Abstract
The primary sequence was established for two lysine tRNA isoacceptors which differ in abundance during development in Bacillus subtilis. Both tRNAs shared the same primary sequence but differed in the degree of post-transcriptional modification in the anticodon loop. The earlier eluting species, tRNA lys 1, had an unmodified C in position 32 and a mixture of N-[9-beta-ribofuranosyl) purin-6-ylcarbamoyl]-L-threonine, t6A, and N-[(9-beta-D-ribofuranosyl-2-methylthio-purin-6-yl)carbamoyl]threonine, ms2t6A, in position 37. The later eluting species, tRNA Lys 3, which is the more efficient in protein synthesis, had a modified C in position 32 and only ms2t6A in position 37. The possibility exists that modification to make a more efficient tRNA species may be part of a functional interaction between the translational and transcriptional changes that are part of the differentiation process in B. subtilis.
Full text
PDF



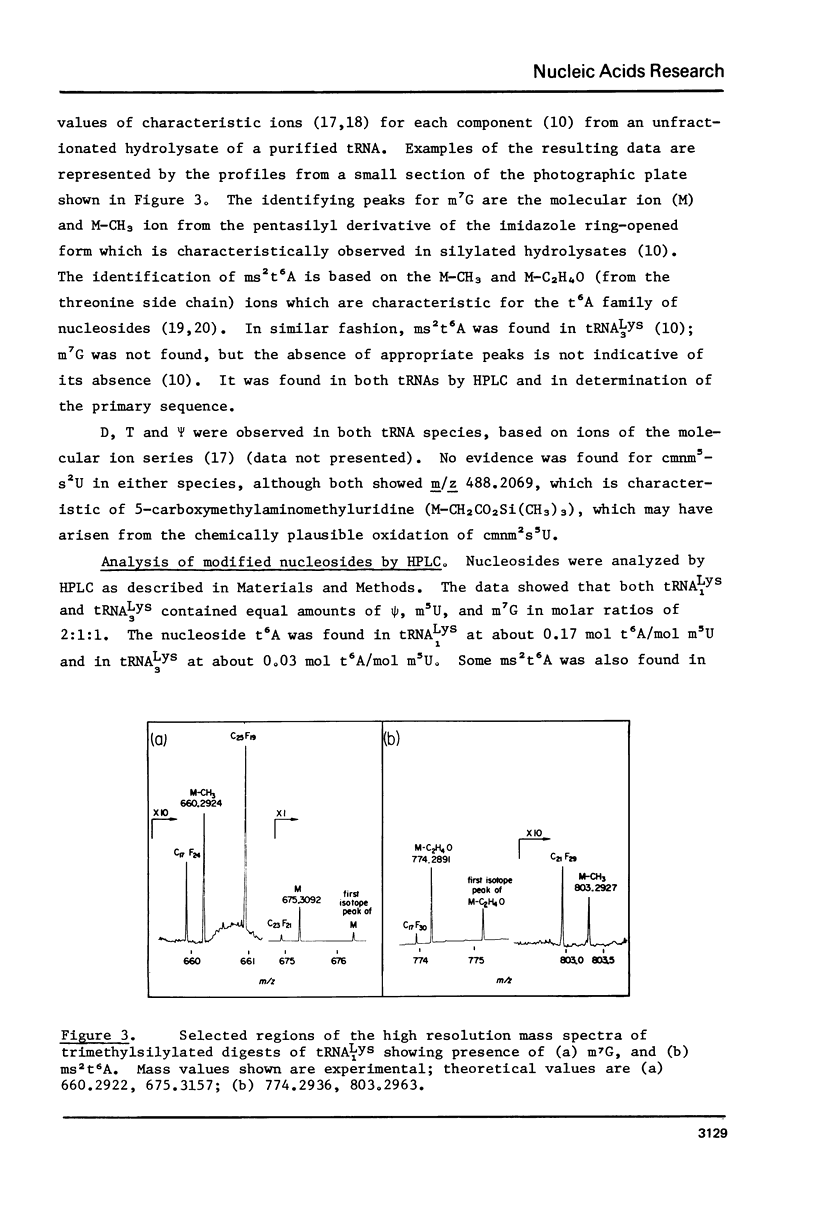



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arceneaux J. L., Sueoka N. Two species of Bacillus subtilis tyrosine transfer ribonucleic acid. Biological properties and alteration in their relative amounts during growth. J Biol Chem. 1969 Nov 10;244(21):5959–5966. [PubMed] [Google Scholar]
- Chia L. L., Randerath K., Randerath E. Base analysis of ribopolynucleotides by tritium incorporation following analytical polyacrylamide gel electrophoresis. Anal Biochem. 1973 Sep;55(1):102–113. doi: 10.1016/0003-2697(73)90295-9. [DOI] [PubMed] [Google Scholar]
- Doi R. H., Kaneko I. Transfer RNA patterns of Bacillus subtilis during sporulation and growth. Cold Spring Harb Symp Quant Biol. 1966;31:581–582. doi: 10.1101/sqb.1966.031.01.075. [DOI] [PubMed] [Google Scholar]
- FRAENKEL-CONRAT H., SINGER B., TSUGITA A. Purification of viral RNA by means of bentonite. Virology. 1961 May;14:54–58. doi: 10.1016/0042-6822(61)90131-3. [DOI] [PubMed] [Google Scholar]
- Gupta R. C., Randerath K. Rapid print-readout technique for sequencing of RNA's containing modified nucleotides. Nucleic Acids Res. 1979 Aug 10;6(11):3443–3458. doi: 10.1093/nar/6.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., Kobayashi Y. Suppression of temperature-sensitive sporulation of a Bacillus subtilis elongation factor G mutant by RNA polymerase mutations. J Bacteriol. 1978 Dec;136(3):883–893. [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Kato M., Sugisaki H., Nishimura S. Nucleotide sequence of starfish initiator tRNA. Nucleic Acids Res. 1979 Aug 10;6(11):3459–3469. doi: 10.1093/nar/6.11.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Mita T., Nishimura S. Nucleotide sequence of cytoplasmic initiator tRNA from Tetrahymena thermophila. Nucleic Acids Res. 1981 Sep 25;9(18):4557–4562. doi: 10.1093/nar/9.18.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarini R. A. Differences in lysine-sRNA from spore and vegetative cells of Bacillus subtillis. Proc Natl Acad Sci U S A. 1966 Jul;56(1):185–190. doi: 10.1073/pnas.56.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey J. A., Lawson A. M., Tsuboyama K., Krueger P. M., Stillwell R. N. Mass spectrometry of nucleic acid components. Trimethylsilyl derivatives of nucleotides, nucleosides, and bases. J Am Chem Soc. 1968 Jul 17;90(15):4182–4184. doi: 10.1021/ja01017a062. [DOI] [PubMed] [Google Scholar]
- Menichi B., Arnold H. H., Heyman T., Dirheimer G., Keith G. Primary structure of Bacillus subtilis tRNAsTyr. Biochem Biophys Res Commun. 1980 Jul 16;95(1):461–467. doi: 10.1016/0006-291x(80)90760-3. [DOI] [PubMed] [Google Scholar]
- Stanley J., Vassilenko S. A different approach to RNA sequencing. Nature. 1978 Jul 6;274(5666):87–89. doi: 10.1038/274087a0. [DOI] [PubMed] [Google Scholar]
- Trowsdale J., Sheflett M., Hoch J. A. New cluster of ribosomal genes in Bacillus subtilis with regulatory role in sporulation. Nature. 1978 Mar 9;272(5649):179–181. doi: 10.1038/272179a0. [DOI] [PubMed] [Google Scholar]
- Vold B. S. Analysis of isoaccepting transfer ribonucleic acid species of Bacillus subtilis: chromatographic differences between transfer ribonucleic acids from spores and cells in exponential growth. J Bacteriol. 1973 Feb;113(2):825–833. doi: 10.1128/jb.113.2.825-833.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vold B. S. Post-transcriptional modifications of the anticodon loop region: alterations in isoaccepting species of tRNA's during development in Bacillus subtilis. J Bacteriol. 1978 Jul;135(1):124–132. doi: 10.1128/jb.135.1.124-132.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Ishikura H. Nucleotide sequence of a lysine tRNA from Bacillus subtilis. Nucleic Acids Res. 1977 Dec;4(12):4291–4303. doi: 10.1093/nar/4.12.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Ishikura H. The presence of N-[(9-beta-D-ribofuranosyl-2-methylthiopurin-6-yl)carbamoyl]threonine in lysine tRNA1 from Bacillus subtilis. J Biochem. 1981 May;89(5):1589–1591. doi: 10.1093/oxfordjournals.jbchem.a133353. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Murao K., Ishikura H. 5-(carboxymethylaminomethyl)-2-thiouridine, a new modified nucleoside found at the first letter position of the anticodon. Nucleic Acids Res. 1981 Apr 24;9(8):1933–1939. doi: 10.1093/nar/9.8.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]