolism (Czubryt and Olson, 2004; Backs and Olson, 2006; McGee and Hargreaves, 2010; Kehat et al., 2011). Class IIa HDACs share a common structure with a C-terminal catalytic domain and an N-terminal regulatory domain that interacts with transcription factors, coactivators, and corepressors (Verdin et al., 2003; Haberland et al., 2009). The N-terminal regulatory domains of class IIa HDACs contain a set of conserved serine residues that control their subcellular localization and confer signal responsiveness to downstream target genes (Grozinger and Schreiber, 2000; McKinsey et al., 2000, 2001). Phosphorylation of these serine residues creates binding sites for the 14-3-3 chaperone protein, which escorts phospho-HDACs from the nucleus to the cytoplasm, allowing the activation of HDAC target genes. Direct oxidation also results in accumulation of HDACs in the cytosol (Ago et al., 2008).
In the nucleus, HDAC4 functions as a repressor of myocyte enhancer factor-2 (MEF2) and serum response factor (SRF), transcription factors that regulate muscle and stress-responsive genes as well as genes that maintain cardiomyocyte integrity (Edmondson et al., 1994; Naya et al., 1999; Passier et al., 2000; Niu et al., 2005; Paroni et al., 2007). Interaction of MEF2 or SRF with class II HDACs silences the expression of target genes of these transcription factors (Miska et al., 1999; Paroni et al., 2007). Mice lacking MEF2D, the predominant MEF2 isoform in the adult heart, display normal cardiac function but are protected against stress-induced cardiac remodeling (Kim et al., 2008). In contrast, SRF is crucial for cardiac function, such that its deletion results in cardiomyopathy (Parlakian et al., 2005). How HDAC4 discriminates between MEF2 and SRF in different settings is unclear. Gene deletion studies in mice revealed that class IIa HDACs are key regulators of tissue growth and development (Haberland et al., 2009). HDAC5 and HDAC9 repress cardiomyocyte hypertrophy (Zhang et al., 2002; Chang et al., 2004), HDAC4 represses chondrocyte hypertrophy (Vega et al., 2004), and HDAC7 regulates vascular integrity (Chang et al., 2006). Mice lacking HDAC4 die perinatally as a result of premature ossification, which prevented us from studying the potential role of HDAC4 in adult hearts.
Recently, we and others demonstrated that CaMKII specifically signals to HDAC4 to promote pathological cardiac remodeling (Backs et al., 2006, 2008, 2009; Little et al., 2007; McKinsey, 2007; Zhang et al., 2007; Métrich et al., 2010). Here, we describe a novel mechanism by which catecholaminergic downstream mediators regulate cardiac transcription through regulated proteolysis of HDAC4. Our findings identify HDAC4 as a common target of CaMKII and PKA. However, whereas CaMKII induces cytosolic accumulation of HDAC4 and de-represses HDAC4 target genes, PKA promotes the generation of an N-terminal HDAC4 cleavage product (HDAC4-NT), which acts as a CaMKII-insensitive repressor that selectively inhibits myocyte enhancer factor 2 (MEF2) but not SRF, antagonizing cardiac remodeling but not affecting cardiomyocyte survival. Thus, HDAC4 functions as an integrator of two opposing signal transduction pathways that govern cardiac gene expression.
Results
PKA represses MEF2 activity
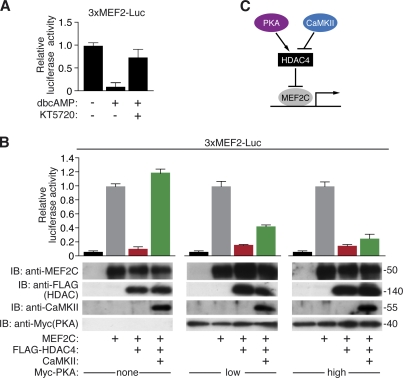
Signaling by β-AR agonists activates CaMKII and other calcium-dependent protein kinases, which promote MEF2 activity by phosphorylating class II HDACs and triggering their export from the nucleus. This regulatory mechanism plays a key role in the control of cardiac hypertrophy and stress responsiveness of cardiomyocytes. Because β-AR agonists also activate PKA, we sought to determine the potential influence of PKA signaling on MEF2 activity. Therefore, we transfected the cardiomyocyte-like cell line H9c2 with an MEF2-responsive reporter (3×MEF2-Luc) and activated PKA by 1 mM dibutyryl cAMP (dbcAMP). As shown in Fig. 1 A, dbcAMP inhibited MEF2 activity, and PKA inhibition by KT5720 blunted the repressive effect of dbcAMP.
Figure 1.
PKA represses MEF2 activity. (A) H9c2 myocytes were transfected with the 3×MEF2-Luc reporter and stimulated with 1 mM dbcAMP in the absence or presence of the PKA inhibitor KT5720 (2 µM). Firefly luciferase was normalized to renilla luciferase expression (driven by a cytomegalovirus promoter). The experiment was performed in triplicates. Similar results were obtained in three different experiments. (B) COS cells were transfected with the 3×MEF-Luc reporter and expression plasmids encoding 100 ng MEF2C, 10 ng FLAG-HDAC4, and 100 ng CaMKII in the absence and presence of low (25 ng) or high (50 ng) amounts of Myc-PKA expression plasmid, as indicated. Firefly luciferase was normalized to renilla luciferase expression. Western blots showing expression of MEF2C, FLAG-HDAC4, CaMKII, and Myc-PKA in the same sample as used for the reporter assay are shown below the reporter assay quantification. IB, immunoblot. Molecular mass is indicated in kilodaltons. The experiment was performed in duplicates. Similar results were obtained in three different experiments. (A and B) Error bars indicate ± SEM. (C) A model showing antagonistic effects of PKA and CaMKII on HDAC4 and MEF2 activity.
To further investigate the potential impact of PKA signaling on MEF2 activity and the relationship between PKA and CaMKII signaling, we tested the effects of these kinases on MEF2 in the presence of HDAC4 in transfected COS cells. As previously reported (Backs et al., 2006), CaMKII signaling relieved MEF2C from repression by HDAC4 (Fig. 1 B). Strikingly, PKA dose dependently overcame the stimulatory effect of CaMKII on MEF2 activity, suggesting that the PKA and CaMKII signaling pathways act in an opposing manner to modulate the MEF2-HDAC4 axis (Fig. 1 C).
PKA-dependent proteolytic processing of HDAC4
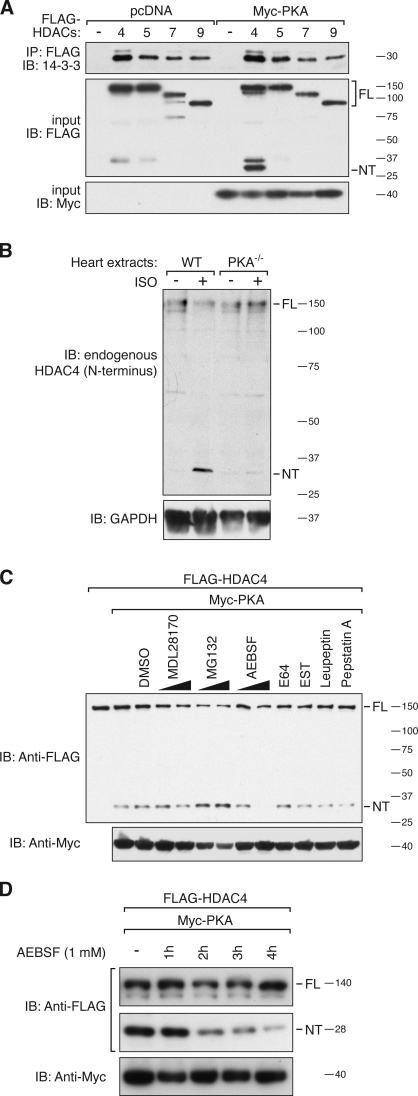
CaMKII phosphorylates three sites in the N-terminal regulatory domains of HDAC4 and other class IIa HDACs, creating binding sites for 14-3-3 chaperone protein, which mediates nuclear export (Backs et al., 2006). To begin to explore the mechanistic basis of the inhibitory effect of PKA on MEF2, we investigated whether PKA altered 14-3-3 binding to HDAC4 and the closely related HDAC5, 7, and 9 (Fig. 2 A). Unlike CaMKII, the catalytic subunit of PKA did not increase 14-3-3 binding to these HDACs. Strikingly, however, we observed an N-terminal fragment of HDAC4 of ∼28 kD when coexpressed with PKA. We refer to this HDAC4 cleavage product as HDAC4-NT. HDAC4-NT production was highly reproducible and was confirmed by antibodies directed against the N terminus of HDAC4, such as anti–HDAC4 N-18 (Santa Cruz Biotechnology, Inc.). Occasionally, we detected another peptide of higher molecular mass, but this was also seen without kinase coexpression. We also tested whether CaMKII could induce limited proteolysis of HDAC4 or HDAC5, but no N-terminal cleavage products were observed (Fig. S1 A). In contrast to HDAC4-NT, no C-terminal cleavage product was detected with a C-terminal HDAC4-Myc tag, suggesting that the C-terminal part of HDAC4 was unstable (Fig. S1 B)
Figure 2.
PKA-dependent proteolysis of HDAC4. (A) Coimmunoprecipitation assay with lysates from COS cells expressing FLAG-HDAC4, -HDAC5, -HDAC7, and -HDAC9 (MEF2-interacting transcription repressor). Extracts were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted (IB) with an antibody directed against endogenous 14-3-3 protein (top). The effect of Myc-PKA was tested. Input proteins were detected with antibodies directed against FLAG and Myc, identifying an N-terminal FLAG-HDAC4 cleavage product (NT) of ∼28 kD. (B) Western blot analysis of cardiac extracts from WT and Prkaca knockout (PKA−/−) mice that were treated for 4 h with or without Iso (60-µg/g body weight, 15 µg/g per hour). The membrane was analyzed with an antibody directed against the N terminus of HDAC4 (N-18). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected as a loading control. (C and D) COS cells were transfected with FLAG-HDAC4 and Myc-PKA. HDAC4-FL and -NT were detected by Western blot analysis using an antibody directed against FLAG. The effects of inhibitors of calpains (5 and 20 µM MDL28170 and 50 µM EST), the proteasome (10 and 25 µM MG132), cysteine proteases (10 µM E64 and 100 µM leupeptin), aspartic proteases (10 µM pepstatin A), and serine proteases (100 and 400 µM AEBSF) on HDAC4-NT production (C) and different treatment periods of AEBSF to determine the half-life of posttranslationally produced HDAC4-NT (D) are shown. Molecular mass is indicated in kilodaltons.
Next, we tested whether endogenous HDAC4 was proteolytically processed in response to β-AR–dependent PKA activation in vivo. We treated wild-type (WT) mice and mice lacking the catalytic α subunit of PKA (Skålhegg et al., 2002), the major PKA catalytic isoform in the heart, with four sequential injections of the adrenergic agonist isoproterenol (Iso) over a period of 4 h (60 µg/g in total). Western blot analysis of cardiac extracts using an antibody to the N terminus of HDAC4 detected a basal level of HDAC4-NT under control conditions and a pronounced increase after Iso treatment (Fig. 2 B). The appearance of HDAC4-NT in response to Iso treatment was accompanied by a decrease in full-length (FL) HDAC4 protein. In PKA-null mice, Iso-induced HDAC4-NT was markedly diminished, confirming that PKA is not only sufficient but also necessary for HDAC4-NT production in response to adrenergic activation of PKA signaling.
We attempted to characterize the PKA-dependent HDAC4 protease using a series of protease inhibitors. AEBSF, a serine protease inhibitor, prevented PKA-mediated HDAC4 cleavage, whereas inhibitors of calpain, caspases, the proteasome, cysteine proteases, and aspartic proteases had no effect on HDAC4-NT production (Figs. 2 C and S1 C). AEBSF inhibits a broad range of serine proteases and typically exerts cytotoxic effects at concentrations >400 µM. AEBSF effectively inhibited HDAC4-NT production at a concentration of 250 µM (Fig. S1 D). The FL HDAC4 protein has been reported to display a half-life of ∼8 h (Liu et al., 2004). To examine the stability of HDAC4-NT, we treated COS cells, in which HDAC4-NT was already produced, with an effective dose of AEBSF and found that HDAC4-NT disappeared with a half-life of ∼2 h (Fig. 2 D).
To verify whether HDAC4 is cleaved in the cytosol or the nucleus, we used two mutants that show mutually exclusive localization to these compartments. Cytosolic HDAC4, in which the NLS (amino acids 247–289) was deleted (HDAC4 ΔNLS), was cleaved to the same degree as a nonphosphorylatable nuclear HDAC4 mutant (HDAC4–Ser-246/467-Ala [2×S/A]) in which Ser246 and Ser467 were replaced by an alanine, implying that cleavage occurs in both compartments (Fig. S1 E).
Identification of the HDAC4 cleavage site
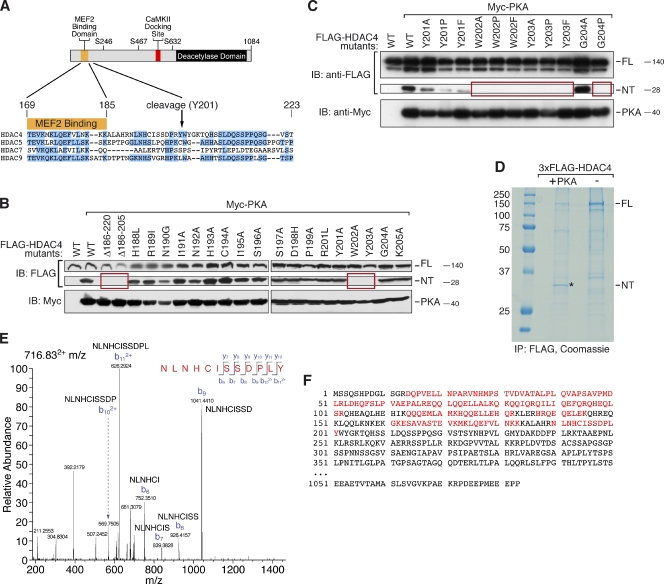
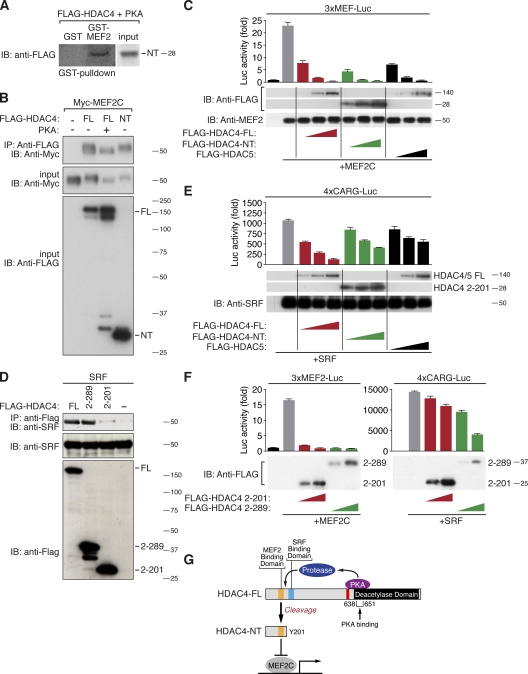
The structure of HDAC4 is schematized in Fig. 3 A. To pinpoint the cleavage site, we generated deletion mutants of HDAC4. HDAC4 Δ186–220 and Δ186–205 were resistant to PKA-mediated cleavage, suggesting that the cleavage site lies between amino acids 186 and 205. We then mutated each individual amino acid between amino acids 188 and 205 and found that amino acids 202 and 203 were essential for the cleavage event (Fig. 3 B). We also mutated amino acids 201 and 204 to residues with distinct biochemical properties and found that Gly204, when mutated to a proline but not to an alanine, prevented cleavage (Fig. 3 C).
Figure 3.
Identification of the HDAC4 cleavage site. (A) A schematic of the HDAC4 protein and the amino acid sequence surrounding Tyr201. The sequence of this HDAC4 domain is aligned with other HDACs. The cleavage site lies 15 amino acids after the MEF2-binding domain in a unique region of HDAC4. Conserved amino acids are highlighted in blue. (B and C) Western blot analysis of lysates from COS cells expressing FLAG-HDAC4-WT and different FLAG-HDAC4 mutants (as indicated) together with Myc-PKA. HDAC4-FL and -NT expression was detected with an antibody directed against FLAG and PKA expression with an antibody directed against Myc. Mutants that fail to be cleaved are boxed in red. IB, immunoblot. (D) An anti-FLAG immunoprecipitate (IP) of lysates of COS cells that expressed 3×FLAG-HDAC4 with and without PKA was analyzed by SDS-PAGE and Coomassie staining. HDAC4-NT (marked with an asterisk in the PKA lane) was identified based on its sole production in the presence of PKA. This band was excised from the gel, and tryptic peptides were analyzed. (E and F) MS analysis of the most C-terminal peptide obtained by tryptic digestion of immunoprecipitated HDAC4-NT (R200L) is shown (E). All HDAC4 peptides that were identified are marked in red (F). Molecular mass is indicated in kilodaltons.
To identify the exact HDAC4 cleavage site, we immunoprecipitated and purified N-terminally 3×FLAG-tagged HDAC4 from an anti-FLAG affinity column and eluted it with 3×FLAG competitor peptide. For this experiment, we used the HDAC4 R200L mutant that was as efficiently cleaved as WT HDAC4. By using this mutant, we could detect protein fragments around amino acids 200–203. Only when coexpressed with PKA did we observe a Coomassie blue–stained band of ∼28 kD after SDS-PAGE (Fig. 3 D). Tryptic digestion and mass spectrometry (MS) of this band identified a peptide ending at amino acid 201 (Fig. 3, E and F). MS of the corresponding region of the control lane lacking PKA yielded no HDAC4 peptides. Based on the mutational analysis in combination with the results of MS, we conclude that the amino acid boundary between Tyr201 and Trp202 defines the PKA-mediated cleavage site of HDAC4, with Trp202, Tyr203, and Gly204 being critical for proteolysis.
Mapping of a critical PKA-binding site on HDAC4
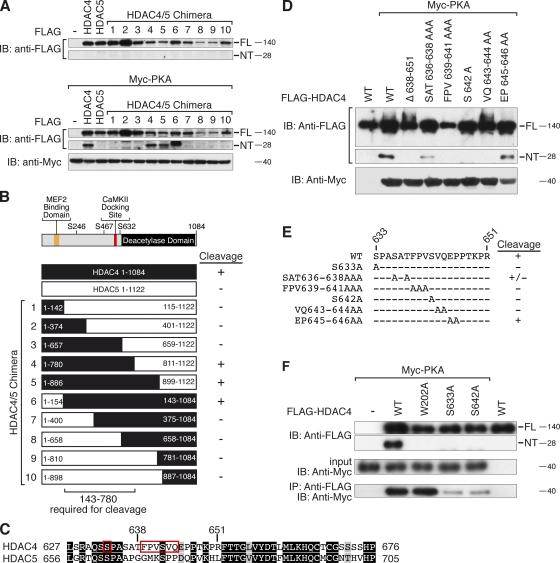
To begin to map the domain of HDAC4 that conferred responsiveness to PKA and consequent proteolytic cleavage, we created a series of HDAC4/HDAC5 chimeric proteins (Fig. 4, A and B). Only those chimeric proteins containing portions of HDAC4 from amino acid 143 to 780 were able to give rise to HDAC4-NT in response to PKA. Thus, besides the actual cleavage site after Tyr201, other domains within this region must be crucial for HDAC4-NT production. Sequence comparison revealed that a region before amino acid 780 contains a polypeptide sequence (amino acids 636–651) that is present only in HDAC4 and not in other class IIa HDACs (Fig. 4 C). Deletion of this sequence or mutation of key residues (HDAC4 639–644 FPVSVQ) within this region prevented HDAC4-NT production in response to PKA (Fig. 4, D and E). We also tested other amino acids around this region and found that Ser633, which lies adjacent to a CaMKII phosphorylation site (Ser632), was required for the PKA effect. Although neither Ser633 nor Ser642 matches the perfect PKA phosphorylation consensus site (RRXSY, with Y being the hydrophobic residue), we performed a series of kinase assays with recombinant GST-HDAC4 fusion proteins (HDAC4 419–739), but we were unable to detect specific phosphorylation events of these sites. It is possible that efficient phosphorylation requires folding events of HDAC4 including N-terminal regions not present in the GST-HDAC4 419–739 fusion protein. However, we found that PKA interacts directly with FL HDAC4 and that the two mutations that prevent PKA-dependent cleavage of HDAC4 markedly diminished the ability of HDAC4 to bind PKA (Fig. 4 F). Thus, we conclude that PKA binding to the central region of HDAC4 renders the N-terminal domain of HDAC4 sensitive to proteolysis. A kinase-dead mutant of PKA (PKA K72H) failed to induce HDAC4-NT production, implying that the PKA requires enzymatic activity (Fig. S2). Because we were unable to identify PKA phosphorylation sites on HDAC4 that mediate proteolysis, we speculate that the HDAC protease may need to be activated by PKA to enable it to cleave HDAC4.
Figure 4.
Identification of a PKA-binding domain on HDAC4. (A and B) FLAG-HDAC4, -HDAC5, and -HDAC4/5 chimeric proteins (as indicated in B) were expressed in COS cells in the presence and absence of Myc-PKA. Expression of FLAG-tagged HDAC4-FL and -NT was detected by Western blot analysis using an antibody directed against FLAG. The schematic (B) summarizes the WT and mutant HDACs that were cleaved. IB, immunoblot. (C and D) Mutational analysis of a unique domain of HDAC4. As indicated in D, PKA was expressed with HDAC4 mutants, in which this domain was deleted or single amino acids were replaced by an alanine. HDAC4-FL and -NT production were detected by Western blot analysis. The amino acids of HDAC4 that were found to be required for HDAC4-NT production are framed in red, and conserved amino acids are highlighted by black (identical residues) and gray (similar residues) boxes (C). (E) Summary of essential amino acids within the central part of HDAC4 required for HDAC4-NT production. The region between amino acid 639 and 644 is required for PKA responsiveness. (F) Coimmunoprecipitation assay with lysates from COS cells expressing FLAG-HDAC4, the indicated FLAG-HDAC4 mutants, and Myc-PKA. Extracts were immunoprecipitated with anti-FLAG antibody and blotted with an antibody directed against Myc. Input proteins were detected with antibodies directed against FLAG and Myc. These data show that Ser633 and Ser642 are required not only for HDAC4-NT production but also for PKA binding. As a control, mutation of the cleavage site (W202A) prevented only HDAC4-NT production but not PKA binding. Molecular mass is indicated in kilodaltons.
HDAC4-NT localizes to the nucleus
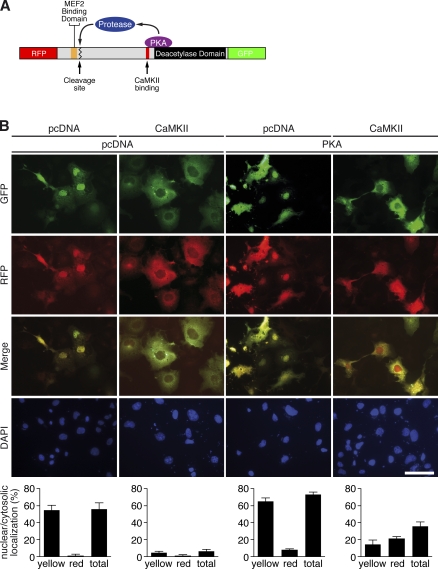
To further determine the subcellular localization of HDAC4-NT, we fused the dsRed monomer fluorescent protein RFP to the N terminus and the EGFP to the C terminus of HDAC4 (Fig. 5 A). Coexpression with CaMKII in COS cells forced the HDAC4 fusion protein to accumulate in the cytosol, where it stained the cytosol yellow. Nuclear localization of RFP-HDAC4-GFP was only very slightly increased when coexpressed with PKA alone. Strikingly, however, in cells coexpressing CaMKII, PKA, and RFP-HDAC4-GFP, ∼20% of the nuclei were red, suggesting that HDAC4 was proteolytically processed and that HDAC4-NT accumulated in the nucleus, where it could act as a transcriptional regulator (Fig. 5 B).
Figure 5.
HDAC4-NT localizes to the nucleus. (A) RFP-HDAC4-GFP was coexpressed with CaMKII, PKA, or both in COS cells. A schematic of the RFP-HDAC4-GFP fusion is shown. (B) Subcellular localization of RFP-HDAC4-GFP in the green and red fluorescence channels is shown. To assess localization of HDAC4-FL versus HDAC4-NT, both channels were merged, identifying red nuclei as a sign for nuclear accumulation of HDAC4-NT. Nuclei are visualized by DAPI staining. The quantitative analysis is shown below the representative images. More than 100 cells per condition were analyzed in three selected fields. The ratio per field was quantified (n = 3). Bar, 25 µm.
HDAC4-NT selectively represses MEF2
Intriguingly, HDAC4-NT contains the MEF2-binding domain (Fig. 3 A). Thus, we tested whether PKA promoted the association of HDAC4-NT with MEF2. Indeed, HDAC4-NT in a lysate of PKA and HDAC4-FL–expressing cells was pulled down with a GST-MEF2C fusion protein (Fig. 6 A). Similarly, Myc-MEF2C coimmunoprecipitated with FLAG-HDAC4 2–201 (NT) (Fig. 6 B). These findings suggested two interesting possibilities: HDAC4-NT might function as a repressor of MEF2 or as a competitor with HDAC4-FL or other class II HDACs, thereby enhancing MEF2 activity. To test these possibilities, we examined whether HDAC4 2–201 (NT) was able to interfere with the activation of an MEF2-dependent luciferase reporter (3×MEF2-Luc). As shown in Fig. 6 C, HDAC4-NT was almost as efficient as HDAC4-FL or HDAC5-FL in repressing MEF2 reporter activity.
Figure 6.
MEF2-selective effects of HDAC4-NT in cardiomyocytes. (A) A pull-down assay with GST or GST-MEF2C and a lysate of COS cells expressing FLAG-HDAC4 and PKA. An anti-FLAG Western blot analysis of the COS cell lysate input and the pull-down experiment is shown. IB, immunoblot. (B) Coimmunoprecipitation assay with lysates from COS cells coexpressing FLAG-tagged HDAC4-FL (in the absence and presence of PKA) or HDAC4-NT and Myc-MEF2C. The immunoprecipitate (IP) was analyzed with an antibody directed against Myc. Input proteins were detected with antibodies directed against FLAG and Myc. These data show that both HDAC4-FL and -NT bind to MEF2C. (C) COS cells were transfected with the 3×MEF-Luc reporter and expression plasmids encoding MEF2C and FLAG-tagged HDAC4-FL, HDAC4-NT, or HDAC5, as indicated. Firefly luciferase activity was normalized to renilla luciferase expression (driven by a cytomegalovirus promoter). Western blots showing expression of FLAG-HDACs and MEF2C in the same sample used for reporter assay are shown below the reporter assay quantification. (D) Coimmunoprecipitation assay with lysates from COS cells expressing FLAG-tagged HDAC4-FL, HDAC4 2–289, and HDAC4 2–201 (NT) together with SRF. The FLAG immunoprecipitate was blotted with an antibody directed against SRF. Input proteins were detected with antibodies directed against SRF and FLAG. These data show that HDAC4-FL and HDAC4 2–289 bind strongly to SRF, but HDAC4 2–201 (NT) does not. (E and F) COS cells were cotransfected with the 4×CARG-Luc reporter and expression plasmids encoding SRF and FLAG-tagged HDAC4-FL, HDAC4-NT (2–201), HDAC5, or HDAC4 2–289, as indicated. Western blots showing expression of FLAG-HDACs and SRF in the same sample used for reporter assay are shown below the reporter assay quantification. Molecular mass is indicated in kilodaltons. (C, E, and F) The experiments were performed in duplicates. Similar results were obtained in three different experiments. (G) A schematic showing the selective interaction and repression of MEF2 by HDAC4-NT.
In addition to inhibiting MEF2, HDAC4 represses SRF-dependent transcription (Davis et al., 2003). Moreover, proteolytic cleavage of HDAC4 at Asp289 is mediated by caspases (Liu et al., 2004; Paroni et al., 2004), and HDAC4 2–289 displayed an increased affinity to SRF (Paroni et al., 2007). However, to our knowledge, the exact SRF-binding domain of HDAC4 was not mapped. In coimmunoprecipitation assays, we found that HDAC4 2–289 bound stronger to SRF than HDAC4 2–201 (NT) (Fig. 6 D), suggesting that the region between 201 and 289 contains a critical part of the SRF-binding domain. Likewise, HDAC4 2–201 (NT) was a relatively inefficient repressor of SRF activity, comparable with the mild repressive function of HDAC5 (Fig. 6 E). Therefore, we compared the repressive potential of HDAC4 2–201 (NT) and 2–289 on MEF2 and SRF reporters. Whereas the longer fragment inhibited both reporters efficiently, the effect of HDAC4 2–201 (NT) on SRF activity was weaker, implying that PKA-dependent proteolytic processing of HDAC4 results in differential regulation of target transcription factors favoring repression of MEF2 (Fig. 6 G).
MEF2-selective effects of HDAC4-NT in cardiomyocytes
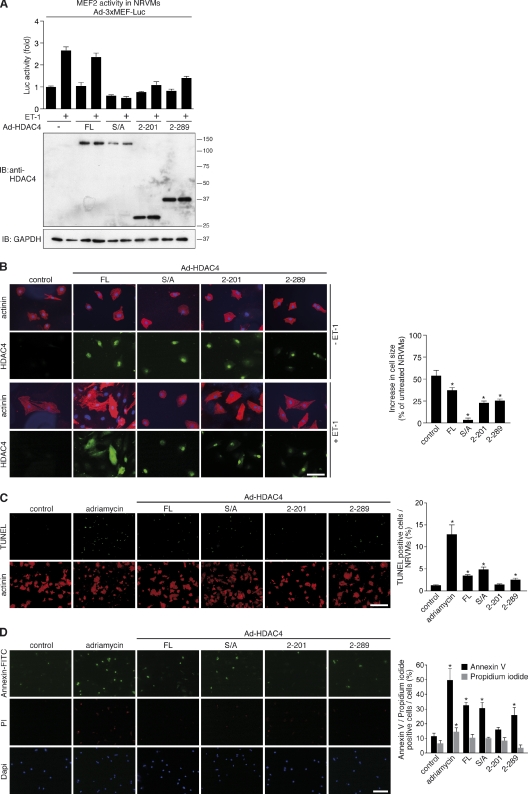
Next, we investigated the effects of HDAC4-NT on neonatal rat ventricular myocytes (NRVMs). First, we activated MEF2 by endothelin-1 (ET1), a potent hypertrophic agonist that drives nuclear export of class II HDACs (McKinsey, 2007). Despite similar expression levels, HDAC4-FL (WT protein) was less efficient in repressing ET1-induced MEF2 activity as compared with HDAC4 2–201 (NT) and 2–289 (Fig. 7 A), confirming that the proteolytic fragments are insensitive to kinases such as CaMKII and PKD, which induce nucleocytoplasmic shuttling of HDAC4-FL. As expected, HDAC4-S/A, a mutant of HDAC4 lacking the three CaMKII/PKD phosphorylation sites, strongly repressed ET1-induced MEF2 activity. Immunocytochemistry of NRVMs showed nuclear localization of HDAC4-S/A, 2–201 (NT), and 2–289 under baseline conditions but also upon stimulation with ET1, confirming the insensitivity of these HDAC4 mutants to upstream kinases (Fig. 7 B). In contrast, HDAC4-FL responded to ET1 signaling and translocated to the cytosol, which is consistent with its effect on MEF2 activity after ET1 stimulation. Sarcomeric α-actinin staining demonstrated that HDAC4-S/A was the strongest repressor of cardiomyocyte size. However, HDAC4-S/A also perturbed myocyte structure. Consistent with its maintained signal responsiveness, HDAC4-FL reduced cardiomyocyte hypertrophy by only 10–20%. HDAC4 2–201 (NT) and 2–289 did not perturb myocyte structure but inhibited cardiomyocyte hypertrophy by 50%. TUNEL and annexin V/propidium iodide (PI) assays revealed that HDAC4-S/A, -FL, and 2–289 induced apoptotic cell death, but HDAC4 2–201 (NT) did not (Fig. 7, C and D). This observation is consistent with the proapoptotic effects of caspase 3, which mediates the production of HDAC4 2–289. Collectively, these data imply that HDAC4-NT (2–201) selectively antagonizes MEF2-dependent cardiomyocyte remodeling and hypertrophy but maintains SRF-mediated myocyte viability.
Figure 7.
HDAC4-NT affects cardiomyocyte hypertrophy but not viability. (A) A luciferase reporter assay in NRVMs that were transduced with adenoviruses (Ad) for 3×MEF2-Luc and HDAC4-FL and mutants, as indicated. Ad–3×MEF2-Luc was cotransduced with adenoviruses encoding the indicated forms of HDAC4 in the presence of 100 nM ET-1. Firefly luciferase expression was normalized to the protein content of each sample. HDAC expression in the same preparation as used for reporter assays was detected using an antibody directed against the N terminus of HDAC4 (N-18) and is shown below the reporter assay quantification. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IB, immunoblot. Molecular mass is indicated in kilodaltons. (B) Immunocytochemistry of NRVMs transduced with Ad–HDAC4-FL, -S/A, 2–201 (NT), or 2–289 using antibodies recognizing sarcomeric α-actinin (red; top) to assess myocyte size and sarcomere organization and FLAG (green; bottom) to determine the subcellular localization of HDAC4-FL and mutants. DAPI staining was used to label nuclei. NRVMs are shown under baseline conditions or after treatment with 100 nM ET-1, as indicated. Quantitative analysis of cardiomyocyte hypertrophy is shown on the right. 51–100 NRVMs were analyzed per condition (n > 50). *, P < 0.05 versus control. Bar, 50 µm. (C) TUNEL assays of NRVMs transduced with Ad–HDAC4-WT, -S/A, 2–201 (NT), or 2–289. Apoptotic nuclei appear in green, and NRVMs were counterstained for sarcomeric α-actinin. Nontransduced NRVMs (control) serve as a negative control, and adriamycin-treated NRVMs serve as a positive control. Quantitative analysis of apoptotic nuclei per NRVMs is shown on the right. More than 55 NRVMs per condition were analyzed in four to eight selected fields. The percentage in each field was quantified (n ≥ 4). *, P < 0.05 versus control. Bar, 200 µm. (D) Annexin V–FITC/PI staining of NRVMs. NRVMs were nontreated (control), treated with adriamycin (positive control), or transduced with Ad–HDAC4-FL, -S/A, 2–201 (NT), or 2–289. Quantitative analysis of annexin V and PI-positive cells per total cell number is shown on the right. 59–144 cells were analyzed per condition in three to five selected fields. The percentage in each field was quantified (n ≥ 3). *, P < 0.05 versus control. Bar, 100 µm.
Discussion
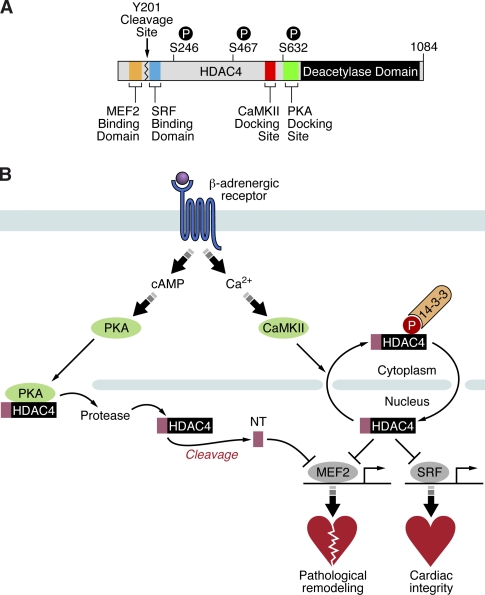
Our results identify HDAC4 as a bipolar integrator of PKA and CaMKII signaling, which impose antagonistic influences on cardiac gene expression (Fig. 8). Whereas CaMKII forces HDAC4 to accumulate in the cytosol, thereby activating MEF2 target genes, PKA triggers proteolytic cleavage of HDAC4, generating a signal-insensitive repressor of MEF2 but not SRF. The differential regulation of HDAC4 function via PKA and CaMKII has implications for many physiological and pathophysiological processes in which PKA and CaMKII are differentially regulated.
Figure 8.
PKA-dependent proteolysis of HDAC4 overcomes CaMKII-mediated MEF2 activation. (A) A schematic of HDAC4 highlighting its domains involved in CaMKII and PKA signaling as well as MEF2 and SRF repression. P, phosphorylated serine. (B) Chronic β-adrenergic stimulation leads, via calcium-dependent mechanisms, to CaMKII activation, which mediates cytosolic accumulation of HDAC4 and activation of MEF2- and SRF-dependent gene programs. In contrast, acute adrenergic stimulation leads to cAMP-dependent PKA activation, which mediates proteolytic processing of HDAC4 to HDAC4-NT. HDAC4-NT acts as a CaMKII-insensitive repressor of MEF2 but not SRF.
Control of HDAC4 targets by opposing kinase pathways
It has been well established that the distribution of class II HDACs between the nucleus and cytoplasm depends on the status of phosphorylation of three serine residues in the N-terminal regulatory domains of these HDACs, which serve as binding sites for 14-3-3 proteins (Verdin et al., 2003; Backs and Olson, 2006; McKinsey, 2007; Haberland et al., 2009). Dephosphorylation of these sites by PP2A antagonizes CaMK signaling, promoting nuclear retention of class II HDACs (Sucharov et al., 2006; Illi et al., 2008; Paroni et al., 2008). We propose that PKA-mediated proteolytic processing of HDAC4 results in a similar effect. However, proteolytic control of HDAC4 is distinct in two important respects. First, HDAC4-NT resists opposing kinase signaling (e.g., by CaMKII or PKD) and persistently represses MEF2 gene programs. In contrast, dephosphorylation is a transient mechanism, continually counteracted by phosphorylation. Second, HDAC4-NT provides another layer of transcription factor specificity because it represses MEF2 more efficiently than SRF. This is in contrast to HDAC4-FL and to caspase-processed HDAC4 (HDAC4 1–289). Moreover, HDAC4-NT would not be predicted to affect the activity of nuclear factor of activated T-cells (NFAT), which binds indirectly via the DnaJ-related factor Mrj to the C-terminal half of HDAC4 (Dai et al., 2005). The regulated proteolysis of HDAC4 therefore allows PKA to selectively repress MEF2 but not SRF or NFAT targets. This is of functional importance because accumulation of both HDAC4-FL and caspase-processed HDAC4 have been shown to induce apoptosis (Paroni et al., 2007), whereas we show here that HDAC4-NT does not efficiently induce apoptosis. Thus, PKA-dependent proteolysis of HDAC4 represents a mechanism of selective cardioprotection against MEF2 target gene activation (Kim et al., 2008).
CaMKII signals selectively to HDAC4 by docking to a domain that is uniquely present in HDAC4 but not in other class IIa HDACs (Backs et al., 2006). Remarkably, we show here that PKA also selectively signals to HDAC4 by docking to another unique domain that actually lies in close proximity to the CaMKII docking site. Why do two downstream kinases in β-AR signaling pathways target the same HDAC and exert opposing functions on HDAC4-dependent cardiac transcription? The answer to this question might lie in the different time courses of PKA versus CaMKII activation. Acute β-AR stimulation activates PKA, but sustained stimulation of β-ARs leads to an uncoupling of adenylyl cyclase from β-ARs, resulting in decreased cAMP levels and PKA inactivation (Grimm and Brown, 2010). Moreover, sustained β-AR stimulation, by mechanisms that are poorly understood, leads to progressive PKA-independent CaMKII activation (Zhu et al., 2003; Wang et al., 2004; Grimm and Brown, 2010). Thus, whereas acute β-AR stimulation would generate high levels of the HDAC4-NT fragment, sustained β-AR stimulation would generate low levels of HDAC4-NT and would translocate the noncleaved HDAC4 to the cytosol. Therefore, HDAC4 represents a transcriptional checkpoint in β-AR signaling, and the balance between PKA and CaMKII activation dictates the consequences on HDAC4-dependent transcription (Fig. 8).
Physiological implications
We speculate that the mechanistic findings of this study explain why repetitive transient elevations of catecholamines—as occur during interval exercise training—would not result in activation of MEF2 target genes. In such settings, HDAC4-NT would be expected to protect the myocardium against the expression of such genes in response to physiological stress conditions. Additionally, the short half-life of HDAC4-NT of ∼2–3 h (as compared with 8 h of HDAC4-FL) would be ideal to bridge such stress situations. During chronic heart failure, CaMKII is typically activated (Wang et al., 2004; Grimm and Brown, 2010), which would result in the cytosolic accumulation of HDAC4-FL (Backs et al., 2006, 2009). We propose that under such conditions, moderate levels of HDAC4-NT are cardioprotective and that the success of β-blocker therapy in heart failure might be explained, at least in part, by its ability to prevent β-AR desensitization, resulting in higher levels of the HDAC4-NT production in response to β-AR stimulation.
It has been suggested that accumulation of HDAC4 in the nucleus may cause cell death in neurons (Bolger and Yao, 2005). Likewise, SRF has been shown to be crucial for cardiac integrity, function, and viability (Parlakian et al., 2005). Thus, in contrast to caspase activation, PKA-mediated proteolysis of HDAC4 may circumvent cell death events as a result of the inability of HDAC4-NT to efficiently repress SRF. Further studies need to clarify whether HDAC4-NT production in certain pathophysiological conditions such as myocardial infarction would be maladaptive or, as proposed here, might even be cardioprotective.
Calcium-dependent signaling via CaMKII and cAMP signaling via PKA play opposing roles in many cellular processes such as chondrocyte differentiation, in which parathyroid hormone-related peptide signaling through PKA keeps chondrocytes in an undifferentiated status, but, noncanonical Wnt signaling, which activates CaMKII, promotes chondrocyte hypertrophy and ossification (Li et al., 2011). Likewise, oocytes are kept in a state of cell cycle arrest by cAMP, whereas CaMKII-γ induces cell cycle resumption (Backs et al., 2010). HDAC4 is highly expressed both in chondrocytes (Vega et al., 2004) and oocytes (Kageyama et al., 2006). Thus, it will be of interest to investigate the role of HDAC-NT production in these and other cell types.
Targets for pharmacological modulation of the HDAC4-MEF2 pathway
In the future, it will be of interest to determine HDAC4-NT levels in different tissues and during different states of cardiovascular diseases. The PKA-dependent HDAC4 protease also represents an interesting therapeutic target. In this study, we attempted to define the nature of the protease by chemical inhibitors. Only serine protease inhibitors were able to prevent PKA-dependent cleavage of HDAC4. The identified cleavage consensus site is unusual and lies in a hydrophobic pocket. It is tempting to speculate that PKA phosphorylation of HDAC4 exposes the cleavage site.
HDAC4-NT is a potent repressor of MEF2 and is unresponsive to CaMKII signaling. Thus, antagonistic kinase-signaling pathways converge on MEF2 to modulate its activity and, as a consequence, the transcriptional program for cardiac remodeling. In contrast to proteolytic processing by caspases, PKA-mediated proteolytic processing of HDAC4 does not reduce myocyte viability by modifying the specificity to target transcription factors. This implies that HDAC4-NT could be cardioprotective in the course of chronic heart failure. Thus, gene therapeutic approaches that aim to overexpress HDAC4 1–201 (NT) could be a novel means of therapeutic cardioprotection and would increase the amount of MEF2-repressive modifiers in the nucleus. Based on the findings of this study, gene transfer of HDAC4 1–201 (NT) would be beneficial over the expression of a nonphosphorylatable constitutive nuclear HDAC mutant in adult hearts, which, as previously described, results in acute cardiac dysfunction (Czubryt et al., 2003). Further in vivo studies are needed to test these promising possibilities.
Materials and methods
Chemical reagents and plasmids
Iso and dbcAMP were purchased from Sigma-Aldrich. AEBSF, KT5720, KN93, MG132, E64, EST, leupeptin, pepstatin A, proteasome inhibitor I, lactacystin, AM114, N-acetyl-leucinyl-leucinyl-norleucinal (ALLN), calpeptin, and Z-VAD(OMe)-fluoromethylketone were obtained from EMD, and MDL28170 was obtained from Enzo Life Sciences. Epitope-tagged derivates of CaMKII and class IIa HDACs containing N-terminal Myc or FLAG tags and C-terminal GFP tags were previously described (Backs et al., 2006). HDAC4-GFP was cloned into pDsRed-Monomer-C1 (Takara Bio Inc.), resulting in an N-terminal RFP and a C-terminal GFP tag. HDAC4-FL, HDAC4 2–201 (NT), and HDAC4 2–289 were cloned into p3×FLAG-myc-CMV-24 (Sigma-Aldrich). Point mutations were introduced with the QuikChange kit (Agilent Technologies). Deletion mutants of HDAC4 were generated by PCR with PfuTurbo polymerase (Agilent Technologies). Myc-PKA (catalytic α subunit of PKA, Prkaca) was a gift from O. Nakagawa (Kyoto University Graduate School of Medicine, Kyoto, Japan), and PKG was a gift from T. Lincoln (College of Medicine, University of South Alabama, Mobile, AL). An adenovirus harboring 3×MEF2-Luc was purchased from Seven Hills Bioreagents.
Mice
Mice lacking the catalytic α subunit of PKA, Prkaca, were previously described (Skålhegg et al., 2002) and purchased from Mutant Mouse Regional Resource Centers.
Cell culture and transfection assays
COS cells were maintained in DME with 10% FBS, 2 mM l-glutamine, and penicillin-streptomycin. Transfection was performed with GeneJammer (Agilent Technologies) according to manufacturer’s instructions. H9c2 cells were transfected with FuGENE HD (Roche) according to manufacturer’s instructions.
Cardiomyocyte isolation and adenoviral infection
NRVMs were isolated from 1–2-d Sprague Dawley rats as previously described (Backs et al., 2006). After isolation, NRVMs were maintained in DME/199 medium (4:1) with 10% FBS, 2 mM l-glutamine, and penicillin-streptomycin. NRVMs were infected 24 h after plating, grown 12 h later in serum-free media for another 4 h, and then stimulated with 100 nM ET1 for the indicated time period. Adenoviruses harboring FLAG-HDAC4-WT and FLAG-HDAC4-S/A were previously described (Backs et al., 2006), and adenoviruses harboring 3×FLAG-HDAC4 2–201 (NT) and 2–289 were generated according to the manufacturer’s instructions (ViraPower Adenoviral Expression System; Invitrogen).
Indirect immunofluorescence
COS cells were grown on glass coverslips, fixed in 4% PFA, permeabilized in 0.1% Triton X-100, and blocked in PBS containing 5% goat serum. A primary antibody against FLAG (monoclonal or rabbit; Sigma-Aldrich) was used at a dilution of 1:200. Secondary antibodies conjugated to either fluorescein or Texas red (Vector Laboratories) were also used at a dilution of 1:200. NRVMs were stained with antibodies directed against sarcomeric α-actinin (Sigma-Aldrich). All images were captured at a magnification of ×40.
Apoptosis assays
TUNEL assays were performed using the in situ cell death detection kit (Roche) according to the manufacturer’s protocol. To quantify the number of apoptotic NRVMs, NRVMs were counterstained with sarcomeric α-actinin, and the total numbers of NRVMs and TUNEL-positive nuclei were counted in 10 low power fields in three independent experiments. For each experimental condition, contiguous visual fields were counted to accumulate data on z100 NRVMs per condition per experiment. For annexin V and PI staining, NRVMs were labeled with annexin V–FITC and PI. Approximately 100 cells were counted per condition in more than three randomly selected fields. Stained cells were normalized to total cell count as judged by DAPI staining. More than 80% of cells were sarcomeric α-actinin positive. Adriamyicn treatment (1 µM for 6 h) was used as a positive control.
Coimmunoprecipitation and immunoblotting
COS and cells and NRVMs were harvested 1 d after transfection or transduction in 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100 supplemented with protease inhibitors (Complete; Roche) and 1 mM PMSF. FLAG-tagged proteins were immunoprecipitated with M2-agarose conjugate (Sigma-Aldrich) and thoroughly washed with lysis buffer. Bound proteins were resolved by SDS-PAGE, transferred to PVDF membranes, and immunoblotted as indicated with either anti-Myc antibody (polyclonal, A-14; Santa Cruz Biotechnology, Inc.), a monoclonal anti-FLAG antibody (M2; Sigma-Aldrich), or a polyclonal anti-14-3-3 antibody (Abcam). HDAC4-NT generated from endogenous HDAC was detected with anti-HDAC4 (polyclonal, N-18). MEF2, SRF, and CaMKII antibodies were obtained from Santa Cruz Biotechnology, Inc. A monoclonal glyceraldehyde 3-phosphate dehydrogenase antibody (Millipore) was used as a loading control.
Reporter assays
COS cells were transiently transfected with vectors for CaMKII, PKA, MEF2C, and a luciferase reporter gene under the control of three copies of an MEF2-binding site (3×MEF2-Luc) or four copies of CARG boxes (4×CARG-Luc). H9c2 cells were transiently transfected with 3×MEF2-Luc, and NRVMs were transduced with Ad–3×MEF2-Luc to detect endogenous MEF2 activity. 24 h after transfection, cells were harvested, and luciferase and β-galactosidase levels were determined.
In-gel tryptic digestion and liquid chromatography tandem MS (MS/MS) analysis
Protein bands were cut from the gel out with a scalpel and reduced, alkylated, and digested with trypsin (Catrein et al., 2005) using a liquid handling system (DigestPro MS; intavis AG). Peptides were extracted from the gel pieces with 50% acetonitrile/0.1% trifluoroacetic acid, concentrated nearly to dryness in a SpeedVac vacuum centrifuge (Concentrator 5301; Eppendorf International), and diluted to a total volume of 30 µl with 0.1% trifluoroacetic acid. 25 µl of the sample was analyzed by a nanoHPLC system (Eksigent) coupled to an electrospray ionization mass spectrometer (LTQ Orbitrap; Thermo Fisher Scientific). The sample was loaded on a C18 trapping column (Inertsil; LC Packings) with a flow rate of 10 µl/min of 0.1% trifluoroacetic acid. Peptides were eluted and separated on an analytical column (75 µm × 150 mm) packed with 3 µm of C18 material (Inertsil) with a flow rate of 200 nl/min in a gradient of buffer A (0.1% formic acid) and buffer B (0.1% formic acid and acetonitrile) for the following: 0–6 min in 3% buffer B, 6–60 min in 3–40% buffer B, and 60–65 min in 60–90% buffer B. The column was connected with a nanoelectrospray ionization emitter (New Objective, Inc.). 1,300 V was applied via liquid junction. One survey scan (resolution of 60,000) was followed by three information-dependent product ion scans in the ion trap. 2+, 3+, and 4+ charged ions were selected for fragmentation.
The uninterpreted MS/MS spectra were searched against a small protein database containing the HDAC4 sequence using Mascot software (Matrix Science). The algorithm was set to use no enzyme specificity, assuming carbamidomethyl as a fixed modification of cysteine and oxidized methionine and deamidation of asparagines and glutamine as variable modifications. Mass tolerance was set to four parts per million and 0.2 D for MS and MS/MS, respectively. The peptide sequences of peptides of potential interest were confirmed by manual evaluation of the fragment spectra.
Online supplemental material
Fig. S1 shows additional biochemical and cell biological observations with regard to proteolytic processing of HDAC4, as shown in Fig. 1. Fig. S2 shows an additional biochemical observation with regard to HDAC4 regulation by PKA, as shown in Fig. 4. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201105063/DC1.
Acknowledgments
We thank Michaela Oestringer, Lonny Jürgensen, Jutta Krebs, Ulrike Oehl, and Siska Hermawan for technical help, Osamu Nakagawa for the PKA expression plasmid, and Thomas Ruppert (Zentrum für Molekulare Biologie der Universität Heidelberg, Heidelberg, Germany) for MS analysis.
J. Backs was supported by grants from the Deutsche Forschungsgemeinschaft (Emmy Noether Programme; BA 2258/2-1), the European Commission (FP7-Health-2010 and MEDIA-261409), the Fritz Thyssen Foundation, and the ADUMED Foundation. E.N. Olson was supported by grants from the National Institutes of Health, the Donald W. Reynolds Cardiovascular Clinical Research Center, the Robert A. Welch Foundation (I-0025), the Fondation Leducq Transatlantic Network of Excellence in Cardiovascular Research Program, and the American Heart Association–Jon Holden DeHaan Foundation.
Footnotes
Abbreviations used in this paper:
- β-AR
- β-adrenergic receptor
- dbcAMP
- dibutyryl cAMP
- FL
- full length
- HDAC
- histone deacetylase
- Iso
- isoproterenol
- MEF2
- myocyte enhancer factor 2
- MS
- mass spectrometry
- NRVM
- neonatal rat ventricular myocyte
- PI
- propidium iodide
- SRF
- serum response factor
- WT
- wild type
References
- Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J.D., Vatner S.F., Sadoshima J. 2008. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 133:978–993 10.1016/j.cell.2008.04.041 [DOI] [PubMed] [Google Scholar]
- Backs J., Olson E.N. 2006. Control of cardiac growth by histone acetylation/deacetylation. Circ. Res. 98:15–24 10.1161/01.RES.0000197782.21444.8f [DOI] [PubMed] [Google Scholar]
- Backs J., Song K., Bezprozvannaya S., Chang S., Olson E.N. 2006. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest. 116:1853–1864 10.1172/JCI27438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backs J., Backs T., Bezprozvannaya S., McKinsey T.A., Olson E.N. 2008. Histone deacetylase 5 acquires calcium/calmodulin-dependent kinase II responsiveness by oligomerization with histone deacetylase 4. Mol. Cell. Biol. 28:3437–3445 10.1128/MCB.01611-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backs J., Backs T., Neef S., Kreusser M.M., Lehmann L.H., Patrick D.M., Grueter C.E., Qi X., Richardson J.A., Hill J.A., et al. 2009. The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc. Natl. Acad. Sci. USA. 106:2342–2347 10.1073/pnas.0813013106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backs J., Stein P., Backs T., Duncan F.E., Grueter C.E., McAnally J., Qi X., Schultz R.M., Olson E.N. 2010. The gamma isoform of CaM kinase II controls mouse egg activation by regulating cell cycle resumption. Proc. Natl. Acad. Sci. USA. 107:81–86 10.1073/pnas.0912658106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bers D.M. 2002. Cardiac excitation-contraction coupling. Nature. 415:198–205 10.1038/415198a [DOI] [PubMed] [Google Scholar]
- Bolger T.A., Yao T.P. 2005. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J. Neurosci. 25:9544–9553 10.1523/JNEUROSCI.1826-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catrein I., Herrmann R., Bosserhoff A., Ruppert T. 2005. Experimental proof for a signal peptidase I like activity in Mycoplasma pneumoniae, but absence of a gene encoding a conserved bacterial type I SPase. FEBS J. 272:2892–2900 10.1111/j.1742-4658.2005.04710.x [DOI] [PubMed] [Google Scholar]
- Chang S., McKinsey T.A., Zhang C.L., Richardson J.A., Hill J.A., Olson E.N. 2004. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24:8467–8476 10.1128/MCB.24.19.8467-8476.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Young B.D., Li S., Qi X., Richardson J.A., Olson E.N. 2006. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 126:321–334 10.1016/j.cell.2006.05.040 [DOI] [PubMed] [Google Scholar]
- Cohn J.N., Levine T.B., Olivari M.T., Garberg V., Lura D., Francis G.S., Simon A.B., Rector T. 1984. Plasma norepinephrine as a guide to prognosis in patients with chronic congestive heart failure. N. Engl. J. Med. 311:819–823 10.1056/NEJM198409273111303 [DOI] [PubMed] [Google Scholar]
- Czubryt M.P., Olson E.N. 2004. Balancing contractility and energy production: The role of myocyte enhancer factor 2 (MEF2) in cardiac hypertrophy. Recent Prog. Horm. Res. 59:105–124 10.1210/rp.59.1.105 [DOI] [PubMed] [Google Scholar]
- Czubryt M.P., McAnally J., Fishman G.I., Olson E.N. 2003. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. USA. 100:1711–1716 10.1073/pnas.0337639100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y.S., Xu J., Molkentin J.D. 2005. The DnaJ-related factor Mrj interacts with nuclear factor of activated T cells c3 and mediates transcriptional repression through class II histone deacetylase recruitment. Mol. Cell. Biol. 25:9936–9948 10.1128/MCB.25.22.9936-9948.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F.J., Gupta M., Camoretti-Mercado B., Schwartz R.J., Gupta M.P. 2003. Calcium/calmodulin-dependent protein kinase activates serum response factor transcription activity by its dissociation from histone deacetylase, HDAC4. Implications in cardiac muscle gene regulation during hypertrophy. J. Biol. Chem. 278:20047–20058 10.1074/jbc.M209998200 [DOI] [PubMed] [Google Scholar]
- Edmondson D.G., Lyons G.E., Martin J.F., Olson E.N. 1994. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 120:1251–1263 [DOI] [PubMed] [Google Scholar]
- Erickson J.R., Joiner M.L., Guan X., Kutschke W., Yang J., Oddis C.V., Bartlett R.K., Lowe J.S., O’Donnell S.E., Aykin-Burns N., et al. 2008. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 133:462–474 10.1016/j.cell.2008.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler M.B., Laser J.A., Hopkins G.L., Minobe W., Bristow M.R. 1986. Assessment of the beta-adrenergic receptor pathway in the intact failing human heart: Progressive receptor down-regulation and subsensitivity to agonist response. Circulation. 74:1290–1302 10.1161/01.CIR.74.6.1290 [DOI] [PubMed] [Google Scholar]
- Grimm M., Brown J.H. 2010. Beta-adrenergic receptor signaling in the heart: Role of CaMKII. J. Mol. Cell. Cardiol. 48:322–330 10.1016/j.yjmcc.2009.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger C.M., Schreiber S.L. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA. 97:7835–7840 10.1073/pnas.140199597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M., Montgomery R.L., Olson E.N. 2009. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10:32–42 10.1038/nrg2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausdorff W.P., Caron M.G., Lefkowitz R.J. 1990. Turning off the signal: Desensitization of beta-adrenergic receptor function. FASEB J. 4:2881–2889 [PubMed] [Google Scholar]
- Illi B., Dello Russo C., Colussi C., Rosati J., Pallaoro M., Spallotta F., Rotili D., Valente S., Ragone G., Martelli F., et al. 2008. Nitric oxide modulates chromatin folding in human endothelial cells via protein phosphatase 2A activation and class II histone deacetylases nuclear shuttling. Circ. Res. 102:51–58 10.1161/CIRCRESAHA.107.157305 [DOI] [PubMed] [Google Scholar]
- Kageyama S., Liu H., Nagata M., Aoki F. 2006. Stage specific expression of histone deacetylase 4 (HDAC4) during oogenesis and early preimplantation development in mice. J. Reprod. Dev. 52:99–106 10.1262/jrd.17044 [DOI] [PubMed] [Google Scholar]
- Kehat I., Accornero F., Aronow B.J., Molkentin J.D. 2011. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J. Cell Biol. 193:21–29 10.1083/jcb.201101046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keteyian S.J., Piña I.L., Hibner B.A., Fleg J.L. 2010. Clinical role of exercise training in the management of patients with chronic heart failure. J. Cardiopulm. Rehabil. Prev. 30:67–76 [DOI] [PubMed] [Google Scholar]
- Kim Y., Phan D., van Rooij E., Wang D.Z., McAnally J., Qi X., Richardson J.A., Hill J.A., Bassel-Duby R., Olson E.N. 2008. The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J. Clin. Invest. 118:124–132 10.1172/JCI33255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ahrens M.J., Wu A., Liu J., Dudley A.T. 2011. Calcium/calmodulin-dependent protein kinase II activity regulates the proliferative potential of growth plate chondrocytes. Development. 138:359–370 10.1242/dev.052324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little G.H., Bai Y., Williams T., Poizat C. 2007. Nuclear calcium/calmodulin-dependent protein kinase IIdelta preferentially transmits signals to histone deacetylase 4 in cardiac cells. J. Biol. Chem. 282:7219–7231 10.1074/jbc.M604281200 [DOI] [PubMed] [Google Scholar]
- Liu F., Dowling M., Yang X.J., Kao G.D. 2004. Caspase-mediated specific cleavage of human histone deacetylase 4. J. Biol. Chem. 279:34537–34546 10.1074/jbc.M402475200 [DOI] [PubMed] [Google Scholar]
- Mangmool S., Shukla A.K., Rockman H.A. 2010. β-Arrestin–dependent activation of Ca2+/calmodulin kinase II after β1–adrenergic receptor stimulation. J. Cell Biol. 189:573–587 10.1083/jcb.200911047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee S.L., Hargreaves M. 2010. Histone modifications and skeletal muscle metabolic gene expression. Clin. Exp. Pharmacol. Physiol. 37:392–396 10.1111/j.1440-1681.2009.05311.x [DOI] [PubMed] [Google Scholar]
- McKinsey T.A. 2007. Derepression of pathological cardiac genes by members of the CaM kinase superfamily. Cardiovasc. Res. 73:667–677 10.1016/j.cardiores.2006.11.036 [DOI] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang C.L., Lu J., Olson E.N. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 408:106–111 10.1038/35040593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang C.L., Olson E.N. 2001. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21:6312–6321 10.1128/MCB.21.18.6312-6321.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MERIT-HF Study Group 1999. Effect of metoprolol CR/XL in chronic heart failure: Metoprolol CR/XL randomised intervention trial in congestive heart failure. Lancet. 353:2001–2007 10.1016/S0140-6736(99)04440-2 [DOI] [PubMed] [Google Scholar]
- Métrich M., Laurent A.C., Breckler M., Duquesnes N., Hmitou I., Courillau D., Blondeau J.P., Crozatier B., Lezoualc’h F., Morel E. 2010. Epac activation induces histone deacetylase nuclear export via a Ras-dependent signalling pathway. Cell. Signal. 22:1459–1468 10.1016/j.cellsig.2010.05.014 [DOI] [PubMed] [Google Scholar]
- Miska E.A., Karlsson C., Langley E., Nielsen S.J., Pines J., Kouzarides T. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099–5107 10.1093/emboj/18.18.5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya F.J., Wu C., Richardson J.A., Overbeek P., Olson E.N. 1999. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development. 126:2045–2052 [DOI] [PubMed] [Google Scholar]
- Niu Z., Yu W., Zhang S.X., Barron M., Belaguli N.S., Schneider M.D., Parmacek M., Nordheim A., Schwartz R.J. 2005. Conditional mutagenesis of the murine serum response factor gene blocks cardiogenesis and the transcription of downstream gene targets. J. Biol. Chem. 280:32531–32538 10.1074/jbc.M501372200 [DOI] [PubMed] [Google Scholar]
- Osadchii O.E. 2007. Cardiac hypertrophy induced by sustained beta-adrenoreceptor activation: Pathophysiological aspects. Heart Fail. Rev. 12:66–86 10.1007/s10741-007-9007-4 [DOI] [PubMed] [Google Scholar]
- Parlakian A., Charvet C., Escoubet B., Mericskay M., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., et al. 2005. Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation. 112:2930–2939 [DOI] [PubMed] [Google Scholar]
- Paroni G., Mizzau M., Henderson C., Del Sal G., Schneider C., Brancolini C. 2004. Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol. Biol. Cell. 15:2804–2818 10.1091/mbc.E03-08-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroni G., Fontanini A., Cernotta N., Foti C., Gupta M.P., Yang X.J., Fasino D., Brancolini C. 2007. Dephosphorylation and caspase processing generate distinct nuclear pools of histone deacetylase 4. Mol. Cell. Biol. 27:6718–6732 10.1128/MCB.00853-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroni G., Cernotta N., Dello Russo C., Gallinari P., Pallaoro M., Foti C., Talamo F., Orsatti L., Steinkühler C., Brancolini C. 2008. PP2A regulates HDAC4 nuclear import. Mol. Biol. Cell. 19:655–667 10.1091/mbc.E07-06-0623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passier R., Zeng H., Frey N., Naya F.J., Nicol R.L., McKinsey T.A., Overbeek P., Richardson J.A., Grant S.R., Olson E.N. 2000. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Invest. 105:1395–1406 10.1172/JCI8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal S., Rajagopal K., Lefkowitz R.J. 2010. Teaching old receptors new tricks: Biasing seven-transmembrane receptors. Nat. Rev. Drug Discov. 9:373–386 10.1038/nrd3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman H.A., Koch W.J., Milano C.A., Lefkowitz R.J. 1996. Myocardial beta-adrenergic receptor signaling in vivo: Insights from transgenic mice. J. Mol. Med. 74:489–495 10.1007/BF00204974 [DOI] [PubMed] [Google Scholar]
- Skålhegg B.S., Huang Y., Su T., Idzerda R.L., McKnight G.S., Burton K.A. 2002. Mutation of the Calpha subunit of PKA leads to growth retardation and sperm dysfunction. Mol. Endocrinol. 16:630–639 10.1210/me.16.3.630 [DOI] [PubMed] [Google Scholar]
- Sucharov C.C., Langer S., Bristow M., Leinwand L. 2006. Shuttling of HDAC5 in H9C2 cells regulates YY1 function through CaMKIV/PKD and PP2A. Am. J. Physiol. Cell Physiol. 291:C1029–C1037 10.1152/ajpcell.00059.2006 [DOI] [PubMed] [Google Scholar]
- Timmins J.M., Ozcan L., Seimon T.A., Li G., Malagelada C., Backs J., Backs T., Bassel-Duby R., Olson E.N., Anderson M.E., Tabas I. 2009. Calcium/calmodulin-dependent protein kinase II links ER stress with Fas and mitochondrial apoptosis pathways. J. Clin. Invest. 119:2925–2941 10.1172/JCI38857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega R.B., Matsuda K., Oh J., Barbosa A.C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J.M., Richardson J.A., et al. 2004. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 119:555–566 10.1016/j.cell.2004.10.024 [DOI] [PubMed] [Google Scholar]
- Verdin E., Dequiedt F., Kasler H.G. 2003. Class II histone deacetylases: Versatile regulators. Trends Genet. 19:286–293 10.1016/S0168-9525(03)00073-8 [DOI] [PubMed] [Google Scholar]
- Wagner S., Ruff H.M., Weber S.L., Bellmann S., Sowa T., Schulte T., Anderson M.E., Grandi E., Bers D.M., Backs J., et al. 2011. Reactive oxygen species-activated Ca/calmodulin kinase II{delta} is required for late INa augmentation leading to cellular Na and Ca overload. Circ. Res. 108:555–565 10.1161/CIRCRESAHA.110.221911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhu W., Wang S., Yang D., Crow M.T., Xiao R.P., Cheng H. 2004. Sustained beta1-adrenergic stimulation modulates cardiac contractility by Ca2+/calmodulin kinase signaling pathway. Circ. Res. 95:798–806 10.1161/01.RES.0000145361.50017.aa [DOI] [PubMed] [Google Scholar]
- Wettschureck N., Offermanns S. 2005. Mammalian G proteins and their cell type specific functions. Physiol. Rev. 85:1159–1204 10.1152/physrev.00003.2005 [DOI] [PubMed] [Google Scholar]
- Zhang C.L., McKinsey T.A., Chang S., Antos C.L., Hill J.A., Olson E.N. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 110:479–488 10.1016/S0092-8674(02)00861-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Kohlhaas M., Backs J., Mishra S., Phillips W., Dybkova N., Chang S., Ling H., Bers D.M., Maier L.S., et al. 2007. CaMKIIdelta isoforms differentially affect calcium handling but similarly regulate HDAC/MEF2 transcriptional responses. J. Biol. Chem. 282:35078–35087 10.1074/jbc.M707083200 [DOI] [PubMed] [Google Scholar]
- Zhu W.Z., Wang S.Q., Chakir K., Yang D., Zhang T., Brown J.H., Devic E., Kobilka B.K., Cheng H., Xiao R.P. 2003. Linkage of beta1-adrenergic stimulation to apoptotic heart cell death through protein kinase A-independent activation of Ca2+/calmodulin kinase II. J. Clin. Invest. 111:617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]