Abstract
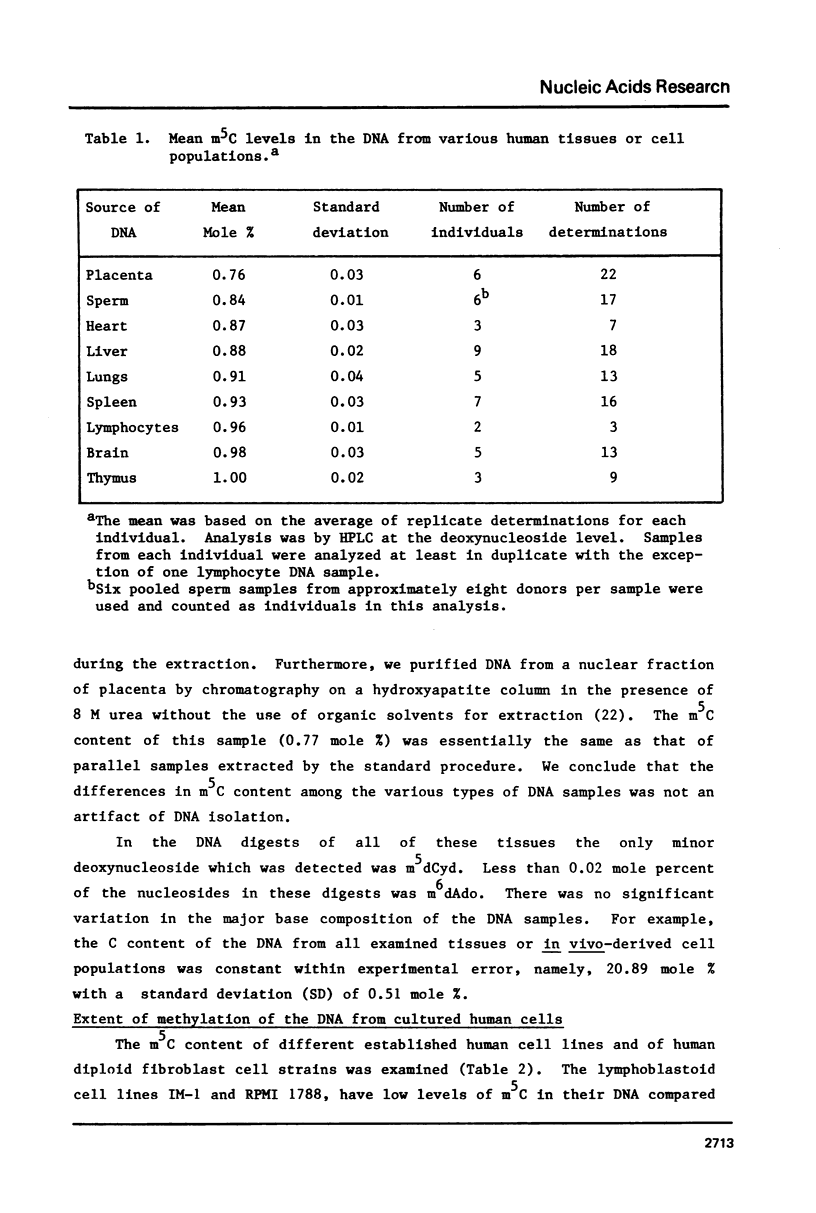
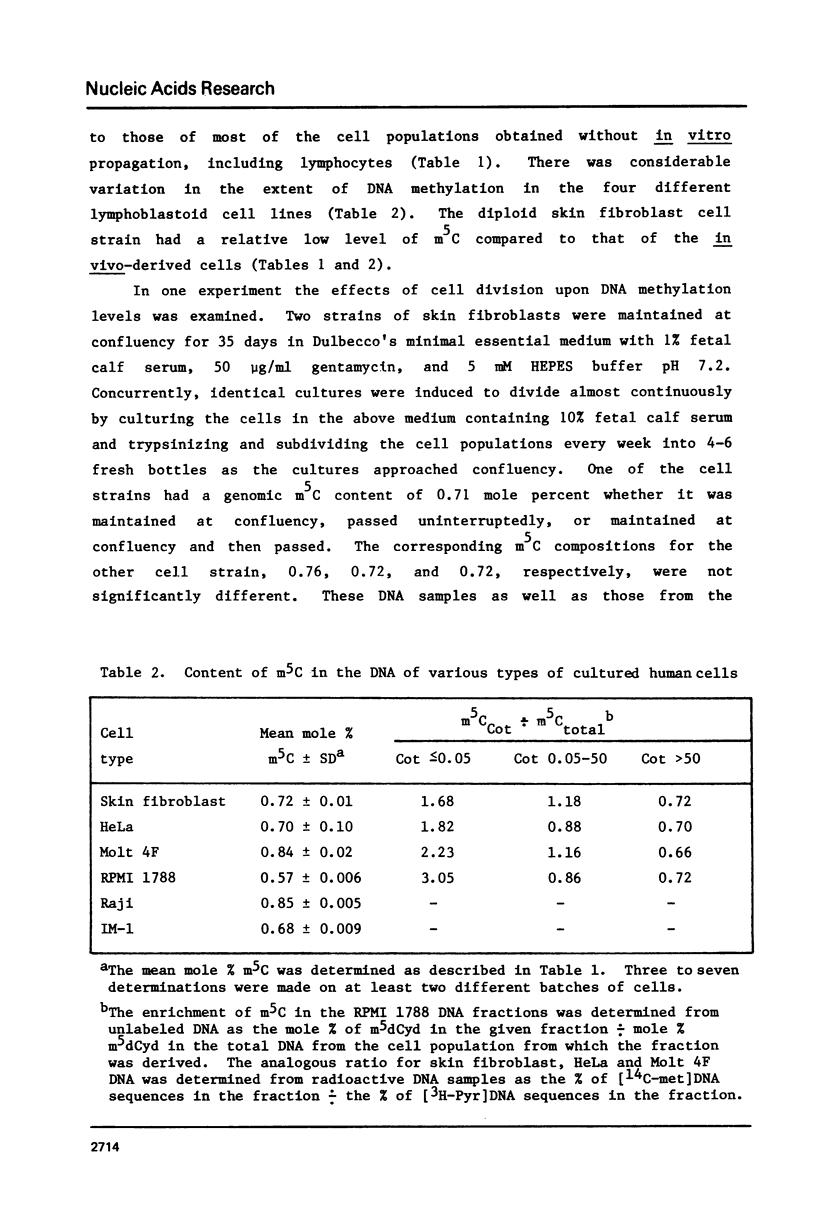
Analysis of the total base composition of DNA from seven different normal human tissues and eight different types of homogeneous human cell populations revealed considerable tissue-specific and cell-specific differences in the extent of methylation of cytosine residues. The two most highly methylated DNAs were from thymus and brain with 1.00 and 0.98 mole percent 5-methylcytosine (m5C), respectively. The two least methylated DNAs from in vivo sources were placental DNA and sperm DNA, which had 0.76 and 0.84 mole percent m5C, respectively. The differences between these two groups of samples were significant with p less than 0.01. The m5C content of DNA from six human cell lines or strains ranged from 0.57 to 0.85 mole percent. The major and minor base composition of DNA fractionated by reassociation kinetics was also determined. The distribution of m5C among these fractions showed little or no variation with tissue or cell type with the possible exception of sperm DNA. In each case, nonrepetitive DNA sequences were hypomethylated compared to unfractionated DNA.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BORENFREUND E., FITT E., BENDICH A. Isolation and properties of deoxyribonucleic acid from mammalian sperm. Nature. 1961 Sep 30;191:1375–1377. doi: 10.1038/1911375a0. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H., Gehring C. A. Methylated and unmethylated ribosomal RNA genes in the mouse. J Mol Biol. 1981 Oct 15;152(1):1–17. doi: 10.1016/0022-2836(81)90092-9. [DOI] [PubMed] [Google Scholar]
- Bird A., Taggart M., Macleod D. Loss of rDNA methylation accompanies the onset of ribosomal gene activity in early development of X. laevis. Cell. 1981 Nov;26(3 Pt 1):381–390. doi: 10.1016/0092-8674(81)90207-5. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D., Clayton D. A. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J Biol Chem. 1974 Dec 25;249(24):7991–7995. [PubMed] [Google Scholar]
- Brown I. R., Church R. B. Transcription of nonrepeated DNA during mouse and rabbit development. Dev Biol. 1972 Sep;29(1):73–84. doi: 10.1016/0012-1606(72)90045-0. [DOI] [PubMed] [Google Scholar]
- Chikaraishi D. M., Deeb S. S., Sueoka N. Sequence complexity of nuclear RNAs in adult rat tissues. Cell. 1978 Jan;13(1):111–120. doi: 10.1016/0092-8674(78)90142-3. [DOI] [PubMed] [Google Scholar]
- Compere S. J., Palmiter R. D. DNA methylation controls the inducibility of the mouse metallothionein-I gene lymphoid cells. Cell. 1981 Jul;25(1):233–240. doi: 10.1016/0092-8674(81)90248-8. [DOI] [PubMed] [Google Scholar]
- Drahovsky D., Boehm T. L., Kreis W. Distribution pattern and enzymic hypermethylation of inverted repetitive DNA sequences in P815 mastocytoma cells. Biochim Biophys Acta. 1979 Jun 20;563(1):28–35. doi: 10.1016/0005-2787(79)90004-2. [DOI] [PubMed] [Google Scholar]
- EDWARDS J. L., KLEIN R. E. Cell renewal in adult mouse tissues. Am J Pathol. 1961 Apr;38:437–453. [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Sarafyan L. P., Simpson N., Downing A. Interaction of normal and unusually modified microbial DNA with cultured mammalian cells. Breakdown and reincorporation vs. uptake of polymerized DNA. Biochim Biophys Acta. 1978 Jan 26;517(1):43–54. doi: 10.1016/0005-2787(78)90032-1. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Fabricant J. D., Wagner E. F., Auer B., Schweiger M. 5-methylcytosine content of DNA during differentiation in mouse teratocarcinoma cells. Exp Cell Res. 1979 Nov;124(1):25–29. doi: 10.1016/0014-4827(79)90253-2. [DOI] [PubMed] [Google Scholar]
- Ford J. P., Coca-Prados M., Hsu M. T. Enzymatic analysis of 5-methylcytosine content in eukaryotic DNA. Study of intracellular Simian Virus 40 DNA. J Biol Chem. 1980 Aug 25;255(16):7544–7547. [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Grouse L., Chilton M. D., McCarthy B. J. Hybridization of ribonucleic acid with unique sequences of mouse deoxyribonucleic acid. Biochemistry. 1972 Feb 29;11(5):798–805. doi: 10.1021/bi00755a019. [DOI] [PubMed] [Google Scholar]
- Hahn W. E., Laird C. D. Transcription of nonrepeated DNA in mouse brain. Science. 1971 Jul 9;173(3992):158–161. doi: 10.1126/science.173.3992.158. [DOI] [PubMed] [Google Scholar]
- Jones R. E., DeFeo D., Piatigorsky J. Transcription and site-specific hypomethylation of the delta-crystallin genes in the embryonic chicken lens. J Biol Chem. 1981 Aug 10;256(15):8172–8176. [PubMed] [Google Scholar]
- Kappler J. W. The 5-methylcytosine content of DNA: tissue specificity. J Cell Physiol. 1971 Aug;78(1):33–36. doi: 10.1002/jcp.1040780106. [DOI] [PubMed] [Google Scholar]
- Kratzer P. G., Chapman V. M. X chromosome reactivation in oocytes of Mus caroli. Proc Natl Acad Sci U S A. 1981 May;78(5):3093–3097. doi: 10.1073/pnas.78.5.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo K. C., McCune R. A., Gehrke C. W., Midgett R., Ehrlich M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 1980 Oct 24;8(20):4763–4776. doi: 10.1093/nar/8.20.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre J. N., Becker F. F. 5-Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun. 1979 Apr 13;87(3):698–705. doi: 10.1016/0006-291x(79)92015-1. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov G. G., Yaneva J. N., Markova N. G., Ivanov I. G. Specificity of transcription of single-copy DNA in different rat tissues. Int J Biochem. 1981;13(1):121–124. doi: 10.1016/0020-711x(81)90146-4. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Meinke W., Goldstein D. A., Hall M. R. Rapid isolation of mouse DNA from cells in tissue culture. Anal Biochem. 1974 Mar;58(1):82–88. doi: 10.1016/0003-2697(74)90444-8. [DOI] [PubMed] [Google Scholar]
- Mohandas T., Sparkes R. S., Shapiro L. J. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981 Jan 23;211(4480):393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- Rae P. M., Barnett T. R., Babbitt D. G. Factors influencing the yield of satellite DNA in extractions from Drosophila virilis and Drosophila melanogaster adults and embryos. Biochim Biophys Acta. 1976 May 3;432(2):154–160. doi: 10.1016/0005-2787(76)90157-x. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Romanov G. A., Vanyushin B. F. Methylation of reiterated sequences in mammalian DNAs. Effects of the tissue type, age, malignancy and hormonal induction. Biochim Biophys Acta. 1981 Apr 27;653(2):204–218. doi: 10.1016/0005-2787(81)90156-8. [DOI] [PubMed] [Google Scholar]
- Rubery E. D., Newton A. A. DNA methylation in normal and tumour virus-transformed cells in tissue culture. I. The level of DNA methylation in BHK21 cells and in BHK21 cells transformed by polyoma virus (PyY cells). Biochim Biophys Acta. 1973 Sep 28;324(1):24–36. doi: 10.1016/0005-2787(73)90247-5. [DOI] [PubMed] [Google Scholar]
- Sawecka J., Kornacka L., Malec J. Heterogeneity of DNA methylation in murine L5178Y lymphoblasts. Experientia. 1979 Sep 15;35(9):1166–1167. doi: 10.1007/BF01963264. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Deininger P. L. Sequence organization of the human genome. Cell. 1975 Nov;6(3):345–358. doi: 10.1016/0092-8674(75)90184-1. [DOI] [PubMed] [Google Scholar]
- Shen C. K., Maniatis T. Tissue-specific DNA methylation in a cluster of rabbit beta-like globin genes. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6634–6638. doi: 10.1073/pnas.77.11.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm K. S., Taylor J. H. Distribution of 5-methylcytosine in the DNA of somatic and germline cells from bovine tissues. Nucleic Acids Res. 1981 Sep 25;9(18):4537–4546. doi: 10.1093/nar/9.18.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F., Mazin A. L., Vasilyev V. K., Belozersky A. N. The content of 5-methylcytosine in animal DNA: the species and tissue specificity. Biochim Biophys Acta. 1973 Mar 28;299(3):397–403. doi: 10.1016/0005-2787(73)90264-5. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M., Koshland M. E. Expression of the J chain gene during B cell differentiation is inversely correlated with DNA methylation. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4907–4911. doi: 10.1073/pnas.78.8.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]


