Abstract
We consider the properties of the Fst measure of genetic divergence between an admixed population and its parental source populations. Among all possible populations admixed among an arbitrary set of parental populations, we show that the value of Fst between an admixed population and a specific source population is maximized when the admixed population is simply the most distant of the other source populations. For the case with only two parental populations, as a function of the admixture fraction, we further demonstrate that this Fst value is monotonic and convex, so that Fst is informative about the admixture fraction. We illustrate our results using example human population-genetic data, showing how they provide a framework in which to interpret the features of Fst in admixed populations.
Keywords: admixture, allele frequencies, Fst
1. Introduction
The well-known “fixation index” Fst quantifies the extent to which a polymorphic population is subdivided into subpopulations (Wright (1951), Excoffier (2001), Rousset (2001), Balding (2003), Holsinger and Weir (2009)). In a definition due to Nei (1973, 1987), Fst is defined in terms of the expected heterozygosity of the overall population and the mean expected heterozygosity across the subpopulations:
Definition The expected heterozygosity in a population for a given locus with I distinct alleles is defined as , where pi is the frequency of allele i.
Definition At a given locus, the fixation index, Fst, is defined as Fst = (Ht − Hs)/Ht, where Ht is the expected heterozygosity of the overall population, and Hs is the mean expected heterozygosity across subpopulations.
Assuming that the subpopulations have equal contribution to the total population, Ht is computed by pooling the various subpopulations in equal proportion, and Hs is calculated by weighting the various subpopulations equally. Recall that Fst always lies between 0 and 1, and that Fst is 0 if and only if Ht = Hs, meaning that the pooled population is unstructured. Fst increases as the genetic differentiation between the various subpopulations increases, and the theoretical maximum of 1 is reached if and only if each subpopulation is entirely monomorphic (and homozygous).
We consider the fixation index in the context of admixture, where one population arises from the amalgamation of multiple populations, typically after a long period of relative isolation for the founding groups. Admixture scenarios have been abundant over the course of human history, and for admixture events that have occurred recently, the admixture process has left a detectable signature in the genomes of admixed individuals. For example, the contributions of European, Native American, and African populations to the genetic history of African American (Parra et al. (1998, 2001), Salas et al. (2005), Tang et al. (2006b), Tishkoff et al. (2009), Zakharia et al. (2009), Bryc et al. (2010a)) and Hispanic and Mestizo (Bonilla et al. (2004), Seldin et al. (2007), Tang et al. (2007), Wang et al. (2008), Risch et al. (2009), Silva-Zolezzi et al. (2009), Bryc et al. (2010b)) populations have been the focus of much investigation. Genetic studies of admixture have further been of interest not only for what they reveal about population history, but also because admixed populations can be used in locating disease-associated genomic regions through methods that search in admixed individuals for regions of the genome with excess ancestry from the ancestral population in which a disease is more prevalent (reviewed by McKeigue (2005), Reich and Patterson (2005), Smith and O’Brien (2005), Seldin (2007), Buerkle and Lexer (2008), Zhu et al. (2008),Winkler et al. (2010)).
In an admixture setting, we provide a theoretical framework for explaining the properties of Fst between admixed populations and their parental populations. We examine the values of Fst between pairs of populations, in which one member of the pair is an admixed population and the other is one of its parental source populations. After introducing notation and an example dataset in Section 2, in Section 3, we prove our main theorem, which concerns the value of Fst between a population formed by admixture of K founding populations and a specific one of the K founding populations. We show that for any K ≥ 2, considering all admixture combinations for a given set of founding populations, this Fst expression is maximized when the admixed population is in fact one of the other founding populations. In Section 4, we then consider the special case of K = 2 founding populations, proving in Section 4.1 that Fst is monotonic and convex in the admixture coefficient. Section 4.2 uses microsatellite genotype data on Mestizo populations to demonstrate that Fst values predicted using our theoretical results closely match observed Fst values. Section 4.3 then suggests an estimator of admixture on the basis of Fst. Finally, in Section 5, we summarize the main results and discuss their broader implications.
2. Notation and data
We use a simplified form of the Fst value between two populations that have the same contribution to the total population and that when pooled together produce a polymorphic population. Denote by p1i the frequency of allele i in population 1 and by p2i the frequency of allele i in population 2. We then have
| (1) |
Table 1 summarizes the notation for the scenario in which K ≥ 2 founding populations give rise to an admixed population. Denote by ri the frequency of allele i in the admixed population. Let γk represent the admixture fraction corresponding to population k, so that fraction γk of the ancestry of the admixed population derives from source population k. The admixed population then has
where γk ≥ 0 for 1 ≤ k ≤ K and . We assume throughout the paper that there exists at least one pair of founding populations, k and ℓ, for which . This assumption corresponds to an assumption that the founding populations do not all have identical allele frequencies.
Table 1.
Notation.
| Type of quantity | Symbol | Description | |
|---|---|---|---|
| Indices | i = 1,…, I | Index over alleles | |
| k = 1,…, K | Index over populations | ||
| Allele frequencies | pki | Frequency of allele i in population k | |
| Vector of allele frequencies for population k | |||
| ri | Frequency of allele i in the admixed population | ||
| Admixture fractions | γk | Admixture fraction for population k | |
| γ̲ | Vector of admixture fractions | ||
The example scenarios that we consider use a subset of the data from Wang et al. (2008), consisting of genotypes of 249 Mestizos, 160 Europeans, 463 Native Americans, and 123 Africans at 678 autosomal microsatellite loci. The Mestizo samples provide an example of admixture primarily between European and Native American founding populations, and to a lesser extent, African populations. In our analyses, we focus on the European and Native American contributions, treating the full Mestizo sample as an admixed population and the European and Native American samples as its founding populations. In each of the population samples, except where otherwise specified (in particular, in Section 4.2), we treat the sample allele frequencies from Wang et al. (2008) as the parametric allele frequencies.
3. General case: K founding populations
Our goal in this section is to examine Fst to a specific founding population over the space of admixture vectors possible for a population admixed among a given collection of K founding populations. We begin by providing an expression for Fst between the admixed population and a specific founding population. Without loss of generality, we investigate Fst between population 1 and the admixed population. Using eq. 1 and viewing Fst as a function of two allele frequency vectors, we have
| (2) |
In Theorem 2, we obtain a result concerning the maximum over admixed populations of Fst between the admixed population and an arbitrarily chosen founding population. We first prove a preliminary result involving Fst between one population and a population formed by admixture of two other populations.
Lemma 1 (Three-population lemma). Denote by the vectors corresponding to the allele frequencies of three populations. Consider a population formed by admixture between populations 2 and 3, where γ ∈ [0, 1] represents the admixture fraction for population 2. Then
Proof. We start by applying eq. 1 to calculate Fst between population 1 and the population formed by admixture of populations 2 and 3. We also introduce some additional variables, δ13i = p1i − p3i, δ23i = p2i − p3i, and τ13i = p1i + p3i, for 1 ≤ i ≤ I, to simplify the notation. Then
| (3) |
We denote the function by G(γ), to emphasize the fact that we aim to maximize this quantity with respect to γ. To calculate the derivative of G(γ), we first do some preliminary calculations:
Putting these results together, we obtain
The denominator is always positive, so we focus on the numerator to see where it is greater or less than 0. We denote half the numerator by E(γ), and note that E(γ) is a polynomial in γ, of degree at most 2. We denote the coefficients of γ2, γ, and 1 in E(γ), by a, b, and c, respectively, and calculate them individually:
| (4) |
We next show that E(γ) is increasing or decreasing on [0, 1]. If a = 0, then E(γ) is linear, and the claim is trivial (a and b cannot both be zero, because a = b = 0 implies that populations 2 and 3 have identical allele frequencies). Suppose a ≠ 0. We show that the position of the vertex of the parabola E(γ) = aγ2 + bγ + c, or −b/(2a), is not in (0, 1). We first note that , so that b ≥ 0. Consequently, if −b/(2a) > 0, then a < 0 and . If 0 < −b/(2a) < 1, then we also have , which means that . This inequality clearly does not hold, because . As a result, −b/(2a) ∉ (0, 1), so that the extrema of E(γ) on [0, 1] must occur at γ = 0 and γ = 1. It follows that E(γ) is either increasing or decreasing for γ ∈ (0, 1).
To demonstrate that E(γ) is increasing, we show that E(1) > E(0):
We note that by the assumption that populations 2 and 3 do not have identical allele frequencies, and . This inequality is strict by this same assumption, as population 1 cannot have identical allele frequencies to both populations 2 and 3. Consequently, E(1) > E(0), and E(γ) is increasing on γ ∈ [0, 1]. Note that if populations 2 and 3 were switched, so that we instead considered the value of rather than , then E(1) − E(0) would not change, and we would reach the same conclusion that E(1) > E(0) and E(γ) is increasing on the interval.
Three possibilities exist for the location of 0: E(1) > E(0) ≥ 0, E(1) > 0 > E(0) or 0 ≥ E(1) > E(0). Because E(γ) only differs from by a positive factor, these three possibilities correspond to three possibilities for the shape of G(γ) on γ ∈ [0, 1]: G(γ) is increasing, G(γ) is decreasing up to a point, then increasing, or G(γ) is decreasing. In each of these three cases, it follows that
| (5) |
The result of this lemma is illustrated on a data example in Figure 1, in which log10[G(γ)] is plotted against γ. In this case, population 1 represents a Mestizo admixed population, and populations 2 and 3 represent European and Native American populations, respectively. For each of twenty loci considered, as in the lemma, the maximum of the function is located either at γ = 0 or at γ = 1.
Figure 1.
Fst between a population and a hypothetical second population that is admixed between two other populations. log10[G(γ)], seen as a function of γ, is plotted against γ, where G(γ) is , (eq. 3). Populations 1, 2, and 3 represent populations of Mestizo, European, and Native American descent, respectively, and are based on allele frequencies estimated from Wang et al. (2008). Twenty randomly selected loci are considered, with each curve representing a different locus. The dots indicate the maxima for individual curves. In accordance with Lemma 1, for γ ∈ [0, 1], the maximal value of log10[G(γ)], and therefore of G(γ), is always at γ = 0 or γ = 1.
We now use the three-population lemma to prove that for any K, Fst between a population formed by admixture of K founding populations and a specific one of those founding populations is maximized when the admixed population is in fact one of the other K − 1 founding populations.
Theorem 2. Denote by the vectors corresponding to the allele frequencies of K populations. Consider a population formed by admixture between the K populations, where γk ∈ [0, 1] represents the admixture fraction for population k, for 1 ≤ k ≤ K, such that . Then
| (6) |
Proof. We prove this result by induction on K.
- Step 1: K = 2. Taking in place of , respectively, Lemma 1 already demonstrates the result in the case that K = 2:
where, for simplicity, we replace γ1 by γ and γ2 by 1 − γ. - Step 2: K → K + 1. We now show that if the result in eq. 6 holds for K populations, then it also holds for K + 1 populations (with γK+1 > 0):
where the last step follows by the inductive hypothesis, eq. 6, for the case with K populations. The expression has the form , with γK/(γK + γK+1) taking on the role of γ. Consequently, using Lemma 1,
so that
Thus, the induction is complete, and we have shown that the result in eq. 6 holds for arbitrary K.
The theorem is sensible, in that considering all possible populations admixed among a given collection of source populations, the most “distant” populations from source population 1 are combinations that do not include ancestry from population 1. The theorem demonstrates that the most distant admixed population according to Fst is precisely one of the remaining source populations, it is not a nontrivial mixture of those source populations, either with each other or with population 1.
An interesting corollary of Theorem 2 is that given a set of K founding populations, considering all admixed populations that can be constructed from those founding populations, the value of Fst between an admixed population and the founding population from which it is maximally distant, or maxk∈{1,…,K} , is bounded above by the maximal Fst among pairs of founding populations, or maxk,ℓ∈{1,…,K} . This result is obtained by simply noting that given k, as a result of the theorem, is bounded above by maxℓ∈{1,…,K} , and by then taking the maximum over k. Thus, according to the Fst measure, an admixed population can be no more distant from any of its founding populations than the two most distant among the founding populations are from each other.
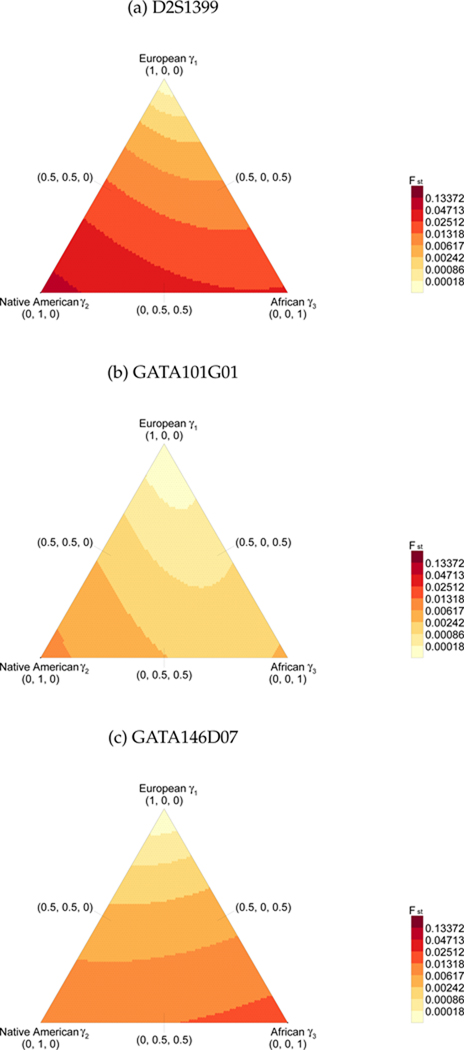
Three examples illustrating the theorem with K = 3 are presented in Figure 2. Sample allele frequencies for three genetic loci in three populations — European, Native American, and African — are used as , respectively. The triangular region shown in the figure for a given locus represents the space of possible admixture vectors (γ1, γ2, γ3). For each locus, the maximal value of Fst between the admixed population and population 1 occurs either at the corner represented by γ2 = 1 or at the corner represented by γ3 = 1, as established in the theorem.
Figure 2.
Fst between a population and hypothetical admixtures of that population with two other populations. , seen as a function of γ̲, is plotted for all possible values of γ̲ (eq. 2). Populations 1, 2, and 3 represent populations of European, Native American, and African descent, respectively, and are based an allele frequencies estimated from Wang et al. (2008). Three loci are considered. In each triangle, the admixture fractions γ1, γ2, and γ3 vary along the three axes, and darker colors correspond to higher values of Fst. In accordance with Theorem 2, considering all possible γ̲, the maximal value of Fst is always at γ2 = 1 or γ3 = 1. (A) Locus D2S1399. (B) Locus GATA101G01. (C) Locus GATA146D07.
4. Special case: Two founding populations
We now consider the special case in which only two founding populations give rise to an admixed population. We can simplify the notation of the previous section, so that γ = γ1, 1 − γ = γ2, τi = p1i + p2i, and δi = p1i − p2i. Using eq. 1, the Fst value between population 1 and the admixed population can be written as
| (7) |
Thus, in this case, Fst is a function only of the admixture fraction, γ, and the sums and differences of the allele frequencies of the two founding populations. The K = 2 case has fewer parameters than the general case of arbitrary K, and it is therefore possible to more precisely examine the properties of Fst as a function of the single admixture coefficient γ.
We first note that it was shown in the proof of Theorem 2 that the maximal Fst between population 1 and the admixed population is obtained when the admixed population is in fact population 2, so that γ = 0:
4.1. Fst is monotonic and convex in the admixture coefficient
For the case with two founding populations, it is of interest to determine whether Fst behaves in a predictable way as a function of γ. We now show that Fst between a founding population and the admixed population is monotonic in the admixture coefficient.
Theorem 3. As a function of γ, is decreasing for γ ∈ [0, 1].
Proof. Let α = 1 − γ. We can use portions of the proof of Lemma 1, with in the role of . Following the proof of Lemma 1, for , E(α) = aα2 + bα + c. Plugging in α = 0, E(α) = c. Noting in eq. 4 that δ13i = 0 when , we get E(0) = 0. We then have E(1) > E(0) ≥ 0, so that G(α) is increasing on α ∈ [0, 1]. As a result, is decreasing in γ on γ ∈ [0, 1].
The theorem supports the intuitive perspective that increasing the admixture fraction from source population 2 increases the genetic divergence of an admixed population from source population 1. We can in fact prove a stronger result. Not only is monotonic in γ, we can also show that it is convex as a function of the admixture fraction.
Theorem 4. As a function of γ, is convex for γ ∈ [0, 1].
Proof. It suffices to demonstrate that the second derivative of as a function of γ is nonnegative. We insert in place of , respectively, in the proof of Lemma 1. Thus, we aim to show that . We have:
To verify that the second derivative of G(γ) is nonnegative, we need only show that is nonnegative. Consider the following expressions:
We note some relationships between these expressions:
| (8) |
| (9) |
Using eq. 9,
| (10) |
Differentiating the individual expressions and then combining them,
We now differentiate eq. 10 and use the expressions above, obtaining:
The last step follows because E2(γ) ≥ 0, as E2(γ) corresponds to four times the (nonnegative) heterozygosity of the pooled population consisting of population 1 and the admixed population with allele frequency vector . By applying eqs. 8 and 9 and simplifying, we obtain:
because both terms are nonnegative.
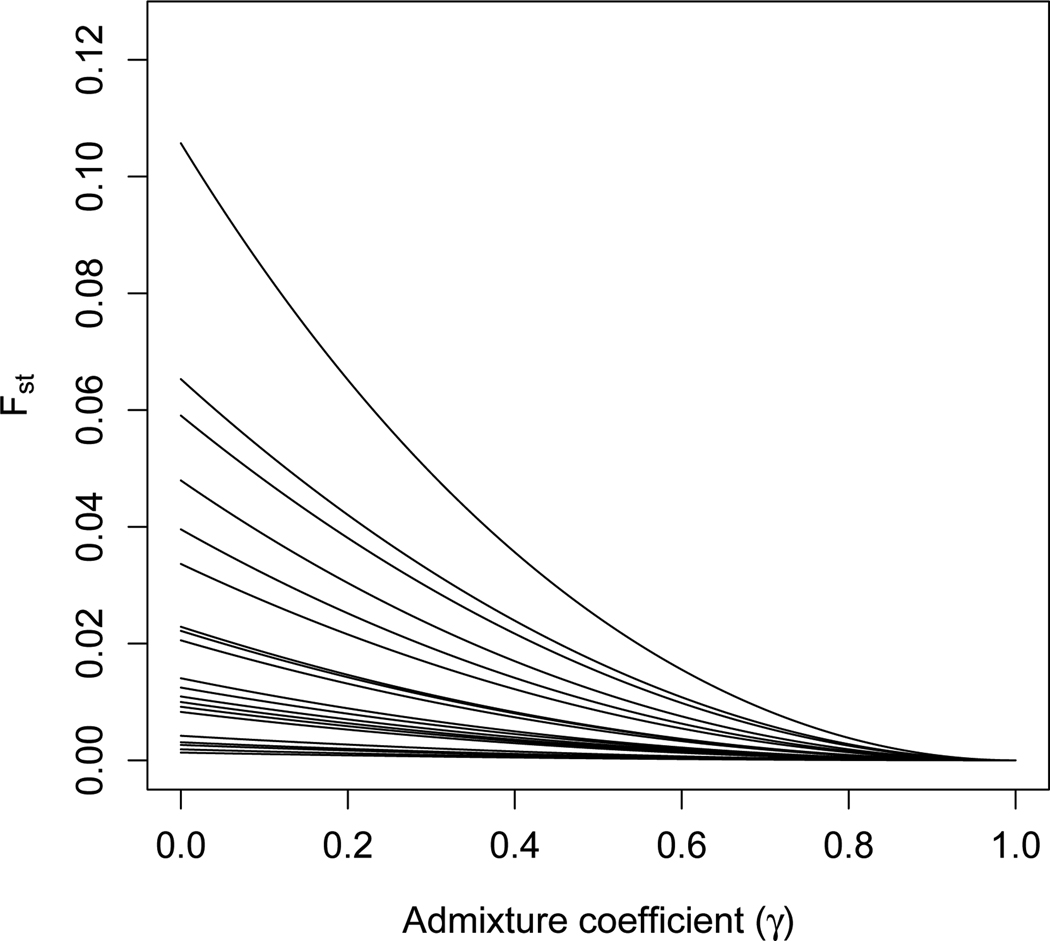
An illustration of Theorems 3 and 4 appears in Figure 3. The same twenty loci from Figure 1 are used; populations 1 and 2 are the European and Native American populations, respectively. For each locus, the Fst value between population 1 and a population formed by the admixture of populations 1 and 2 can be seen to be decreasing and convex in γ ∈ [0, 1], where γ is the admixture fraction for population 1.
Figure 3.
Fst between a population and a hypothetical admixture of that population with a second population. , seen as a function of γ, is plotted against γ (eq. 7). Populations 1 and 2 represent populations of European and Native American descent, respectively, and are based on allele frequencies estimated from Wang et al. (2008). The same twenty randomly selected loci as in Figure 1 are considered, with each curve representing a different locus. In accordance with Theorems 3 and 4, Fst is always decreasing and convex in γ.
4.2. Comparison of predicted Fst to observed Fst
When allele frequencies are available on both the admixed population and the founding populations, we are able to calculate the observed Fst value between a specific founding population and a population formed by admixture of multiple founding populations. For example, it is possible to calculate the observed Fst between an African-American population and a putative African founding population, or between a Mestizo population and a putative European founding population. In practice, the true founding populations are not precisely known, no longer exist, or may not have data available, so that in general, only an approximation is possible.
In such cases, our results provide a way of predicting the Fst value between a population formed by an admixture of multiple founding populations and a specific founding population, on the basis of measured allele frequencies and admixture coefficients. The predicted Fst value can be calculated when the allele frequencies and the admixture coefficients are available or can be estimated for the founding populations. Estimation of the admixture fractions at a given locus for the various founding populations can be achieved via maximum likelihood (Millar (1987)) or other techniques.
For the Wang et al. (2008) data, we estimated the fraction of European ancestry in the Mestizo population at each of the 678 loci, treating the European and Native American populations as founding populations. This approach followed the procedure of Schroeder et al. (2009), with all of the various subgroups in the Mestizo sample of Wang et al. (2008) pooled together (indeed the admixture estimates are the same as those used in the “Combined admixed sample” analysis in Table 1 of Schroeder et al. (2009)). Following Schroeder et al. (2009), for any allele present in at least one individual in the Mestizo population but not present in both founding populations, and for each founding population that did not possess the allele, a single copy of the allele was artificially added to that ancestral population. Sample allele frequencies that were then obtained for Europeans and Native Americans were treated as true allele frequencies for use in the maximum likelihood inference of the European admixture proportion, assuming Hardy-Weinberg equilibrium in the admixed population. Maximum likelihood estimates were obtained numerically and were used to obtain the predicted Fst according to eq. 7.
The observed and predicted Fst values for individual loci are compared in Figure 4. In general, we find that the observation closely matches the prediction. In most cases (549 of 678 loci), however, the prediction provides an underestimate of the observed value. This systematic underestimation might arise from the use of estimated rather than true values to obtain the prediction; in particular, the prediction relies on both the estimated allele frequencies and the maximum likelihood estimate of γ obtained from the same data used to estimate the allele frequencies.
Figure 4.
Predicted and observed Fst. (A) The predicted and observed Fst values between an admixed Mestizo population and a European founding population are plotted against the European admixture fraction γ in the Mestizo population, estimated by maximum likelihood. The prediction is based on eq. 7, using the European and Native American allele frequencies estimated from Wang et al. (2008) as , respectively, together with the maximum likelihood estimate of γ. The observation is based on Fst estimated from eq. 1, inserting estimated allele frequencies from Wang et al. (2008) on European and Mestizo populations. (B) The observed Fst value is plotted against the predicted Fst value. The identity line is shown in gray. In both panels, each point represents one of the 678 loci used. The correlation coefficient between the predicted and observed Fst values is 0.978.
4.3. An admixture estimator on the basis of Fst
As an alternative to use of an estimated admixture coefficient to predict Fst, an observed Fst value between a population formed by admixture of two founding populations and a specific founding population can be used as a way of estimating the admixture fraction. In the case of two founding populations, the quadratic equation in eq. 7 can be solved to provide an estimator in the style of the method of moments. This approach is reasonable, as the monotonicity result in Theorem 3 indicates that for fixed allele frequencies, γ is identifiable from Fst. The resulting estimator is non-parametric, in that it does not make assumptions on the form of the probability distribution of the allele frequencies at a given locus:
| (11) |
It can be shown that γ̂+ ≥ 1 for all possible values of Fst and the δi and τi. Therefore, if γ̂− is between 0 and 1, it is chosen as the estimate. In the case in which γ̂− < 0, 0 is chosen as the estimate, and if γ̂− > 1, 1 is chosen as the estimate.
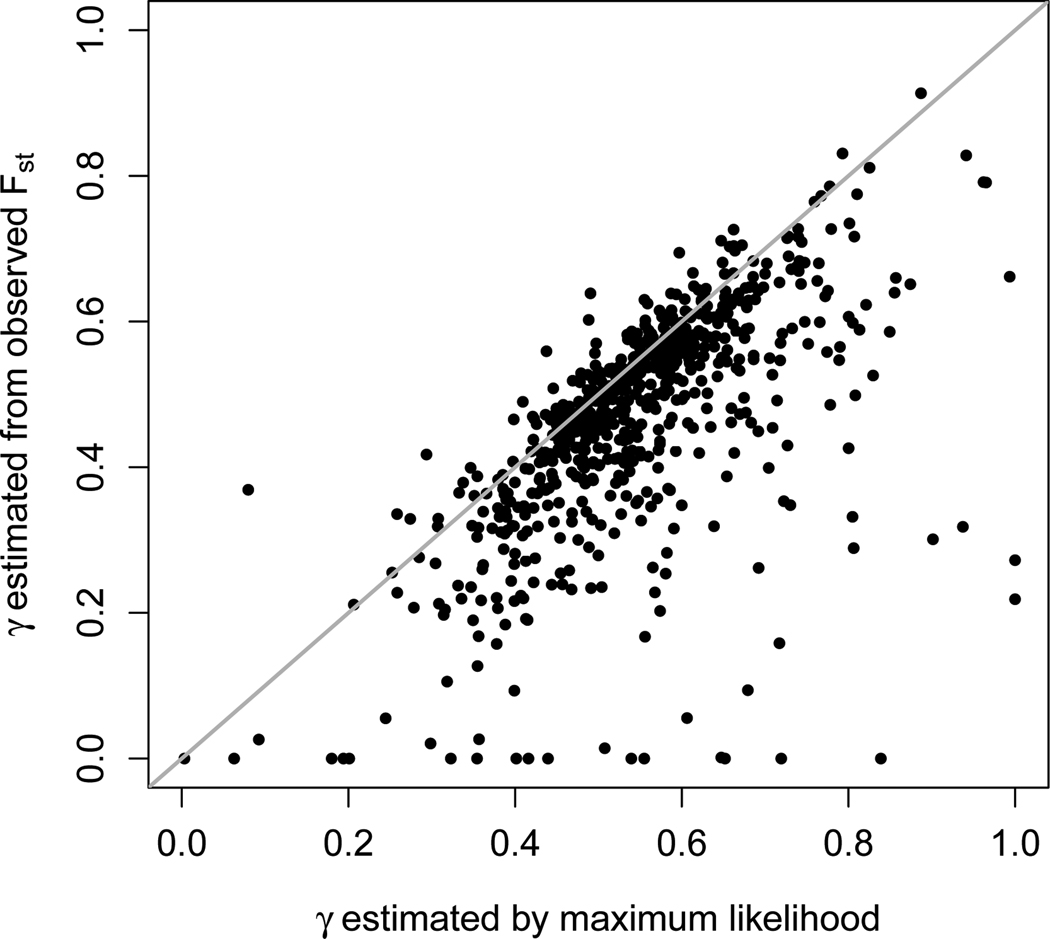
For the example data, Figure 5 presents a plot of the estimate of γ from the observed Fst versus the maximum likelihood estimate of γ, with Europeans and Native Americans as populations 1 and 2, and with Mestizos as the admixed population. The correlation between the estimates from these two methods is 0.618, and in general, the moment estimator produces smaller estimates than the maximum likelihood method, including several estimates of zero in cases where maximum likelihood obtains a positive value.
Figure 5.
Admixture estimates obtained from observed Fst (eq. 11) versus estimates obtained by maximum likelihood. The plot represents a scenario in which a European and a Native American population are the founding populations and a Mestizo population is the admixed population, and allele frequencies estimated from Wang et al. (2008) on all three populations are used to estimate γ, the European admixture fraction in the Mestizo population. The identity line is shown in gray, and each point represents one of the 678 loci used. The correlation coefficient between the two sets of estimates is 0.618.
5. Discussion
In this paper, we have considered the Fst measure in the context of admixed populations. We have explored the Fst value between a population formed by the admixture of K founding populations and one of those founding populations. In the general case of arbitrary K ≥ 2, we have demonstrated that this value is maximized when the admixed population is in fact one of the other founding populations. In the particular case of K = 2, this Fst value is monotonic and convex in the admixture fraction. We have also provided a formula for predicting Fst in an admixed population on the basis of the estimated admixture coefficient and the allele frequencies in the founding populations, producing very similar values to those observed in an empirical example utilizing the data of Wang et al. (2008).
Further, we discussed a non-parametric method of estimating the admixture fraction from the observed Fst values, and we compared it to the maximum likelihood method. In general, the non-parametric estimator is useful primarily for the purpose of illustrating the close relationship between Fst and the admixture coefficient, and its statistical properties are likely to be poorer than those of modern genome-based approaches to admixture estimation (Falush et al. (2003), Hoggart et al. (2004), Tang et al. (2006a), Sankararaman et al. (2008) Alexander et al. (2009), Price et al. (2009), Engelhardt and Stephens (2010)). However, as a straightforward formula that is calculated from quantities that are easily obtained, it provides a convenient approach when a computationally simple initial estimate is desirable.
Our results can provide a basis for interpreting Fst in admixed populations. In particular, in the case in which K = 2, the monotonicity and convexity of Fst in the admixture coefficient imply that Fst is informative about the level of admixture, and vice versa. This relationship can be a useful starting point for measurement of admixture, and a comparison of observed and predicted Fst values can be used as an initial check on the extent to which estimates of the admixture fraction obtained by maximum likelihood or other algorithms are sensible.
We note several limitations of our work. First, our admixture model does not involve a mechanistic evolutionary process, considering only the linear combination of allele frequencies that occurs when an admixed population is produced instantaneously from a set of source populations. Second, we examine admixture only at the population level, disregarding variation that might exist in admixture levels across individuals within a population. Third, as in many methods for analysis of admixed populations, we caution that our work presumes that the source populations for a given admixed population have been correctly specified. It is encouraging, however, that in spite of these concerns, the predicted Fst values generally agree with the values observed in our empirical example. As illustrated by the results presented here, further analysis of the properties of Fst in an admixture setting will continue to facilitate the understanding of population-genetic issues in the context of admixture research.
Acknowledgments
Support for this work was provided by NIH grants R01 GM081441 and T32 GM074906, NSF grant BCS-1024627, the Burroughs Wellcome Foundation, and the Johns Hopkins Sommer Scholar Program. We also thank two anonymous reviewers for comments that significantly improved the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Research. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balding DJ. Likelihood-based inference for genetic correlation coefficients. Theoretical Population Biology. 2003;63:221–230. doi: 10.1016/s0040-5809(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, Ferrell RE, Hoggart CL, McKeigue PM, Shriver MD. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Annals of Human Genetics. 2004;68:139–153. doi: 10.1046/j.1529-8817.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo J-M, Wambebe C, Tishkoff SA, Bustamante CD. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proceedings of the National Academy of Sciences of the United States of America. 2010a;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K, Velez C, Karafet T, Moreno-Estrada A, Reynolds A, Auton A, Hammer M, Bustamante CD, Ostrer H. Genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proceedings of the National Academy of Sciences of the United States of America. 2010b;107:8954–8961. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerkle CA, Lexer C. Admixture as the basis for genetic mapping. Trends in Ecology and Evolution. 2008;23:686–694. doi: 10.1016/j.tree.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Engelhardt BE, Stephens M. Analysis of population structure: a unifying framework and novel methods based on sparse factor analysis. PLoS Genetics. 2010;6 doi: 10.1371/journal.pgen.1001117. e1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L. Analysis of population subdivision. chapter 10. In: Balding DJ, Bishop M, Cannings C, editors. Handbook of Statistical Genetics. Chichester, UK: Wiley; 2001. pp. 271–307. [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. American Journal of Human Genetics. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nature Reviews Genetics. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeigue PM. Prospects for admixture mapping of complex traits. American Journal of Human Genetics. 2005;76:1–7. doi: 10.1086/426949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar RB. Maximum likelihood estimation of mixed stock fishery composition. Canadian Journal of Fisheries and Aquatic Sciences. 1987;44:583–590. [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. American Journal of Human Genetics. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Kittles RA, Argyropoulos G, Pfaff CL, Hiester K, Bonilla C, Sylvester N, Parrish-Gause D, Garveyand WT, Jin L, McKeigue PM, Kamboh MI, Ferrell RE, Pollitzer WS, Shriver MD. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. American Journal of Physical Anthropology. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, Beaty TH, Mathias R, Reich D, Myers S. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000519. e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N. Will admixture mapping work to find disease genes? Philophical Transactions of the Royal Society of London B–Biological Sciences. 2005;360:1605–1607. doi: 10.1098/rstb.2005.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Choudhry S, Via M, Basu A, Sebro R, Eng C, Beckman K, Thyne S, Chapela R, Rodriguez-Santana JR, Rodriguez-Cintron W, Avila PC, Ziv E, Burchard EG. Ancestry-related assortative mating in Latino populations. Genome Biology. 2009;10:R132. doi: 10.1186/gb-2009-10-11-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Inferences from spatial population genetics. chapter 9. In: Balding DJ, Bishop M, Cannings C, editors. Handbook of Statistical Genetics. Chichester, UK: Wiley; 2001. pp. 239–269. [Google Scholar]
- Salas A, Carracedo A, Richards M, Macaulay V. Charting the ancestry of African Americans. American Journal of Human Genetics. 2005;77:676–680. doi: 10.1086/491675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Sridhar S, Kimmel G, Halperin E. Estimating local ancestry in admixed populations. American Journal of Human Genetics. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KB, Jakobsson M, Crawford MH, Schurr TG, Boca SM, Conrad DF, Tito RY, Osipova LP, Tarskaia LA, Zhadanov SI, Wall JD, Pritchard JK, Malhi RS, Smith DG, Rosenberg NA. Haplotypic background of a private allele at high frequency in the Americas. Molecular Biology and Evolution. 2009;26:995–1016. doi: 10.1093/molbev/msp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MF, Tian C, Shigeta R, Scherbarth HR, Silva G, Belmont JW, Kittles R, Gamron S, Allevi A, Palatnik SA, Alvarellos A, Paira S, Caprarulo C, Guillerón C, Catoggio LJ, Prigione C, Berbotto GA, García MA, Perandones CE, Pons-Estel BA, Alarcon-Riquelme ME. Argentine population genetic structure: large variance in Amerindian contribution. American Journal of Physical Anthropology. 2007;132:455–462. doi: 10.1002/ajpa.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MF. Admixture mapping as a tool in gene discovery. Current Opinion in Genetics & Development. 2007;17:177–181. doi: 10.1016/j.gde.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Zolezzi I, Hidalgo-Miranda A, Estrada-Gil J, Fernandez-Lopez JC, Uribe-Figueroa L, Contreras A, Balam-Ortiz E, del Bosque-Plata L, Velazquez-Fernandez D, Lara C, Goya R, Hernandez-Lemus E, Davila C, Barrientos E, March S, Jimenez-Sanchez G. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8611–8616. doi: 10.1073/pnas.0903045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, O’Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nature Reviews Genetics. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- Tang H, Coram M, Wang P, Zhu X, Risch N. Reconstructing genetic ancestry blocks in admixed individuals. American Journal of Human Genetics. 2006a;79:1–12. doi: 10.1086/504302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Jorgenson E, Gadde M, Kardia SLR, Rao DC, Zhu X, Schork NJ, Hanis CL, Risch N. Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Human Genetics. 2006b;119:624–633. doi: 10.1007/s00439-006-0175-4. [DOI] [PubMed] [Google Scholar]
- Tang H, Choudhry S, Mei R, Morgan M, Rodriguez-Cintron W, Burchard EG, Risch NJ. Recent genetic selection in the ancestral admixture of Puerto Ricans. American Journal of Human Genetics. 2007;81:626–633. doi: 10.1086/520769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo J-M, Doumbo O, Ibrahim M, Juma AT, Kotze MJ, Lema G, Moore JH, Mortensen H, Nyambo TB, Omar SA, Powell K, Pretorius GS, Smith MW, Thera MA, Wambebe C, Weber JL, Williams SM. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ray N, Rojas W, Parra MV, Bedoya G, Gallo C, Poletti G, Mazzotti G, Hill K, Hurtado AM, Camrena B, Nicolini H, Klitz W, Barrantes R, Molina JA, Freimer NB, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Dipierri JE, Alfaro EL, Bailliet G, Bianchi NO, Llop E, Rothhammer F, Excoffier L, Ruiz-Linares A. Geographic patterns of genome admixture in Latin American mestizos. PLoS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000037. e1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler CA, Nelson GW, Smith MW. Admixture mapping comes of age. Annual Review of Genomics and Human Genetics. 2010;11:65–89. doi: 10.1146/annurev-genom-082509-141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Annals of Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Zakharia F, Basu A, Absher D, Assimes TL, Go AS, Hlatky MA, Iribarren C, Knowles JW, Li J, Narasimhan B, Sidney S, Southwick A, Myers RM, Quertermous T, Risch N, Tang H. Characterizing the admixed African ancestry of African Americans. Genome Biology. 2009;10:R141. doi: 10.1186/gb-2009-10-12-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Tang H, Risch N. Admixture mapping and the role of population structure for localizing disease genes. Advances in Genetics. 2008;60:547–569. doi: 10.1016/S0065-2660(07)00419-1. [DOI] [PubMed] [Google Scholar]