When fission yeast express mutant cofilin that is inefficient at actin filament severing, cytokinetic contractile ring formation is severely impaired, but ring contraction proceeds efficiently.
Abstract
We created two new mutants of fission yeast cofilin to investigate why cytokinesis in many organisms depends on this small actin-binding protein. These mutant cofilins bound actin monomers normally, but bound and severed ADP-actin filaments much slower than wild-type cofilin. Cells depending on mutant cofilins condensed nodes, precursors of the contractile ring, into clumps rather than rings. Starting from clumped nodes, mutant cells slowly assembled rings from diverse intermediate structures including spiral strands containing actin filaments and other contractile ring proteins. This process in mutant cells depended on α-actinin. These slowly assembled contractile rings constricted at a normal rate but with more variability, indicating ring constriction is not very sensitive to defects in severing by cofilin. Computer simulations of the search-capture-pull and release model of contractile ring formation predicted that nodes clump when the release step is slow, so cofilin severing of actin filament connections between nodes likely contributes to the release step.
Introduction
Cytokinesis, the final stage of cell division, depends on the assembly and constriction of a contractile ring of actin filaments, myosin-II, formins, and many other proteins. Here we focus on the actin binding protein cofilin, which concentrates in the cleavage furrow during cytokinesis in animal cells (Nagaoka et al., 1995) and is required in some way for cytokinesis in a variety of organisms including nematodes (Ono et al., 2003), fruit flies (Gunsalus et al., 1995), frogs (Abe et al., 1996) and fission yeast (Nakano and Mabuchi, 2006).
Analysis of cytokinesis is favorable in fission yeast, thanks to the most complete genetic inventory of participating proteins (Bathe and Chang, 2010; Pollard and Wu, 2010). Cofilin is essential for the viability of fission yeast and depletion of the native protein or replacement of the adf1+ gene with a temperature sensitive mutation (adf1-1) results in defects in contractile ring assembly and stability (Nakano and Mabuchi, 2006; note, Nakano and Mabuchi named the single cofilin gene in fission yeast adf1+ for actin depolymerizing factor, the original name of this family [Bamburg et al., 1980]. We retain this gene name but call the protein cofilin, a later name [Nishida et al., 1984], because these proteins sever but do not depolymerize actin filaments [Andrianantoandro and Pollard, 2006; Pavlov et al., 2007; Chan et al., 2009]). However, these observations of fixed cells did not reveal why contractile ring assembly fails in adf1-1 cells, nor were the cofilin mutants characterized biochemically, leaving open questions about mechanisms.
A simple search-capture-pull and release model explains many features of contractile ring assembly in fission yeast (Vavylonis et al., 2008). The model postulates that myosin-II, located in precursor structures called nodes, captures actin filaments growing from formins in neighboring nodes and pulls the nodes together. Short-lived connections between nodes explain their motions and are required to form contractile rings in computer simulations because stable connections between nodes produce clumps of nodes rather than a continuous ring. Two mechanisms were proposed to break connections between nodes: either myosin-II might dissociate from the connecting actin filaments owing to its low processivity; or the connecting actin filaments might break. Live cell microscopy with fluorescence markers on both myosin-II and actin filaments provided evidence for both mechanisms (Vavylonis et al., 2008).
We tested the hypothesis that the role of cofilin in cytokinesis is to sever actin filaments during assembly of contractile rings. Our initial observations of the temperature-sensitive cofilin adf1-1 strain by fluorescence microscopy of live cells showed that precursors called nodes failed to condense into a contractile ring at the restrictive temperature. The adf1-1 cofilin protein (L57S) was not sufficiently stable to purify, so we created seven new mutations based on temperature-sensitive mutations of budding yeast cofilin (Lappalainen et al., 1997). Two of these mutant cofilins were defective in binding and severing actin filaments. Experiments with these two cofilins in live cells revealed that the actin filament severing by cofilin is crucial for the normal assembly and stability of the contractile ring. Cells depending on a mutant cofilin failed to assemble a contractile ring directly from a broad band of precursor nodes, but most of these cells eventually formed contractile rings via a slow, unreliable process depending on the actin filament cross-linking protein α-actinin, similar to that observed in cells lacking anillin Mid1p. The cofilin mutations had only subtle effects on constriction of contractile rings, in spite of the difficulties during assembly.
Results
Contractile ring defects in cells with a temperature-sensitive cofilin mutation
We used fluorescence microscopy to characterize the phenotype of live adf1-1 cells expressing a triple GFP-tag on the myosin-II regulatory light chain (Rlc1p-3GFP) to mark the contractile ring and its precursors (Vavylonis et al., 2008). At 25°C adf1-1 cells were indistinguishable from wild-type cells. Both strains grew at the same rate. Both concentrated Rlc1p-3GFP in punctate cortical structures called nodes starting 10 min before spindle pole body separation, time zero in the cellular time frame defined by Wu et al. (2003). Beginning at time +1 min, nodes in both strains condensed over 10 min into a contractile ring that began to constrict at the same rate ∼30 min later (Fig. S1 A).
At 36°C wild-type cells completed every stage of cytokinesis normally but twice as fast as at 25°C (Wu et al., 2003) (Fig. S1 B, Video 1), but adf1-1 cells developed contractile ring defects within 5 min of shifting to the restrictive temperature. Shifting adf1-1 cells with broad bands of nodes to 36°C caused the nodes to coalesce into clumps within 10 min rather than forming continuous contractile rings (Fig. S1 C, Video 2).
Interpretation of the phenotype of the adf1-1 mutant required information about the biochemical properties of adf1-1 cofilin, so we attempted to purify recombinant adf1-1 cofilin L57S from bacteria. The protein was insoluble in low salt buffer, which precluded purification.
Localization of cofilin during cytokinesis
GFP-cofilin concentrated with actin filaments in endocytic actin patches and contractile rings (Fig. S1) as observed previously (Nakano and Mabuchi, 2006), but did not accumulate directly in nodes early in the process of cytokinesis (Fig. S1). GFP-cofilin was abundant in the broad band of nodes but spread diffusely between the nodes during contractile ring assembly (Fig. S1, E and F). Overexpression of GFP-cofilin from either the strong ADH promoter or 3nmt1 promoter was required to complement deletion of adf1+ because cells depending on GFP-cofilin expressed from the endogenous adf1 promoter were not viable (unpublished data). Therefore, as in budding yeast (Okreglak and Drubin, 2007), GFP-cofilin is not fully functional in fission yeast.
Construction of new cofilin mutants
We created new cofilin mutants that are useful for both biochemical and cellular studies. We based seven fission yeast cofilin mutants (Table I) on three temperature-sensitive alanine substitution mutants of budding yeast cofilin (Lappalainen et al., 1997). We integrated these mutations into the endogenous cofilin locus in a diploid strain, with one copy of wild-type adf1+. Tetrad analysis showed that haploid cells depending on six of the cofilin mutants were viable even at high temperature (Table I), so these mutations were not temperature sensitive.
Table I.
Cofilin mutants used in this study
| Strain | Corresponding S. cerevisiae strain (point mutations) | Amino acid substitutions | Growth at 25°C | Growth at 36°C | Synthetic lethal with Δaip1 | Synthetic lethal with ts tropomyosin mutant cdc8-27 | Synthetic interaction with α-actinin deletion mutant |
| adf1-1 | None | L57S | Normal | None | Yes | No | N/T |
| adf1-M1 | cof1-5 (D10A E11A) | P10A, E11A | Normal | Normal | Yes | N/T | N/T |
| adf1-M1A | None | E11A | Normal | Normal | N/T | N/T | N/T |
| adf1-M2 | cof1-8 (K23A, K24A, K26A) | K23A, S24A, R26A | Slow | Slow | Yes | No | Synthetic slow growth |
| adf1-M2A | None | K23A | Slow | Slow | N/T | No | N/T |
| adf1-M2B | None | K23A, R26A | Slow | Slow | N/T | N/T | N/T |
| adf1-M3 | None | E132A, K133A | Slow | Slow | Yes | No | N/T |
| adf1-M3A | cof1-22 (E134A, R135A, R138A) | E132A, K133A, R136A | None | None | N/T |
Growth was measured on YE5s agar plates as in Fig. 1. N/T, not tested.
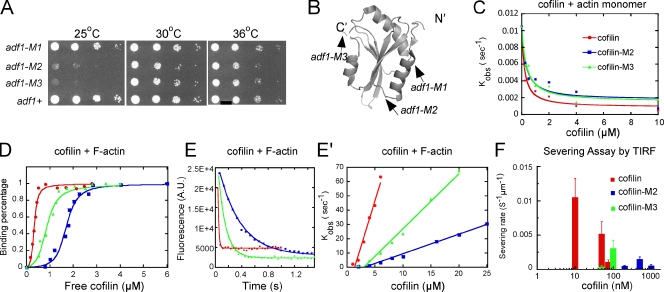
We focused on adf1-M2 and adf1-M3, which had the most serious growth defects (Fig. 1, A and B; Table I). The mutations in adf1-M3 (E132A, K133A) are located near the C terminus of cofilin. The corresponding residues are part of the interface between the twinfillin C-terminal ADH domain and actin monomer in a cocrystal structure (Paavilainen et al., 2008), so the mutations might directly compromise binding. The mutations in adf1-M2 (K23A, S24A, R25A) are in a loop on the opposite side of cofilin and not expected to interact directly with either an actin monomer or subunits in an actin filament (Paavilainen et al., 2008).
Figure 1.
Mutant cofilins with defects in severing actin filaments. (A) Growth of wild-type and mutant strains. Drops with 10-fold serial dilutions were plated on YE5s agar and grown for 4 d at 25°C, or for 3 d at 30 or 36°C. (B) Ribbon diagram of fission yeast cofilin (PDB: 2i2q) rendered by PyMol (DeLano Scientific). Arrows indicate sites of point mutations in adf1-M1 (P10A, E11A), adf1-M2 (K23A, S24A, R25A), and adf1-M3 (E132A, K133A). (C–F) Biochemical characterization of wild-type cofilin (red symbols and lines), cofilin-M2 (blue), and cofilin-M3 (green). (C) Dependence of kobs for dissociation of etheno-ATP from actin monomers in 1 mM MgCl2, 1 mM EGTA, 2 mM TrisCl, and 0.5 mM DTT, pH 8.0 on the concentrations of the three cofilins. The smooth curves are fits of the binding equation (Bryan, 1988; Hawkins et al., 1993) yielding the Kds listed in Table II. (D) Equilibrium binding of a range of cofilin concentrations to 2 µM pyrene-labeled actin filaments in KMEI polymerization buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0) measured by fluorescence after 0.5 h incubation. The fraction bound was calculated from the fluorescence decrease relative to the maximum decrease. The free cofilin concentration was the total cofilin in the reaction minus the cofilin bound to the actin filaments. (E) Time course of 3 µM cofilin binding to 1 µM pyrene-labeled actin filaments in KMEI polymerization buffer measured by the decrease in fluorescence. Fits of a single exponential equation to the data gave observed rate constants (kobs) of 73.6 s−1 (wild type), 3.0 s−1 (cofilin-M2), and 8.9 s−1 (cofilin-M3). (E′) Concentration dependence of kobs for cofilin binding to 2 µM pyrene-labeled actin filaments in KMEI polymerization buffer. Each data point is an average of kobs for at least three reactions. (F) Cofilin concentration dependence of the actin filament–severing rate in TIRF buffer observed by total internal reflection fluorescence (TIRF) microscopy (see Fig. S2 E). Severing of actin filaments by 200 nM cofilin-M3 was negligible. Error bars represent the standard deviations.
Similar to adf1-1 cells at the restrictive temperature (Nakano and Mabuchi, 2006), adf1-M2 and adf1-M3 mutations were synthetically lethal with deletion mutations of actin-interacting protein 1 aip1+ at 25°C (Table I), consistent with the behavior of budding yeast cofilin mutants (Rodal et al., 1999). On the other hand, our cofilin mutants were not synthetically lethal with temperature-sensitive tropomyosin mutant cdc8-27 at the permissive temperature (Table I).
Biochemical characterization of cofilin mutants cofilin-M2 and cofilin-M3
We characterized the biochemical properties of cofilin-M2 and cofilin-M3 purified from bacteria (Fig. S2 A). Cofilin-M3 is slightly more stable to urea denaturation than the wild-type protein, whereas cofilin-M2 is slightly less stable (Table II; Fig. S2 B). Cells grew normally when dependent on cofilin-M1 (Fig. 1 A), a mutant with stability similar to cofilin-M2 (unpublished data). Measurements of the inhibition of nucleotide exchange of monomeric actin (Figs. S2 C and 1 C) showed that wild-type cofilin, cofilin-M2, and cofilin-M3 all have Kds of ∼0.1 µM for binding monomeric actin (Table II). Therefore, the cellular defects caused by these cofilin mutations are unlikely a result from defects in binding to actin monomers.
Table II.
Biochemical properties of mutant cofilins
| Parameter | Cofilin | Cofilin-M2 | Cofilin-M3 |
| Stability: ΔG (kcal mol−1) | 9.10 | 7.91 | 10.32 |
| Mg-ATP-actin monomer affinity, Kd (μM) | 0.08 | 0.11 | 0.17 |
| ADP-actin filament apparent affinity, Ka (μM−1) | 0.15 | 0.012 | 0.08 |
| ADP-actin filament affinity, Kd (μM) | 0.33 | 1.7 | 0.89 |
| ADP-actin filament binding cooperativity, ω | 19.9 | 48.7 | 14.2 |
| ADP-actin filament association rate constant, k+ (µM−1 s−1) | 12.5 | 1.4 | 3.8 |
| ADP-actin filament dissociation rate constant, k- (s−1) | 200 | 4600 | 140 |
| Ratio of k-/k+ (µM) | 16 | 3300 | 37 |
| Optimal severing concentration (nM) | 10 | 500 | 100 |
| Maximum serving rate (events per 1,000 subunits s−1) | 0.032 | 0.009 | 0.012 |
Stability was determined from the dependence of the intrinsic fluorescence on urea concentration (Fig. S2 B). Affinity for ATP-actin monomers was measured from the effect on nucleotide exchange (Fig. 1 C and S2 C). Association equilibrium constants Ka and cooperativity ω were calculated from best fits to equilibrium binding data of Fig. 1 D. Apparent dissociation equilibrium constants Kd were calculated as Kd = 1/(ω*Ka). Association k+ and dissociation k- rate constants for cofilin binding pyrenyl-actin filaments were calculated from kinetic curves of Figs. 1, E–E’, and S2 D. Severing rates were measured by TIRF microscopy (Figs. 1 F and S2 E).
Both cofilin mutants had selective defects in actin filament binding and severing. In equilibrium binding experiments, where cofilin quenched the fluorescence of pyrenyl-actin filaments in proportion to binding (Carlier et al., 1997), plots of fluorescence vs. concentration of free cofilin were sigmoidal (Fig. 1 D). Analysis of these binding data with the De La Cruz application (De La Cruz, 2005) of an unlimited nearest-neighbor cooperativity model (McGhee and von Hippel, 1974; Kowalczykowski et al., 1986) gave apparent Kds of 1.7 µM for cofilin-M2, 0.89 µM for cofilin-M3, and 0.34 µM for wild-type cofilin, the latter close to previous work (Andrianantoandro and Pollard, 2006).
Both cofilins M2 and M3 bound to pyrenyl-actin filaments slower than wild-type cofilin (Fig. 1 E). Cooperativity makes these reactions more complicated than a simple bimolecular reaction, so single exponentials fit the time courses even though the conditions were not pseudo-first order (Chan et al., 2009). Therefore, slopes of the linear plots of observed rate constants vs. cofilin concentration (Fig. 1 E′) are apparent association rate constants (Table II). As noted for wild-type cofilins, the ratios of k−/k+ disagree with the Kds measured under equilibrium conditions and indicate that M2 is more disabled for filament binding than revealed by the other parameters. Cofilin-M2 also dissociates slower from actin filaments than wild-type cofilin and cofilin-M3 (Fig. S2 D).
Real-time observations by total internal reflection fluorescence (TIRF) microscopy (Fig. S2 E) showed that the cofilin mutants were defective at severing actin filaments (Fig. 1 F). The severing activity of cofilin-M2 peaked around 500 nM and cofilin-M3 around 100 nM, whereas severing was optimal with 10 nM wild-type cofilin (Andrianantoandro and Pollard, 2006). Even at its optimal concentration, cofilin-M2 severed filaments far slower than cofilin-M3, which was also slower than wild-type cofilin (Fig. 1 F and Table II).
Effects of low affinity cofilins on contractile ring assembly
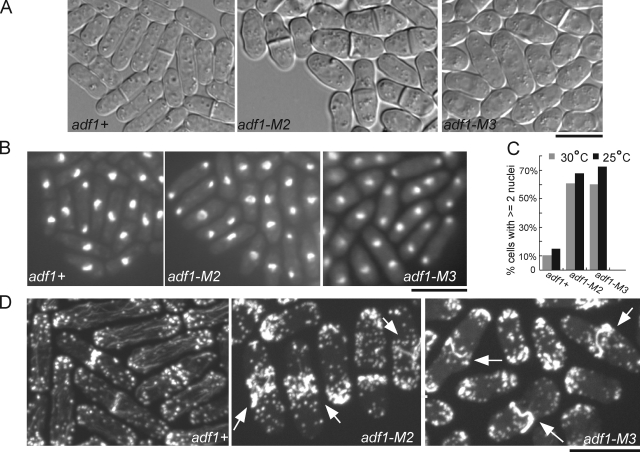
Cells dependent on cofilin-M2 or cofilin-M3 had defects in morphology and septation (Fig. 2 A). Cofilin mutant cells were 30% larger in diameter but about normal in length (Fig. S3, A and B). Many cofilin mutant cells had two or more nuclei, indicating a defect in cell division (Fig. 2, B and C). Staining fixed cells with Bodipy-phallacidin showed thick strands containing actin filaments near the equators of many cofilin mutant cells (Fig. 2 D). Wild-type cells rarely had such strands. These phenotypes are likely to be due to the biochemical defects of these mutant cofilins rather than expression problems because cells expressed cofilin-M2 or cofilin-M3 at levels similar to wild-type cofilin (Fig. S3 C).
Figure 2.
Cytokinesis defects in cells with mutant cofilins. Comparison of wild-type (adf1+), adf1-M2, and adf1-M3 cells. (A) DIC images at 25°C. (B) Fixed cells stained with DAPI showing many mutant cells with two nuclei. (C) Histograms of the fractions of cells with two or more nuclei in samples of >300 cells of each strain grown at 25 or 30°C in two separate experiments, fixed and stained with DAPI. (D) Fixed cells stained with Bodipy-phallacidin showing thick bundles of actin filaments in the middle of many mutant cells (arrows). Bars, 10 µm.
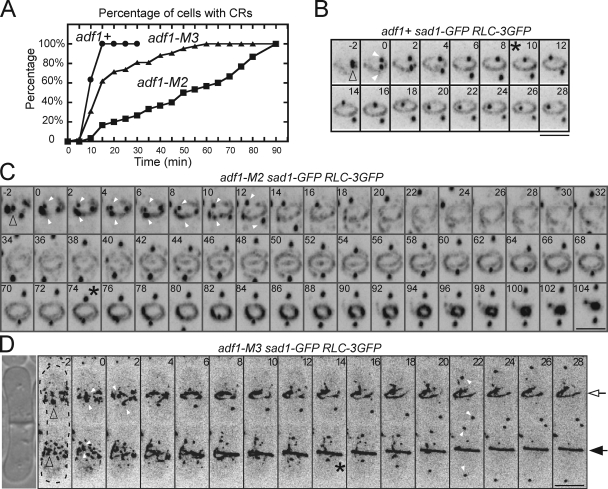
Contractile ring assembly was much slower and more variable in cells depending on mutant cofilins than in wild-type cells (Fig. 3, A–D; Videos 3 and 4). Spindle pole bodies marked with Sad1p-GFP allowed us to align all of the cells on the same time frame (Wu et al., 2003), while Rlc1p-3GFP marked nodes and contractile rings. Wild-type cells condensed broad bands of nodes into rings in 10.2 ± 1.7 min (mean ± SD; n = 31; Fig. 3 B) at 25°C, but adf1-M2 cells required an average of 52.1 ± 29.1 min (n = 30) and adf1-M3 cells took 21.5 ± 18.0 min (n = 42; Table III). 15 min after the separation of SPBs, every wild-type cell had a complete contractile ring compared with just 17% of adf1-M2 cells and 60% of adf1-M3 cells (Fig. 3 A). In spite of these difficulties most mutant cells completed cytokinesis, so <1% of interphase mutant cells had two nuclei. On the other hand, many binucleate cells accumulated during the greatly prolonged cytokinesis (Fig. 2, B and C).
Figure 3.
Contractile ring assembly in strains with cofilin mutations. (A) Time course of the accumulation of complete contractile rings in populations of wild-type (adf1+), adf1-M2, and adf1-M3 cells with the separation of SPBs at time zero. (B–D) Time-lapse series of negative-contrast fluorescence micrographs showing contractile ring assembly in cells expressing Rlc1p-3GFP and the spindle pole body (SPB) marker Sad1-GFP. Maximum intensity projections were titled 45° to show SPBs and contractile rings and their precursors in B and C. Numbers are times in minutes after SPB separation. Empty arrowheads mark SPBs before they split and filled arrowheads mark separated SPBs. Asterisks mark the completion of contractile rings. Bars, 5 µm. (B) This wild-type cell assembled a complete contractile ring by time +10 min (also see Video 3). (C) This adf1-M2 cell split its SPB at time zero and condensed the nodes into clumps between time −2 to +8 min. During the following 70 min, strands containing myosin-II grew from these clumps and assembled a complete ring, which then constricted (also see Video 4). (D) Two conjoined daughter adf1-M3 cells (DIC image shown on the left) underwent the next round of cytokinesis simultaneously but differed in the time to assemble contractile rings: time +14 min for the bottom cell (filled arrow) and time +28 min for the top cell (empty arrow).
Table III.
Cytokinesis defects of cofilin mutant cells
| Cells with two nuclei at 30°C | Average time to assemble a ring | Cells with a complete contractile ring 15 min after separation of SPBs | Average constriction rate ± SD | |
| % | min | % | µm/min | |
| adf1+ | 10 | 10.2 ± 1.7 | 100 | 0.30 ± 0.03 |
| adf1-M2 | 61 | 52.1 ± 29.1 | 17 | 0.32 ± 0.10 |
| adf1-M3 | 60 | 21.5 ± 18.0 | 60 | 0.30 ± 0.08 |
To investigate the defects in contractile ring assembly in cofilin mutant cells, we looked closely at the behavior of the precursors, nodes of proteins, which normally coalesce into a contractile ring (Wu et al., 2006; Vavylonis et al., 2008). Wild-type and cofilin mutant cells initiated contractile rings starting from similar numbers of nodes (Fig. S3 E) located in broad bands around the equator and containing myosin-II, formin Cdc12p, and F-BAR protein Cdc15p. During ring assembly in both wild-type and cofilin mutant cells these nodes became enmeshed in a network of actin filaments (Fig. S3 F).
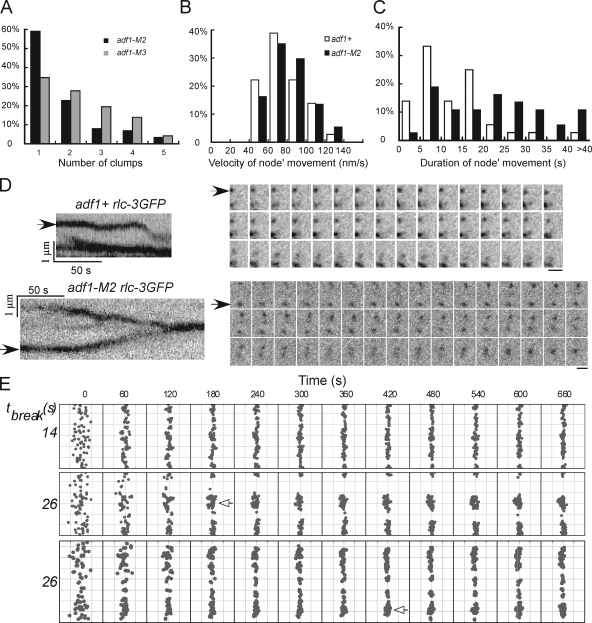
When actin filaments first appeared at time +1 min (Vavylonis et al., 2008), nodes began moving in both wild-type (Fig. 4 A; Video 5) and cofilin mutant cells (Fig. 4, B and C; Videos 6 and 7), resulting in the nodes coalescing into a ring in ∼10 min in wild-type cells (Vavylonis et al., 2008), but forming one or more large, highly fluorescent clump in cofilin mutant cells (Fig. 4, B–E; Videos 6 and 7). We defined clumps as bright accumulations of Rlc1p-3GFP that were stationary for at least 120 s after the time that nodes started to move because nodes and small accumulations of nodes rarely pause >60 s in wild-type cells (Video 5). Nodes formed clumps in adf1-M2 cells in ∼300 s (291 ± 92 s, n = 7) from the point when they started to move. As they grew in size, clumps attracted nearby nodes (Fig. 4, D and E). We counted the numbers of clumps that were stationary for ≥10 min in cofilin mutant cells: 81% of adf1-M2 cells and 62% of adf1-M3 cells had 1 or 2 such clumps, whereas the other cells had more (Fig. 5 A). Nodes in adf1-1 cells also condensed into clumps at the restrictive temperature (Fig. S3 G).
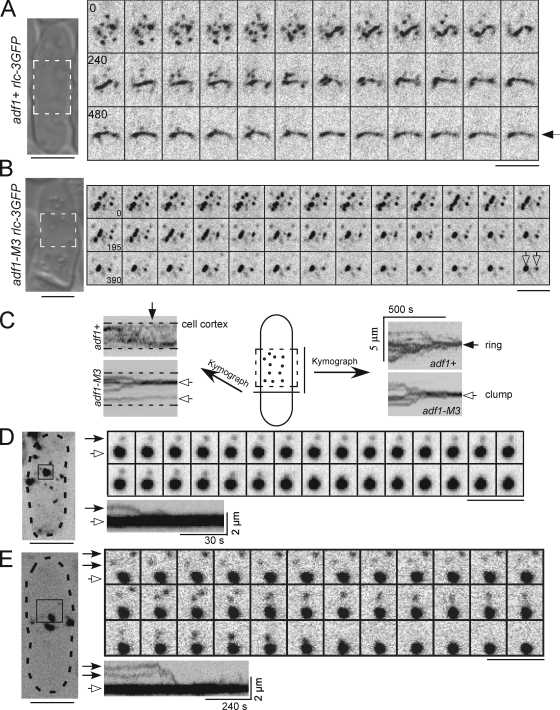
Figure 4.
Node movements in wild-type and cofilin mutant cells. Negative-contrast fluorescence micrographs of cells with nodes marked by Rlcp-3GFP at 25°C. Numbers are time in seconds on an arbitrary time scale. Bars, 5 µm. (A and B) A DIC image of each cell is shown on the left with the region of interest highlighted by dashed lines. Montages show time series of single confocal sections. (A) This wild-type cell condensed nodes into a ring (arrow) in 480 s from the beginning of this series. The interval between each frame is 20 s. Also see Video 5. (B) The nodes in this adf1-M3 cell formed one big clump and a small clump (empty arrows). Also see Videos 6 and 7. (C) Kymographs of node movements in an adf1+ cell (from A) and an adf1-M3 cell (from B). Kymographs were constructed from time series of projections of the top confocal section of the regions of interest (dashed rectangle) onto lines either perpendicular (left) or parallel (right) to the long axis of the cell. Over time nodes formed either a contractile ring (arrow) or clumps (empty arrows). (D and E) Motions of nodes (closed head arrows) relative to clumps (open head arrows) in adf1-M2 cells. Boxes outline regions of interest (ROI) in maximum intensity projections of a Z-series of fluorescence micrographs of whole cells (dashed lines) at the start of the time series. Kymographs were generated as in C. (D) A node merged with a nearby clump of nodes. The ROI was rotated 90° clockwise for viewing. (E) Two nodes merged with each other before merging with a clump of nodes.
Figure 5.
Measurements and simulations of node movements in wild-type and cofilin mutant cells. (A) The numbers of clumps of nodes stationary for ≥10 min in adf1-M2 (black, n = 88) and adf1-M3 (gray, n = 72) cells. (B) Histogram of the distribution of the durations of node movements in wild-type (unfilled, n = 36) and adf1-M2 (black, n = 37) cells. (C) Histogram of the distributions of node velocities in wild-type and adf1-M2 cells. A–C are based on data from single experiments, exacted from multiple image sets acquired independently. (D) Time-lapse fluorescence micrographs of moving nodes in an adf1+ cell (top) and an adf1-M2 cell (bottom). Intervals between frames are 3 s. Left, kymographs of each time lapse series made as in Fig. 4 C. The node in the adf1+ cell (arrow) alternated between pauses and directed movements, while the node in the adf1-M2 cell (arrow) moved for a much longer time. Bars, 1 µm. (E) Snapshots of Monte Carlo simulations of the search-capture-pull and release model of contractile ring assembly (Vavylonis et al., 2008). Top two rows show simulations with a cell radius of 1.8 µm and two values of τbreak, based on node movements in adf1+ (14 s) and adf1-M2 (26 s) cells. The bottom row is a simulation with τbreak = 26 s and the radius of cofilin mutant cells, 2.4 µm. The equator of the cell is filleted open with the circumference vertical for display. The 65 nodes in the broad band are indicated as gray dots. Nodes form clumps (empty arrows), with τbreak = 26 s. The numbers on the top are times in seconds. Representatives from multiple simulations are shown.
We tracked the movements of nodes marked with Rlc1p-3GFP at 1-s intervals in a confocal section of the top of cells at 25°C to understand the defects in cofilin mutant cells. Nodes moved at the same velocity in wild-type and adf1-M2 cells and in adf1-M3 cells (Fig. 5 B and Table IV). However, nodes in cofilin mutant cells moved longer total distances over longer times between pauses (Fig. 5, C and D, Table IV): 27% of nodes in adf1-M2 cells moved for >30 s, compared with only 5% in wild-type cells (Fig. 5 C and Table IV).
Table IV.
Node movements during contractile ring formation
| Durationa | Total distance traveledb | Velocityc | Displacementd | |
| s | μm | nm/s | μm | |
| adf1+ (n = 36) | 14 ± 8 | 1.1 ± 0.9 | 79 ± 21 | 0.33 ± 0.12 |
| adf1-M2 (n = 37) | 26 ± 18*** | 2.0 ± 1.5** | 81 ± 22 | 0.43 ± 0.23* |
| adf1-M3 (n = 43) | 23 ± 17** | 1.8 ± 1.2** | 83 ± 27 | 0.48 ± 0.24*** |
The measurements are mean values ±1 standard deviation. Two-tailed t tests were used to calculate the statistical significance between wild-type and cofilin mutant cells: ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Duration is defined as the time that a node moved continuously between pauses.
Total distance traveled is the cumulative distance that nodes traveled between pauses.
Velocity is the total distance that a node traveled between pauses divided by duration of that movement.
Displacement is the straight line distance between the point where a node started to move and the end point where it stopped.
Monte Carlo simulations of contractile ring assembly by the search-capture-pull and release mechanism (Vavylonis et al., 2008) correctly predicted that long-lived actin filament connections between nodes result in clumping of nodes (Fig. S11 in Vavylonis et al., 2008). The relevant parameter in the model is τbreak, the lifetime of connections between neighboring nodes. In the cells we studied τbreak was estimated to be 14 s in wild-type cells, 26 s in adf1-M2 cells, and 23 s in adf1-M3 cells (Table IV) on the basis of the durations of node movements, as expected if severing of actin filaments connecting neighboring nodes contributes to pauses. In computer simulations of ring assembly (Vavylonis et al., 2008) nodes condensed into rings with few breaks with τbreak = 14 s but formed clumps separated by large gaps in a few hundred seconds with τbreak = 26 s in either the wild-type or larger diameter of cofilin mutants (Fig. 5 E). Fig. S5 illustrates the formation of clumps with larger values of τbreak. Increasing the cell radius by up to 30% with native τbreak of 14 s had only minor effects on the assembly of rings in 600 s (Fig. S4 B).
Transformation of clumps of nodes into contractile rings in cofilin mutant cells
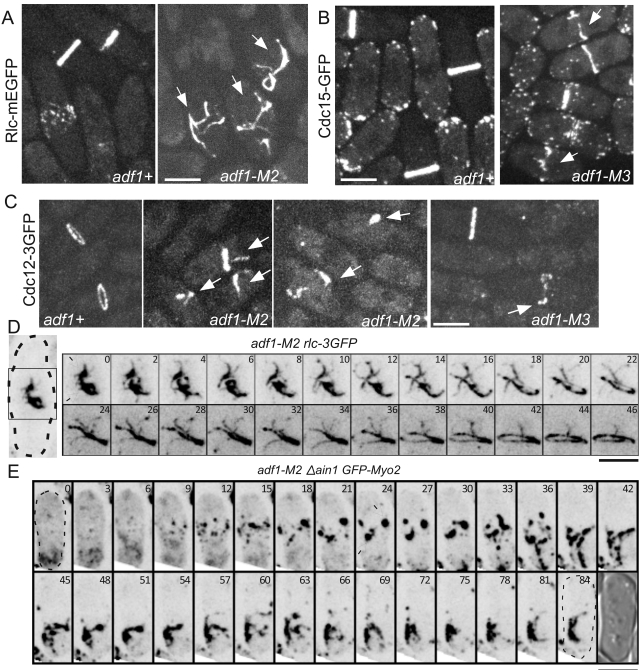
Rather than being a dead end, the clumps of nodes in cofilin mutant cells became organizing centers for contractile ring assembly. Strands composed of contractile ring proteins (actin [Fig. 2 D], myosin-II [Fig. 6 A], Cdc15p [Fig. 6 B], and Cdc12p [Fig. 6 C]) emerged from clumps of nodes and slowly organized into few long bundles (Video 8), which then slowly compacted into a contractile ring (Figs. 3 C and 6 D; Videos 4 and 9).
Figure 6.
Slow unreliable assembly of contractile rings from clumps in cofilin mutant cells. (A–C) Fluorescence micrographs of maximum intensity projections of wild-type and cofilin mutant cells expressing fluorescent proteins fused to contractile ring proteins. These proteins concentrated in strands (arrows) in dividing cells. (A) Rlc1p-mEGFP in wild-type and adf1-M2 cells. (B) GFP-Cdc15p in wild-type and adf1-M3 cells. (C) Cdc12p-3GFP in wild-type, adf1-M2, and adf1-M3 cells. (D and E) Time series of maximum intensity projections of negative-contrast confocal slides; fluorescence micrographs of cells expressing Rlc1p-3GFP. Numbers are times in minutes. (D) This adf1-M2 cell assembled a contractile ring from two clumps of nodes (empty arrows). The region of interest is outlined in the image of the whole cell on the left. (E) This adf1-M2 Δain1 cell failed to assemble stable strands or a ring. The last frame is a DIC micrograph showing the cell at the last time point. Bars, 5 µm.
Contractile ring assembly in cofilin mutant cells was more dependent than wild-type cells on the actin filament cross-linking protein α-actinin. Deletion of the α-actinin ain1+ gene had little effect on cytokinesis in otherwise normal fission yeast cells (Wu et al., 2001; Fig. S5 A). Similar to wild-type cells, Δain1 cells assembled complete contractile rings in 24.7 ± 3.6 min (n = 25) after Myo2 appeared in nodes. However, Δain1 adf1-M2 double-mutant cells are much sicker than either adf1-M2 or Δain1 cells (Fig. S5 B). Although less than 1% of Δain1 or adf1-M2 cells had two interphase nuclei, 7.7% of Δain1 adf1-M2 double-mutant cells had two or more interphase nuclei (Fig. S5 C; n = 225) resulting from failures of cytokinesis. Live cell imaging showed that Δain1 adf1-M2 cells frequently failed to assemble a contractile ring, even after attempting to form rings from strands as in adf1-M2 cells (Fig. 6 E).
Effects of cofilin mutants on contractile ring stability and constriction
In 10% of cofilin mutant cells (4/42) with complete rings, the ring fragmented and degenerated into clumps (Fig. S5 D). Similarly, contractile rings in adf1-1 cells shifted to the restrictive temperature disintegrated within 10 min into puncta about the same size as nodes (Fig. S5 E, Video 10). This was never observed in wild-type cells, so contractile rings are less stable in cofilin mutant cells.
Wild-type cells delayed for ∼30 min (34 ± 7.4 min, n = 8) after completing contractile ring assembly before starting to constrict the ring (Wu et al., 2003), but contractile rings in adf1-M2 cells began to constrict soon (10 ± 4.8 min, n = 11) after they formed (Fig. 3 C; Video 4).
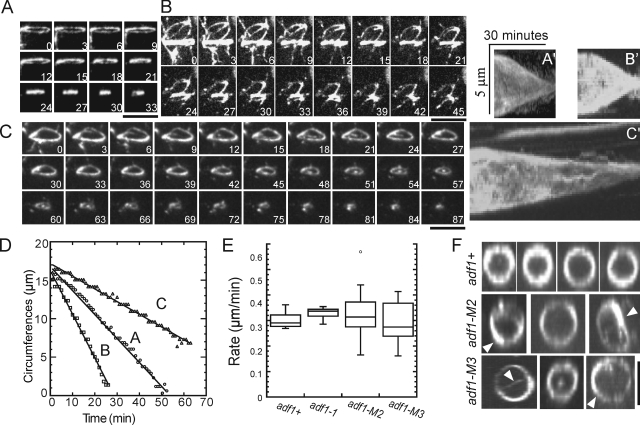
At 25°C the average rate of contractile ring constriction was the same in wild-type, adf1-M2, and adf1-M3 cells (Fig. 7 E and Table III), but the rates of constriction were much more variable in mutant cells (Fig. 7, A–C). Pauses did not account for the slower rates because each ring constricted at a constant rate (Fig. 7 D). Constricting contractile rings were homogenous in wild-type cells but were often irregular in cofilin mutant cells with thick bundles of actin filaments (Fig. 7 F), which persisted throughout constriction.
Figure 7.
Contractile ring constriction in wild-type and cofilin mutant cells. Time courses of contractile ring constriction observed by fluorescence microscopy of adf1-M2, adf1-M3, adf1-1, and wild-type strains expressing Rlc1p-3GFP at 25°C. Bars, 5 µm. (A–C) Time series of maximum intensity projections of fluorescence images of three adf1-M2 rlc1-3GFP cells. Numbers are times in minutes from the beginning of the recordings. (A′–C′) Kymographs of ring constriction by cells A, B, and C constructed by projecting fluorescence of the rings onto a time line. (D) Time course of changes in contractile ring circumference calculated from projection images for cells A–C. Constriction rates: (A) 0.31 µm/min; (B) 0.56 µm/min; (C) 0.16 µm/min. (E) Box plots of contractile ring constriction rates. The horizontal line in the box is the median. The top and bottom of the box include ± 25% from the mean value of the population. The lines extending from the top and bottom of each box mark the minimum and maximum values except for one outlier (displayed as a single point), which exceeded the median ± the inter-quartile value. Average constriction rates were indistinguishable in these four strains: 0.30 ± 0.03 µm/min (wild type, n = 14), 0.33 ± 0.02 µm/min (adf1-1, n = 13), 0.32 ± 0.10 µm/min (adf1-M2, n = 17), and 0.30 ± 0.08 µm/min (adf1-M3, n = 11). Variances were larger for adf1-M2 and adf1-M3 cells than for wild-type cells. (F) Contractile rings reconstructed from Z-series of fluorescence micrographs. Contractile rings in adf1-M2 and adf1-M3 cells were less homogeneous with some thicker bundles (arrowheads) than wild-type adf1+ cells. Bar, 5 µm.
Discussion
Nakano and Mabuchi (2006) established the importance of cofilin for cytokinesis in fission yeast, but left open the questions that we addressed in this paper. In both cofilin temperature-sensitive and depletion mutants, they found defects in the assembly and maintenance of contractile rings, as well as more actin patches than in wild-type cells. They proposed two mechanisms to account for the cytokinesis defects in cofilin mutant cells.
First, they suggested that excess actin patches might deplete the pool of actin monomers available to assemble contractile rings. Although endocytic actin patches in our cofilin mutant cells accumulated more actin filaments than normal (Fig. 2 D), the networks of actin filaments around the equators of many dividing mutant cells were actually denser than in wild-type cells (Fig. S3 F). Thus, the cytokinesis defects in the cofilin mutant cells do not appear to arise from a shortage of actin monomers.
Second, Nakano and Mabuchi proposed that the numerous actin bundles in mitotic cells arose from severing defects in temperature-sensitive cofilin mutant cells at the restrictive temperature. Because cofilins sever but do not depolymerize actin filaments (Andrianantoandro and Pollard, 2006; Pavlov et al., 2007; Chan et al., 2009), we agree that severing is the most likely defect in the temperature-sensitive cofilin mutant cells. To support this interpretation, Nakano and Mabuchi used complementation experiments with yeast cofilins with point mutations previously interpreted to compromise severing by porcine cofilin (Moriyama and Yahara, 1999). However, they did not establish that the point mutations of fission yeast cofilin caused severing defects, an important control because one cannot assume that a particular point mutation will have the same properties in a second cofilin. A further concern was that the assays used by Moriyama and Yahara (1999) are difficult to interpret mechanistically. Severing defects may also explain how a cofilin mutant with normal actin monomer binding caused serious defects in endocytosis in budding yeast (Lappalainen and Drubin, 1997; Lappalainen et al., 1997).
Roles for cofilin in contractile ring assembly, stability, and constriction
Our new cofilin mutants provide a more definitive test of the role of actin filament severing in cytokinesis. Based on our biochemical characterization (Fig. 1), we interpret the effects of our mutant cofilins in cells as due exclusively to slow binding to actin filaments and low severing activity.
Our experiments and computer simulations support the hypothesis that actin filament severing is required for the even condensation of nodes into a homogeneous contractile ring. Nodes formed clumps in cells with defects in actin filament severing like nodes with long-lived actin filament connections in simulations of contractile ring assembly by the search-capture-pull and release mechanism (Vavylonis et al., 2008; Fig. S11 in that paper and Fig. 5 E in this paper). This agreement provides evidence for the importance of the release step in the search-capture-pull and release mechanism and shows that severing of actin filaments by cofilin contributes to the process. Dissociation of the unprocessive myosin-II motor from connecting filaments may also contribute to the release process (Vavylonis et al., 2008). The cofilin mutant cells are 30% wider than wild-type cells. However, simulations show that this alone does not cause defects in contractile ring assembly (Fig. S5 B), as also observed in many fission yeast morphogenesis mutants with large diameters (Toda et al., 1993, 1996).
Surprisingly, actin filament severing is also important for the stability of fully formed contractile rings. Contractile rings in adf1-1 cells broke into small fragments after shifting to the restrictive temperature. Fully formed contractile rings occasionally broke apart before they constricted in adf1-M2 and adf1-M3 cells, but never in wild-type cells. During the interval between ring formation and constriction, formin Cdc12p actively polymerizes actin filaments (Pelham and Chang, 2002), but the mass of polymerized actin in the ring appears to be constant judging from the numbers of actin-binding proteins such as tropomyosin Cdc8p and α-actinin Ain1p (Wu et al., 2003). Thus, a disassembly process must balance ongoing assembly. As observed in vitro (Michelot et al., 2007), cofilin might sever off the older parts of filaments distal to the barbed end where formin catalyzes assembly. Additional work will be required to discover how actin filament severing paradoxically stabilizes contractile rings. One possibility is that severing by cofilin might facilitate cross-linking into stable bundles as observed in vitro (Maciver et al., 1991).
Because contractile rings disassemble as they constrict, and because cofilin was proposed to promote the turnover of actin filaments during ring constriction (Gunsalus et al., 1995; Abe et al., 1996; Somma et al., 2002; Kaji et al., 2003; Ono et al., 2003), we were surprised to find that contractile rings constricted at the same average rates in cells with wild-type or mutant cofilins. The greater variability in constriction rates in mutant cells may result from minor mechanical defects that arise during the assembly of rings from strands of precursors after the initial failure to assemble a complete ring by condensation of a broad band of nodes. Many questions remain about the processes that determine the constriction rates of contractile rings, especially the relative contributions of force generation by actin and myosin-II in the contractile ring relative to growth of the primary septum, which is also required for ring constriction (Le Goff et al., 1999; Liu et al., 2000).
Pathways of contractile ring assembly
Our observations of live cells revealed the source of the equatorial actin bundles in cofilin mutant cells (Nakano and Mabuchi, 2006). The mechanism of contractile ring assembly is robust, so if problems arise owing to stress or defects in any of the components including cofilin, cells attempt to assemble a contractile ring from the available functional proteins and to complete cytokinesis. Cells depending on cofilin adf1-M2 first try to coalesce nodes into a contractile ring, but when this fails, most mutant cells slowly assemble a contractile ring through a remarkable diversity of intermediate structures including spiral strands containing actin filaments and other contractile ring proteins that often anneal to form a ring. We think that wild-type and mutant cells use the same underlying biochemical reactions, but cofilin mutations make the process more dependent on actin filament cross-linking by α-actinin Ain1p (Wu et al., 2001), even though the single mutant strains are viable and assemble rings (Fig. S5 A).
Assembly of contractile rings from strands of actin filaments in adf1-M2 cells appears to be similar to strategies used by fission yeast confronted with a variety of challenges, including deletions of anillin Mid1p or polo kinase Plo1p (Bähler et al., 1998; Hachet and Simanis, 2008; Huang et al., 2008). Ring assembly is even less reliable without anillin Mid1p because myosin-II does not concentrate in nodes around the equator and rings assemble from widely distributed strands containing myosin-II (Hachet and Simanis, 2008; Huang et al., 2008). Ring assembly in cofilin mutants is also similar to the leading cable mechanism proposed for contractile ring assembly in wild-type fission yeast (Chang et al., 1997; Mishra and Oliferenko, 2008; Bathe and Chang, 2010), including dependence on α-actinin (Hachet and Simanis, 2008; Coffman et al., 2009). Thus, cells with prominent leading cables may have been stressed in some way, as hypothesized by others (Hachet and Simanis, 2008; Coffman et al., 2009).
A mechanism to monitor contractile ring assembly
The ability of adf1-M2 cells to persist in the cytokinesis phase for more than an hour as they assemble a physically closed contractile ring shows that fission yeast have a mechanism to delay both septum formation and constriction of bundles of contractile ring precursors composed of actin filaments, myosin-II, and α-actinin until the ring is complete. The ability to sense the completion of a ring explains why few cofilin mutant cells have more than two nuclei during interphase despite the defects that prolong cytokinesis. Cells with bgs1 (cps1) mutations compromising septum deposition and contractile ring constriction also exhibit a safeguard mechanism (Liu et al., 2000). The ability of the hypothetical mechanism to monitor the integrity of the contractile ring and delay entry into the next round of mitosis may explain the slow growth and thicker septa of cofilin mutant cells.
Role of cofilin in animal cell cytokinesis
Cytokinesis in animal cells also depends on cofilins, but the contributions of cofilin have not been investigated in detail. Animal cells deficient in cofilin form clumps of actin filaments rather than complete rings (Gunsalus et al., 1995; Giansanti et al., 1999; Somma et al., 2002; Ono et al., 2003) and have difficulties completing cleavage (Kaji et al., 2003; Ono et al., 2003). These problems may have causes in common with the defects we observed in fission yeast. For example, unstable contractile rings may result in regression of furrows, as observed with exposure to cytochalasin (Carter, 1967) and with mutations in many different genes (Mackay et al., 1998; Powers et al., 1998; Schumacher et al., 1998; Jantsch-Plunger et al., 2000; D’Avino et al., 2004). Live cell microscopy will be required to appreciate these mechanisms.
Materials and methods
Strains and culture conditions
We subcloned cofilin cDNA into pFA6a-KanMX6 to make pFA6a-ADF1-KanMX6. Then we made point mutations of the cofilin cDNA by site-directed mutagenesis (QuikChange II; Agilent Technologies) and used it to replace the ura4+ cassette in the diploid strain adf1/Δadf1::ura4+ through PCR-based integration (Bähler et al., 1998). Each resulting strain adf1/Δadf1::adf1*-KanMX6 was sporulated on SPA5s plates and dissected into at least 20 tetrads. The spores Δadf1::adf1*-KanMX6 were selected with 100 µg/ml G418 (Geneticin; American Bioanalytical). Cofilin loci of the positive clones were sequenced to verify the mutations. Cofilin mutant strains were backcrossed several times with the wild-type strain to confirm that phenotypes were stable.
Fluorescence microscopy
Yeast cells were cultured by standard methods. To visualize nuclei and cell walls, cells were fixed with cold 70% ethanol and stained with 1 µg/ml DAPI (Sigma-Aldrich) or 40 µg/ml Calcofluor (Sigma-Aldrich) and images were taken with a CCD camera (Orca-ER; Hamamatsu Photonics) on a microscope (model IX-71; Olympus) equipped for epifluorescence. To visualize actin filaments, cells were fixed with 4% formaldehyde and permeabilized with 1% Triton X-100 before staining with 860 µM Bodipy-phallacidin (Invitrogen). For imaging live cells, actively growing cultures in YE5s media with OD between 0.4 and 0.5 were harvested by spinning the cells at 3,000 rpm for 1 min and washing briefly three times with EMM5s before resuspending, applying to a 25% gelatin pad made with EMM5s, and sealing under a coverslip. Live cells were imaged with a 100x Plan Apochromat objective lens (NA 1.40) on a microscope (model IX-71; Olympus) equipped with either a confocal spinning disk unit (Ultra-view RS; PerkinElmer) and an Orca-ER camera (Hamamatsu Photonics) or with a CSU X-1 spinning disk confocal unit (Yokogawa) and an Ixon EMCCD camera (Andor). Images were processed using ImageJ (National Institutes of Health, Bethesda MD) with free available plug-ins. Unless specified, live cells were all imaged at 25°C. A heated objective lens collar (Omegalux) was used for imaging live cells at 36°C. We tracked the positions of the centers of mass of nodes manually in time-lapse movies of live cells at 1-s intervals with the MtrackJ plug-in for ImageJ (http://www.imagescience.org/meijering/software/mtrackj) starting from the initial movement until the node merged with another node, a clump of nodes, or a contractile ring. We imaged the nodes in a single confocal section every second, manually tracked their centers of mass during quick bursts of movement, and calculated average velocities along these paths.
Protein purification and biochemistry
Actin was purified from an acetone powder of chicken breast muscle by one cycle of polymerization and depolymerization followed by gel filtration in Buffer G (2 mM TrisCl, pH 8.0, 0.5 mM ATP, 0.5 mM DTT, 0.1 mM CaCl2, and 1 mM NaN3; MacLean-Fletcher and Pollard, 1980). Ca-ATP-actin was converted to Mg-ATP-actin by incubating for at least 2 min at 25°C after adding 50 µM MgCl2 and 0.2 mM EGTA to buffer G. Purified actin filaments were labeled on cysteine-374 with pyrene iodoacetamide (Invitrogen), depolymerized, clarified and gel filtrated on Sephacryl S-300 (GE Healthcare; Pollard, 1984). Purified actin was labeled on cysteine-374 with Oregon Green 488 iodoacetamide mixed isomers (Invitrogen), depolymerized, and purified by chromatography on columns of DEAE-Sepharose fast flow and Sephacryl S-300 (Kuhn and Pollard, 2005). Mutant cofilins were expressed from the plasmid pMW172 in BL21(DE3) PlyS cells (EMD) and purified like wild-type cofilin (Andrianantoandro and Pollard, 2006). Bacteria were induced with 1 µM IPTG at 37°C for 5 h, lysed by sonication, and clarified. Protein was precipitated with 70% ammonium sulfate, resuspended in buffer D (10 mM TrisCl, 1 mM NaN3, 1 mM EDTA, 250 mM NaCl, and 2 mM DTT, pH 8.0) and gel filtered on a 400-ml column of Sephacryl S-200. Peak cofilin fractions were dialyzed against DEAE buffer (25 mM TrisCl, pH 8.4, and 2 mM DTT) loaded on a DEAE-Sepharose column and eluted with a 500-ml gradient of 0–500 mM NaCl in DEAE buffer.
Assay for stability
Samples of 5 µM cofilins were mixed with 0–8 M urea in 10 mM TrisCl (pH 7.4) at room temperature for 1 h. Fluorescence emission spectra were recorded from 310 to 380 nm with excitation at 295 nm with a PTI Alphascan fluorimeter (Photo Technology International). The data were analyzed by the linear extrapolation method (Pace, 1986), which assumes a linear relationship between free energy of unfolding (DG0u,water) and urea concentration ([urea]). A two-state model relates the observed fluorescence signal (Yobs) to the unfolding equilibrium of the native and denatured states (KU = [D]/[N]):
YN and YD are observed fluorescence of the protein at the native and denatured state. T is the absolute temperature, and R is the gas constant.
Binding of cofilin to actin filaments
The experiments were performed in KMEI polymerization buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0) with filaments polymerized from Mg-ATP-actin monomers. We measured binding of cofilin and cofilin mutants to pyrenyl-actin filaments by quenching of the fluorescence using a PTI Alphascan fluorimeter (Photo Technology International) with excitation at 365 nm and emission at 410 nm (Andrianantoandro and Pollard, 2006). To measure the time course of binding, the reactants were mixed in a stop-flow apparatus (Kin-Tek). To measure equilibrium binding, the reactants were mixed manually in a 96-well plate for 30 min before the fluorescence was read with a plate reader (Spectra Max Gemini XPS; Molecular Devices).
Binding of cofilin to actin monomers
We measured the affinity of cofilin and cofilin mutants for actin monomers in 1 mM MgCl2, 1 mM EGTA, 2 mM TrisCl, and 0.5 mM DTT, pH 8.0, by the inhibition of nucleotide exchange (Andrianantoandro and Pollard, 2006). Etheno-ATP-actin was prepared by removing free ATP from Mg-ATP-actin with AX1-X4 (Bio-Rad Laboratories) resin in a Spin-X mini column (Corning) before incubating the actin with 500 µM etheno-ATP (Invitrogen) for 12 min at 25°C. Excess etheno-ATP was removed by running the solution through AX1-X4 resin a second time in a Spin-X column. The exchange reaction was initiated by manually mixing 1 µM Mg-etheno-ATP-actin with 500 µM ATP and a range of concentrations of cofilin. The time course of the dissociation of etheno-ATP from actin was measured by the decrease in fluorescence with excitation at 360 nm and emission at 410 nm.
Actin filament–severing assay
We observed severing of actin filaments by cofilin or cofilin mutants by total internal reflection fluorescence microscopy (TIRF) (Andrianantoandro and Pollard, 2006). A sample of 2 µM 40% labeled Oregon green 488–labeled Ca-ATP-actin was exchanged with 0.2 mM EGTA, 0.05 mM MgCl2 for 2 min at 25°C and then polymerized in 1x TIRF buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole, pH 7.0, 100 mM DTT, 0.2 mM ATP, 15 mM glucose, 0.5% methyl cellulose [15 centipoise at 2%], 20 µg/ml catalase [Sigma-Aldrich], and 100 µg/ml glucose oxidase [Sigma-Aldrich; Kuhn and Pollard, 2005]). Flow cells were preincubated with 1 vol of 200 nM skeletal muscle myosin inactivated with 1 mM N-ethylmaleimide (NEM) for 1 min, washed with 2 vol of HS-TBS (50 mM TrisCl, pH 7.6, and 600 mM NaCl), 2 vol of 2% bovine serum albumin (BSA) in HS-TBS, and 2 vol of 1x TIRF buffer. Then 15 µl of Mg-ATP actin in TIRF buffer was loaded into the flow cell by capillary action. After the attached actin filaments polymerized for appropriately 5 min, the solution in the flow cell was replaced with 15 µl of cofilin in 1x TIRF buffer. Fluorescence was excited by prism-style TRIF with an argon laser (Series 43; CVI Melles Griot). Images were acquired with a microscope (model IX-71; Olympus) with a 60x Plan Apochromat (NA 1.4) objective lens with an Orca ER camera (Hamamatsu Photonics). Each data point is the average of three independent experiments.
Computer simulations
The computer simulations were done with SCPR Controller, written by Dimitrios Vavylonis (Vavylonis et al., 2008) with the following parameters: number of nodes = 65; number of filaments per node = 2; fraction of nodes with formins = 1; actin polymerization rate Vpol = 74 subunits/s; capture radius = 0.15 µm; short range repulsion = 0.15 µm; force (F) produced by myosin II = 12 pN, based on F=V*ζ, V is velocity of the nodes, ζ is the friction coefficient; friction = 400 pN*s/µm; broad band half width = 0.8 µm; cell radius = 1.87 µm (wild-type cells and adf1-1 cell) or 2.43 µm (adf1-M2 and adf1-M3 cells); simulation time interval dt = 0.5 s; node connection lifetime τbreak = variable seconds; lifetime of actin filaments τturnover = τbreak. Each simulation was run at least five times with the same set of parameters.
Online supplemental material
Figure S1 shows contractile ring assembly in temperature-sensitive cofilin mutant adf1-1 cells and localization of cofilin. Figure S2 shows biochemical characterization of mutant cofilin proteins. Figure S3 shows characterization of cofilin mutant adf1-M2 and adf1-M3 cells. Figure S4 shows Monte Carlo simulations of the search-capture-pull and release model of contractile ring assembly with variations in cell diameter and τbreak. Figure S5 shows that actinin is important for cofilin mutant cells to assemble contractile rings and cofilin contributes to the stability of the rings. Video 1 is a time-lapse movie of fluorescence micrographs of a wild-type cell expressing Rlc1p-3GFP at 36°C. Video 2 is a time-lapse movie of fluorescence micrographs of an adf1-1 cell expressing Rlc1p-3GFP at 36°C. Video 3 is a time-lapse movie of fluorescence micrographs of an adf1+ cell expressing Rlc1p-3GFP and SPB marker Sad1-mEGFP at 25°C. Video 4 is a time-lapse movie of fluorescence micrographs of an adf1-M2 cell expressing Rlc1p-3GFP and SPB marker Sad1-mEGFP at 25°C. Video 5 is a time-lapse movie of negative-contrast fluorescence micrographs of a wild-type cell expressing Rlc1p-3GFP at 25°C. Video 6 is a time-lapse movie of negative-contrast fluorescence micrographs of an adf1-M3 cell expressing Rlc1p-3GFP at 25°C. Video 7 is a time-lapse movie of negative-contrast fluorescence micrographs of an adf1-M3 cell expressing Rlc1p-3GFP at 25°C. Video 8 is a time-lapse movie of negative-contrast fluorescence micrographs of an adf1-M3 cell expressing Rlc1p-TdTomato and GFP-CHD. Video 9 is a time-lapse movie of negative-ontrast fluorescence micrographs of an adf1-M2 cell expressing Rlc1p-3GFP. Video 10 is a time-lapse movie of fluorescence micrographs of an adf1-1 cell expressing GFP-myo2. Table S1 lists the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201103067/DC1.
Acknowledgments
The authors thank David Taylor, Cambria Alpha, and Charmaine Chan for contributing preliminary data; Chad McCormick for writing ImageJ plug-ins for the image analysis; Naomi Courtemanche for analysis of the data from the protein stability experiment; Dimitrios Vavylonis for discussion of the results and providing the software for computer simulations; Kentaro Nakano, Jian-Qiu Wu, and Dannel McCollum for strains; and members of our laboratory for helpful discussions. We acknowledge the use of the MtrackJ plug-in for ImageJ, developed by Dr. Erik Meijering at Biomedical Imaging Group Rotterdam of the Erasmus MC - University Medical Center Rotterdam, Netherlands.
This work was supported by National Institutes of Health research grant GM-026132.
Footnotes
Abbreviations used in this paper:
- ADF
- actin depolymerizing factor
- SPB
- spindle pole body
- TIRF
- total internal reflection fluorescence
- TS
- temperature sensitive
References
- Abe H., Obinata T., Minamide L.S., Bamburg J.R. 1996. Xenopus laevis actin-depolymerizing factor/cofilin: a phosphorylation-regulated protein essential for development. J. Cell Biol. 132:871–885 10.1083/jcb.132.5.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T.D. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 24:13–23 10.1016/j.molcel.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Bähler J., Steever A.B., Wheatley S., Wang Y., Pringle J.R., Gould K.L., McCollum D. 1998. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J. Cell Biol. 143:1603–1616 10.1083/jcb.143.6.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg J.R., Harris H.E., Weeds A.G. 1980. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 121:178–182 10.1016/0014-5793(80)81292-0 [DOI] [PubMed] [Google Scholar]
- Bathe M., Chang F. 2010. Cytokinesis and the contractile ring in fission yeast: towards a systems-level understanding. Trends Microbiol. 18:38–45 10.1016/j.tim.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan J. 1988. Gelsolin has three actin-binding sites. J. Cell Biol. 106:1553–1562 10.1083/jcb.106.5.1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.F., Laurent V., Santolini J., Melki R., Didry D., Xia G.X., Hong Y., Chua N.H., Pantaloni D. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136:1307–1322 10.1083/jcb.136.6.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S.B. 1967. Effects of cytochalasins on mammalian cells. Nature. 213:261–264 10.1038/213261a0 [DOI] [PubMed] [Google Scholar]
- Chan C., Beltzner C.C., Pollard T.D. 2009. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19:537–545 10.1016/j.cub.2009.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Drubin D., Nurse P. 1997. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137:169–182 10.1083/jcb.137.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Nile A.H., Lee I.J., Liu H., Wu J.Q. 2009. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol. Biol. Cell. 20:5195–5210 10.1091/mbc.E09-05-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino P.P., Savoian M.S., Glover D.M. 2004. Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J. Cell Biol. 166:61–71 10.1083/jcb.200402157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz E.M. 2005. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. J. Mol. Biol. 346:557–564 10.1016/j.jmb.2004.11.065 [DOI] [PubMed] [Google Scholar]
- Giansanti M.G., Bonaccorsi S., Gatti M. 1999. The role of anillin in meiotic cytokinesis of Drosophila males. J. Cell Sci. 112:2323–2334 [DOI] [PubMed] [Google Scholar]
- Gunsalus K.C., Bonaccorsi S., Williams E., Verni F., Gatti M., Goldberg M.L. 1995. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 131:1243–1259 10.1083/jcb.131.5.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O., Simanis V. 2008. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 22:3205–3216 10.1101/gad.1697208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins M., Pope B., Maciver S.K., Weeds A.G. 1993. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 32:9985–9993 10.1021/bi00089a014 [DOI] [PubMed] [Google Scholar]
- Huang Y., Yan H., Balasubramanian M.K. 2008. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J. Cell Biol. 183:979–988 10.1083/jcb.200806151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V., Gönczy P., Romano A., Schnabel H., Hamill D., Schnabel R., Hyman A.A., Glotzer M. 2000. CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 149:1391–1404 10.1083/jcb.149.7.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji N., Ohashi K., Shuin M., Niwa R., Uemura T., Mizuno K. 2003. Cell cycle-associated changes in Slingshot phosphatase activity and roles in cytokinesis in animal cells. J. Biol. Chem. 278:33450–33455 10.1074/jbc.M305802200 [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S.C., Paul L.S., Lonberg N., Newport J.W., McSwiggen J.A., von Hippel P.H. 1986. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 25:1226–1240 10.1021/bi00354a006 [DOI] [PubMed] [Google Scholar]
- Kuhn J.R., Pollard T.D. 2005. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88:1387–1402 10.1529/biophysj.104.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P., Drubin D.G. 1997. Cofilin promotes rapid actin filament turnover in vivo. Nature. 388:78–82 10.1038/40418 [DOI] [PubMed] [Google Scholar]
- Lappalainen P., Fedorov E.V., Fedorov A.A., Almo S.C., Drubin D.G. 1997. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 16:5520–5530 10.1093/emboj/16.18.5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff X., Woollard A., Simanis V. 1999. Analysis of the cps1 gene provides evidence for a septation checkpoint in Schizosaccharomyces pombe. Mol. Gen. Genet. 262:163–172 10.1007/s004380051071 [DOI] [PubMed] [Google Scholar]
- Liu J., Wang H., Balasubramanian M.K. 2000. A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J. Cell Sci. 113:1223–1230 [DOI] [PubMed] [Google Scholar]
- Maciver S.K., Wachsstock D.H., Schwarz W.H., Pollard T.D. 1991. The actin filament severing protein actophorin promotes the formation of rigid bundles of actin filaments crosslinked with alpha-actinin. J. Cell Biol. 115:1621–1628 10.1083/jcb.115.6.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay A.M., Ainsztein A.M., Eckley D.M., Earnshaw W.C. 1998. A dominant mutant of inner centromere protein (INCENP), a chromosomal protein, disrupts prometaphase congression and cytokinesis. J. Cell Biol. 140:991–1002 10.1083/jcb.140.5.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean-Fletcher S.D., Pollard T.D. 1980. Viscometric analysis of the gelation of Acanthamoeba extracts and purification of two gelation factors. J. Cell Biol. 85:414–428 10.1083/jcb.85.2.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J.D., von Hippel P.H. 1974. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 86:469–489 10.1016/0022-2836(74)90031-X [DOI] [PubMed] [Google Scholar]
- Michelot A., Berro J., Guérin C., Boujemaa-Paterski R., Staiger C.J., Martiel J.L., Blanchoin L. 2007. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 17:825–833 10.1016/j.cub.2007.04.037 [DOI] [PubMed] [Google Scholar]
- Mishra M., Oliferenko S. 2008. Cytokinesis: catch and drag. Curr. Biol. 18:R247–R250 10.1016/j.cub.2008.01.029 [DOI] [PubMed] [Google Scholar]
- Moriyama K., Yahara I. 1999. Two activities of cofilin, severing and accelerating directional depolymerization of actin filaments, are affected differentially by mutations around the actin-binding helix. EMBO J. 18:6752–6761 10.1093/emboj/18.23.6752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka R., Abe H., Kusano K., Obinata T. 1995. Concentration of cofilin, a small actin-binding protein, at the cleavage furrow during cytokinesis. Cell Motil. Cytoskeleton. 30:1–7 10.1002/cm.970300102 [DOI] [PubMed] [Google Scholar]
- Nakano K., Mabuchi I. 2006. Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol. Biol. Cell. 17:1933–1945 10.1091/mbc.E05-09-0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E., Maekawa S., Sakai H. 1984. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry. 23:5307–5313 10.1021/bi00317a032 [DOI] [PubMed] [Google Scholar]
- Okreglak V., Drubin D.G. 2007. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178:1251–1264 10.1083/jcb.200703092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K., Parast M., Alberico C., Benian G.M., Ono S. 2003. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J. Cell Sci. 116:2073–2085 10.1242/jcs.00421 [DOI] [PubMed] [Google Scholar]
- Paavilainen V.O., Oksanen E., Goldman A., Lappalainen P. 2008. Structure of the actin-depolymerizing factor homology domain in complex with actin. J. Cell Biol. 182:51–59 10.1083/jcb.200803100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C.N. 1986. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 131:266–280 10.1016/0076-6879(86)31045-0 [DOI] [PubMed] [Google Scholar]
- Pavlov D., Muhlrad A., Cooper J., Wear M., Reisler E. 2007. Actin filament severing by cofilin. J. Mol. Biol. 365:1350–1358 10.1016/j.jmb.2006.10.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R.J., Chang F. 2002. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 419:82–86 10.1038/nature00999 [DOI] [PubMed] [Google Scholar]
- Pollard T.D. 1984. Polymerization of ADP-actin. J. Cell Biol. 99:769–777 10.1083/jcb.99.3.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Wu J.Q. 2010. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 11:149–155 10.1038/nrm2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J., Bossinger O., Rose D., Strome S., Saxton W. 1998. A nematode kinesin required for cleavage furrow advancement. Curr. Biol. 8:1133–1136 10.1016/S0960-9822(98)70470-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal A.A., Tetreault J.W., Lappalainen P., Drubin D.G., Amberg D.C. 1999. Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145:1251–1264 10.1083/jcb.145.6.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J.M., Golden A., Donovan P.J. 1998. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 143:1635–1646 10.1083/jcb.143.6.1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma M.P., Fasulo B., Cenci G., Cundari E., Gatti M. 2002. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell. 13:2448–2460 10.1091/mbc.01-12-0589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Shimanuki M., Yanagida M. 1993. Two novel protein kinase C-related genes of fission yeast are essential for cell viability and implicated in cell shape control. EMBO J. 12:1987–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Niwa H., Nemoto T., Dhut S., Eddison M., Matsusaka T., Yanagida M., Hirata D. 1996. The fission yeast sts5+ gene is required for maintenance of growth polarity and functionally interacts with protein kinase C and an osmosensing MAP-kinase pathway. J. Cell Sci. 109:2331–2342 [DOI] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.Q., Hao S., O’Shaughnessy B., Pollard T.D. 2008. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 319:97–100 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Wu J.Q., Bähler J., Pringle J.R. 2001. Roles of a fimbrin and an alpha-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol. Biol. Cell. 12:1061–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.Q., Kuhn J.R., Kovar D.R., Pollard T.D. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 5:723–734 10.1016/S1534-5807(03)00324-1 [DOI] [PubMed] [Google Scholar]
- Wu J.Q., Sirotkin V., Kovar D.R., Lord M., Beltzner C.C., Kuhn J.R., Pollard T.D. 2006. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 174:391–402 10.1083/jcb.200602032 [DOI] [PMC free article] [PubMed] [Google Scholar]