CENP-C provides a link between existing CENP-A chromatin and the proteins required for new CENP-A nucleosome assembly.
Abstract
Eukaryotic chromosomes segregate by attaching to microtubules of the mitotic spindle through a chromosomal microtubule binding site called the kinetochore. Kinetochores assemble on a specialized chromosomal locus termed the centromere, which is characterized by the replacement of histone H3 in centromeric nucleosomes with the essential histone H3 variant CENP-A (centromere protein A). Understanding how CENP-A chromatin is assembled and maintained is central to understanding chromosome segregation mechanisms. CENP-A nucleosome assembly requires the Mis18 complex and the CENP-A chaperone HJURP. These factors localize to centromeres in telophase/G1, when new CENP-A chromatin is assembled. The mechanisms that control their targeting are unknown. In this paper, we identify a mechanism for recruiting the Mis18 complex protein M18BP1 to centromeres. We show that depletion of CENP-C prevents M18BP1 targeting to metaphase centromeres and inhibits CENP-A chromatin assembly. We find that M18BP1 directly binds CENP-C through conserved domains in the CENP-C protein. Thus, CENP-C provides a link between existing CENP-A chromatin and the proteins required for new CENP-A nucleosome assembly.
Introduction
Accurate chromosome segregation is essential for the faithful distribution of the genome during cell division. In mitosis, each chromosome assembles a multiprotein complex called the kinetochore that serves as the primary binding site for microtubules of the mitotic spindle. The kinetochore mediates the bipolar attachment of paired chromosomes to the mitotic spindle, monitors proper chromosome–spindle attachment through the mitotic checkpoint, and couples spindle forces to chromosome segregation at anaphase (Rieder and Salmon, 1998; Cleveland et al., 2003; Cheeseman and Desai, 2008). The assembly site for the kinetochore is a specialized chromatin domain called the centromere that is characterized by the incorporation of the histone H3 variant CENP-A (centromere protein A) into centromeric nucleosomes (Palmer et al., 1987, 1991; Sullivan et al., 1994). CENP-A chromatin recruits a collection of ∼20 proteins called the constitutive centromere-associated network (CCAN) that are bound to the chromosome throughout the cell cycle and serve as the site for mitotic kinetochore assembly. Mutation or loss of either CENP-A or proteins of the CCAN results in kinetochore formation defects and chromosome missegregation (Foltz et al., 2006; McClelland et al., 2007; Cheeseman et al., 2008; Hori et al., 2008; Amano et al., 2009).
Vertebrate centromeres occur at a single region along the length of the chromosome that is maintained through mitotic and meiotic divisions. Human centromeric chromatin assembles on long stretches of repetitive α-satellite DNA. In rare cases, de novo centromere formation can occur outside of α-satellite–containing DNA sequences, resulting in the formation of stably maintained neocentromeres marked by CENP-A nucleosomes. This suggests that the determinant of centromere position is the site of CENP-A nucleosome incorporation into chromatin and not DNA sequence (Voullaire et al., 1993; Barry et al., 1999; Carroll and Straight, 2006; Allshire and Karpen, 2008). CENP-A nucleosome assembly into chromatin occurs during a specific time window in the cell cycle, during telophase/G1 in somatic cells and after anaphase chromosome segregation in embryos (Jansen et al., 2007; Schuh et al., 2007; Bernad et al., 2011). How the preexisting centromere directs the local assembly of new CENP-A nucleosomes during G1 is not known.
Several protein complexes that govern CENP-A assembly and maintenance at centromeres have been identified through genetic and biochemical studies in yeasts, flies, worms, and humans. A study of chromosome missegregation mutants in Schizosaccharomyces pombe discovered the mis16 and mis18 genes, which when mutated lead to a loss of Cnp1 (S. pombe CENP-A) from centromeres (Hayashi et al., 2004). S. pombe mis16 is homologous to the histone chaperones RbAp46 and 48, and RbAp46/48 depletion from human cells using RNAi leads to defects in CENP-A assembly (Hayashi et al., 2004; Dunleavy et al., 2009). Two homologues of mis18 have been identified in vertebrates, Mis18-α and Mis18-β, as well as an additional Mis18-α–Mis18-β binding protein, M18BP1 (Fujita et al., 2007). A homologue of M18BP1, knl-2 (kinetochore-null 2), was discovered through RNAi screening for chromosome segregation defects in worms (Maddox et al., 2007). Depletion of either Mis18-α from human cells or KNL-2 from worm embryos prevented CENP-A assembly at centromeres (Fujita et al., 2007; Maddox et al., 2007). Immunoprecipitation experiments from yeast and human cells have shown that S. pombe Mis18 can coprecipitate Mis16, and both human Mis18-α and M18BP1 can coprecipitate RbAp46/48 (Fujita et al., 2007; Hayashi et al., 2008; Lagana et al., 2010). Furthermore, RbAp46/48 copurifies with CENP-A in affinity purification experiments from both human and fly cells (Furuyama et al., 2006; Dunleavy et al., 2009). Immunoprecipitation of KNL-2 from Caenorhabditis elegans embryos also precipitates CENP-A–containing chromatin, but the human Mis18 complex components do not appear to directly associate with soluble or nucleosomal CENP-A (Hayashi et al., 2004; Fujita et al., 2007; Maddox et al., 2007; Carroll et al., 2009; Lagana et al., 2010).
HJURP (Holliday junction–recognizing protein)/Scm3 is a CENP-A chaperone protein that binds directly to soluble CENP-A and is required for CENP-A chromatin assembly. First identified as a suppressor of CSE4 (S. cerevisiae CENP-A) mutants (Chen et al., 2000), the Scm3 protein was shown to interact directly with Cse4 and was required for Cse4 association with yeast centromeres (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007). The S. pombe homologue of Scm3 is also required for Cnp1 localization to centromeres in fission yeast and was shown to interact with Cnp1 and with Mis16 and Mis18, thus providing another molecular link between the Mis18 complex of proteins and CENP-A (Pidoux et al., 2009; Williams et al., 2009). In human cells, affinity purification of a soluble (nonchromatin associated) human CENP-A complex identified four proteins (CENP-A, histone H4, NPM1 [nucleophosmin 1], and HJURP) that stably associate and function as a chaperone complex for CENP-A (Dunleavy et al., 2009; Foltz et al., 2009). HJURP contains a region of homology to the yeast Scm3 proteins (the Scm3 domain), and HJURP appears to function analogously to Scm3 in fungi (Sanchez-Pulido et al., 2009), as RNAi-mediated depletion of HJURP from human cells or depletion of Xenopus laevis HJURP from frog egg extracts causes defects in CENP-A assembly akin to the defects resulting from Scm3 mutation in yeast (Dunleavy et al., 2009; Foltz et al., 2009; Bernad et al., 2011).
Regulating the localization of the Mis18 complex and the HJURP chaperone complex proteins is likely to be an important step in achieving spatial and temporal control of the CENP-A assembly process. In human cells, the Mis18 complex and the HJURP chaperone complex are targeted to centromeres during late anaphase/telophase and remain associated with centromeres during G1 phase, the time window during which new CENP-A assembly occurs (Fujita et al., 2007; Maddox et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009; Silva and Jansen, 2009). The Mis18 complex localizes to centromeres before HJURP (Foltz et al., 2009), and HJURP fails to localize to centromeres in cells depleted of the Mis18 complex (Barnhart et al., 2011). It has been proposed that Mis18 complex proteins may “prime” the centromere so that it is competent to load new CENP-A histone molecules (Fujita et al., 2007). However, the molecular interactions that target the Mis18 complex and the HJURP complex to centromeres are unknown.
Here, we use an in vitro system for CENP-A assembly on sperm chromatin in extracts of Xenopus eggs that recapitulates the cell cycle dependence, HJURP dependence, and Mis18 complex requirements for CENP-A assembly in cells. Using this in vitro system, we demonstrate that CENP-C is required to target the Mis18 complex protein M18BP1 to Xenopus sperm centromeres in metaphase. In the absence of CENP-C, M18BP1 and HJURP targeting to centromeres is disrupted, and new CENP-A assembly into centromeric chromatin is inhibited. We find that CENP-C interacts directly with M18BP1 and that this interaction requires both the conserved CENP-C motif and the cupin dimerization domain. Our experimental results demonstrate a novel role for CENP-C in recruiting CENP-A nucleosome assembly proteins to the centromere to ensure centromere propagation.
Results
Xenopus HJURP (xHJURP) promotes CENP-A assembly in a cell-free system
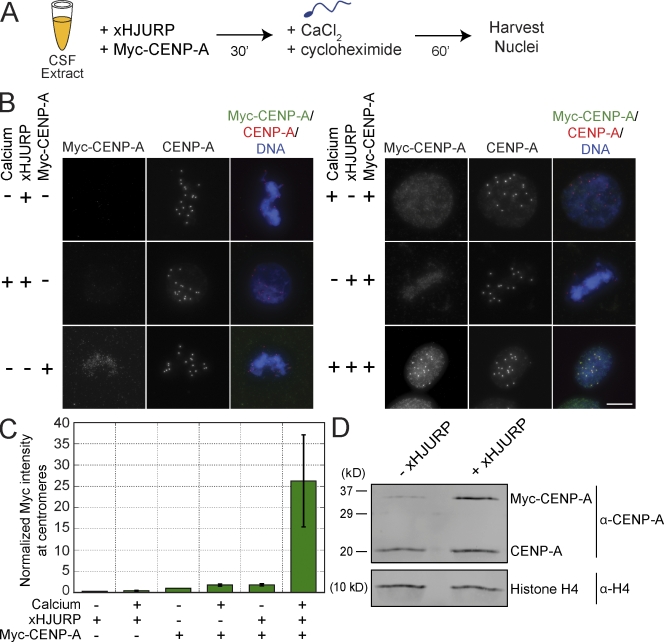
To study the assembly of CENP-A chromatin, we developed an in vitro system for CENP-A assembly in Xenopus egg extracts similar to a recently described method (Bernad et al., 2011). Xenopus sperm chromatin contains CENP-A, and when that chromatin is added to metaphase egg extracts, functional centromeres and kinetochores assemble at the preexisting centromere (Desai et al., 1997; Milks et al., 2009). To follow new CENP-A assembly in egg extracts, we translated myc epitope–tagged CENP-A in the extracts and then added Xenopus sperm chromatin and calcium to drive the extract from metaphase into interphase. The translation of myc–CENP-A in the extract produced a fivefold excess of myc–CENP-A to the endogenous CENP-A (Fig. S1 A) but did not result in detectable myc–CENP-A at sperm centromeres as assayed by immunofluorescence (Fig. 1, A–C; Bernad et al., 2011). We cloned the Xenopus homologue of HJURP (xHJURP) and tested its ability to stimulate myc–CENP-A assembly into centromeric chromatin. We found that adding xHJURP to the extracts and releasing the extract from metaphase arrest caused a 26 ± 11–fold stimulation in centromeric myc–CENP-A compared with extracts lacking exogenously added xHJURP and calcium (Fig. 1, B and C). Western blotting confirmed that the stimulation of myc–CENP-A assembly was not caused by variation in myc–CENP-A protein levels in the extract (Fig. S1 B). Myc–CENP-A was productively incorporated into chromatin as indicated by its resistance to salt extraction (Fig. S1 C), and myc–CENP-A assembly was independent of DNA replication (Fig. S1, D and E).
Figure 1.
CENP-A assembly in Xenopus extracts requires xHJURP addition and mitotic exit. (A) Schematic of xHJURP-mediated CENP-A assembly assay in Xenopus egg extract. (B) Representative images from xHJURP-mediated CENP-A assembly assay. The staining for myc–CENP-A, total CENP-A, or the merge of myc–CENP-A, total CENP-A, and DNA is indicated above the images. Calcium, xHJURP, or myc–CENP-A addition is shown to the left of each image row. Bar, 10 µm. (C) Quantification of myc–CENP-A fluorescence intensity at centromeres for the assembly reactions represented in B, normalized to the metaphase control sample without xHJURP but with myc–CENP-A RNA added (−/−/+). Mean per pixel intensity at centromeres was quantified, and >200 centromeres were quantified per condition. Error bars show SEM; n = 4. (D) Western blot of chromatin fractions from CENP-A assembly reactions performed in the presence (+) of calcium, Myc-CENP-A, and xHJURP (+/+/+) or in the presence of calcium and Myc-CENP-A but lacking (−) xHJURP (+/−/+). Histone H4 was used as a loading control. n = 3.
We added xHJURP-FLAG to extracts and found that it localized to centromeres in interphase, similar to the endogenous protein (Fig. S2 A). Western blotting with an xHJURP-specific antibody demonstrated that addition of xHJURP to 1.6-fold the levels of HJURP present in the extract was sufficient to promote myc–CENP-A assembly (Fig. S2, B and C). In contrast to a previous study (Bernad et al., 2011), we did not detect xHJURP-FLAG at metaphase centromeres (Fig. S2 A), and supplementing the extract with human HJURP (hHJURP) did not stimulate myc–CENP-A assembly (Fig. S2, D–H).
We measured the relative amounts of myc–CENP-A and endogenous CENP-A in chromatin after HJURP-dependent CENP-A assembly. We recovered interphase nuclei from the assembly reactions, extracted the nuclei with 300 mM salt to remove nonnucleosomal CENP-A, and then Western blotted for CENP-A. We observed a twofold increase in total CENP-A in chromatin upon addition of calcium and xHJURP (Fig. 1 D), indicating that CENP-A is not over assembled under these conditions. We found that xHJURP addition led to increased incorporation of both the endogenous CENP-A and myc–CENP-A into chromatin. The ratio of newly assembled myc–CENP-A to CENP-A in chromatin corresponded to the ratio of myc–CENP-A to CENP-A in the extract (3.1:1 and 3.2:1, respectively), indicating that exogenously added xHJURP stimulates assembly of endogenous and myc-tagged CENP-A with equal efficiency (Fig. 1 D). When we omitted myc–CENP-A from the assembly reactions, we detected a 1.2-fold increase in the levels of CENP-A in chromatin without added xHJURP (Fig. S3, A–C) and a 1.7-fold increase in chromatin-incorporated CENP-A with xHJURP addition (Fig. S3, B and C).
We tested whether xHJURP, CENP-A, and/or sperm chromatin must be present in the extract during mitotic exit to assemble CENP-A. We added xHJURP, myc–CENP-A, and sperm chromatin to extracts in metaphase or interphase followed by release of the metaphase extract into interphase. CENP-A assembled into chromatin regardless of the timing of xHJURP, myc–CENP-A, and sperm chromatin addition (Fig. S4, A–D), indicating that xHJURP, myc–CENP-A, and sperm chromatin do not have to be present in the extract as it exits mitosis.
Characterization of Xenopus M18BP1
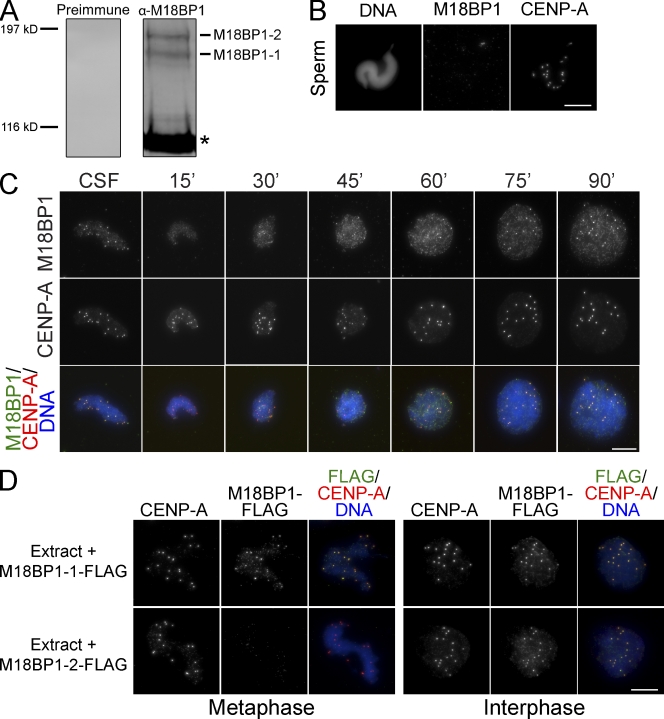
In human somatic cells and C. elegans, new CENP-A assembly requires the M18BP1/KNL-2 protein (Fujita et al., 2007; Maddox et al., 2007). To understand the functions of M18BP1 in CENP-A assembly, we cloned the Xenopus M18BP1 gene (xM18BP1) and found that two closely related isoforms of M18BP1 (xM18BP1-1 and xM18BP1-2) are expressed in Xenopus eggs. An antibody to a conserved region of xM18BP1 recognized both isoforms in frog egg extracts (Fig. 2 A). Unlike CENP-A, xM18BP1 was not retained in Xenopus sperm chromatin (Fig. 2 B). We determined the timing of M18BP1 association with centromeres in somatic cells and in egg extracts using immunofluorescence. We found that xM18BP1 localized to centromeres throughout mitosis in somatic cells and to sperm centromeres in metaphase egg extracts (Fig. 2 C and Fig. S5 A). Upon release of the egg extract from metaphase into interphase, xM18BP1 was lost from centromeres within 30 min and then reaccumulated at centromeres 60–75 min after calcium addition (Fig. 2 C).
Figure 2.
Characterization of Xenopus M18BP1. (A) Western blot of Xenopus egg extract with preimmune sera (Preimmune) or affinity-purified rabbit antibody raised against Xenopus M18BP1 (α-M18BP1). α-M18BP1 recognizes both isoforms of xM18BP1, and both isoforms of xM18BP1 are present in egg extract. The asterisk denotes a cross reacting band. (B) M18BP1 is not present on Xenopus sperm chromatin. After decondensation with Xenopus Nap1, Xenopus sperm chromatin shows staining for CENP-A but not M18BP1. (C) Time course of M18BP1 localization in the Xenopus egg extract. Xenopus sperm chromatin was incubated in metaphase extract for 30 min, and sperm chromatin was stained for M18BP1 at various time points after release from metaphase arrest. Time (minutes) after release from metaphase is indicated above each image column. M18BP1, CENP-A, or merged M18BP1, CENP-A, and DNA staining are indicated on the left. (D) M18BP1-1 assembles at sperm centromeres in metaphase extract, and both M18BP1 isoforms assemble at sperm centromeres in interphase extract. Metaphase extracts are shown on the left, and interphase extracts are shown on the right. The FLAG epitope–tagged M18BP1 isoform that was added to the extract is indicated on the left. Immunostaining is shown above the images. Bars, 10 µm.
Our antibody recognized both isoforms of xM18BP1; therefore, to assay the localization of each isoform separately, we expressed FLAG epitope–tagged versions of each in metaphase and interphase extracts. We found that only M18BP1-1 localized to metaphase centromeres (Fig. 2 D), similar to C. elegans KNL-2, which is present at centromeres throughout mitosis (Maddox et al., 2007). Both M18BP1-1 and M18BP1-2 localized to interphase centromeres (Fig. 2 D), similar to the human M18BP1 and S. pombe Mis18 proteins, which localize to centromeres during late mitosis and G1 (Fujita et al., 2007; Maddox et al., 2007).
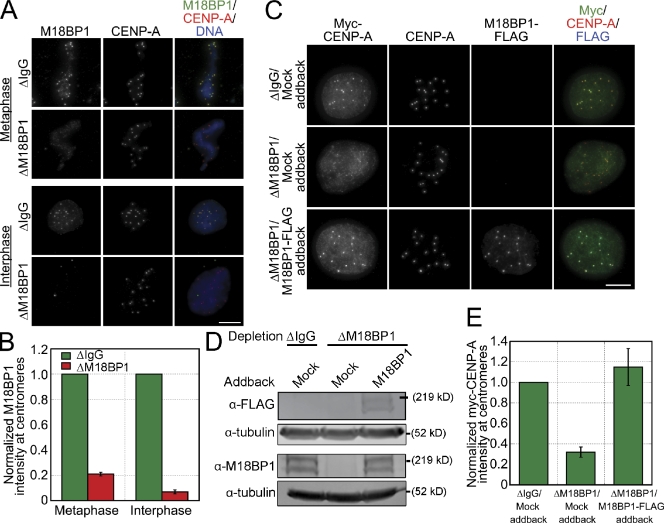
We determined the function of xM18BP1 in CENP-A assembly in vitro by depleting both isoforms from egg extracts and then measuring myc–CENP-A incorporation into chromatin. In this and all subsequent CENP-A assembly reactions, myc–CENP-A, xHJURP, and calcium were added to control and depleted extracts (+/+/+ condition, as described in Fig. 1 B). We were able to deplete ≥90% of both isoforms from egg extracts, resulting in a 79 ± 1.4% reduction in M18BP1 levels at metaphase centromeres and a 93 ± 1.6% reduction in M18BP1 levels at interphase centromeres. (Fig. 3, A and B). In M18BP1-depleted extracts, we found that myc–CENP-A assembly was reduced by 68 ± 5% relative to mock-depleted extracts (Fig. 3, C–E). To determine whether the defect in myc–CENP-A assembly was specific to the loss of M18BP1, we complemented the depleted extracts with endogenous levels of in vitro translated FLAG-tagged M18BP1-1 and M18BP1-2 proteins and found that myc–CENP-A assembly was fully restored (115 ± 18%; Fig. 3, C–E). Collectively, these data indicate that xM18BP1 is required for CENP-A assembly in Xenopus egg extract, as it is in other systems.
Figure 3.
Depletion of M18BP1 inhibits xHJURP-mediated CENP-A assembly in extract. (A) M18BP1 depletion from egg extract inhibits M18BP1 localization to centromeres. Xenopus sperm chromatin was incubated in mock- or M18BP1-depleted metaphase and interphase extracts and stained for M18BP1 and CENP-A. The depletion condition and cell cycle stage are listed to the left of the images, and the localized proteins are indicated above the images. (B) Quantification of M18BP1 fluorescence intensity at centromeres (normalized to the levels in mock-depleted extracts) after M18BP1 depletion. Quantification was performed as described in Fig. 1. Error bars show SEM; n = 3. (C) M18BP1 is required for CENP-A assembly in Xenopus extract. Representative images from CENP-A assembly assays performed using either mock- or M18BP1-depleted extracts that were supplemented with mock or M18BP1-FLAG IVT proteins. The depletion and addback conditions are listed to the left of each image row, and the immunolocalized proteins are listed above. (D) Representative Western blots of extracts from CENP-A assembly assays described in C. The top blots show the levels of the FLAG-tagged M18BP1 isoforms added back to the extract. The bottom blots show the total M18BP1 levels in the extract. Tubulin was used as a loading control. (E) Quantification of myc–CENP-A fluorescence intensity at centromeres for CENP-A assembly reactions described in C, which were normalized to the levels in mock-depleted extracts. Quantification was performed as described in Fig. 1. Error bars show SEM; n = 3. Bars, 10 µm.
CENP-C is required for the cell cycle–dependent localization of M18BP1 to centromeres
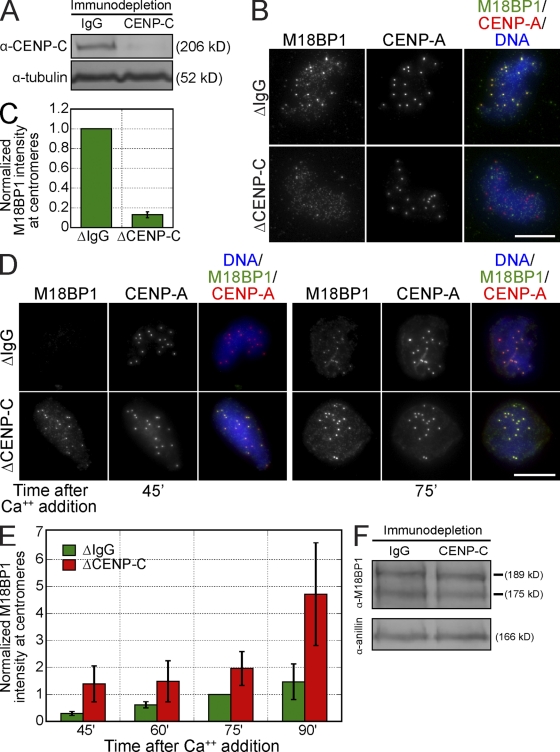
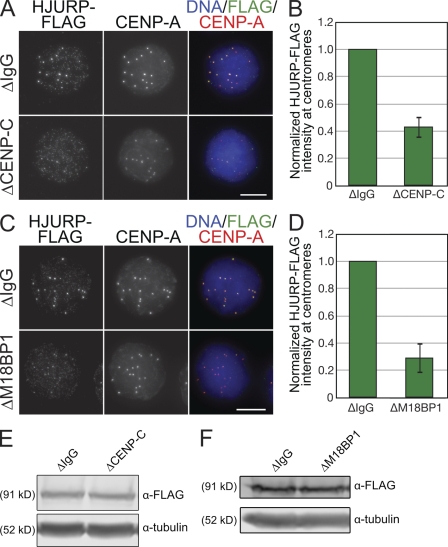
M18BP1 targeting to centromeres is thought to be important for new CENP-A assembly, but how it is recruited to centromeres is unknown (Maddox et al., 2007). Immunoprecipitation of C. elegans KNL-2 coprecipitates CENP-A chromatin, but human M18BP1 does not appear to directly bind CENP-A nucleosomes (Maddox et al., 2007; Carroll et al., 2009). However, CENP-C directly binds CENP-A nucleosomes and is required for new CENP-A assembly (Erhardt et al., 2008; Carroll et al., 2010); thus, we tested whether CENP-C might recruit M18BP1 to centromeres. We depleted CENP-C from egg extracts (Fig. 4 A) and found that CENP-C depletion reduced M18BP1 levels at metaphase centromeres by 87 ± 3% (Fig. 4, B and C). M18BP1-1, but not M18BP1-2, localized to metaphase centromeres, and we found that CENP-C depletion only affected M18BP1-1–FLAG targeting to the centromere (Fig. S5 B). We confirmed by Western blotting that the loss of M18BP1 at centromeres was not caused by codepletion of M18BP1 with CENP-C (Fig. 4 F).
Figure 4.
CENP-C is required for M18BP1 assembly at metaphase centromeres in Xenopus egg extract. (A) Representative Western blot of mock- and CENP-C–depleted extracts. CENP-C depletion led to a >90% reduction in CENP-C levels. Tubulin was used as a loading control. (B) CENP-C depletion prevented M18BP1 assembly at centromeres in metaphase. Xenopus sperm chromatin was incubated in mock- or CENP-C–depleted metaphase extracts and stained for M18BP1 and CENP-A. The depletion conditions are listed to the left of the images, and immunolocalized proteins are listed above. (C) Quantification of M18BP1 fluorescence intensity at metaphase centromeres (normalized to the levels in mock-depleted extracts) after CENP-C depletion. Quantification was performed as described in Fig. 1. Error bars show SEM; n = 3. (D) CENP-C depletion led to premature, increased association of M18BP1 at interphase centromeres. Xenopus sperm chromatin was incubated in mock- or CENP-C–depleted extracts and stained for M18BP1 at various time points after release from metaphase arrest. Depletion conditions are listed to the left of the images, time (minutes) after calcium addition is listed below the images, and immunolocalized proteins are listed above. (E) Quantification of M18BP1 fluorescence intensity at centromeres in mock- and CENP-C–depleted extracts at various time points after release from metaphase arrest as described in D. Values are normalized to the levels in mock-depleted extracts 75 min after calcium addition. Quantification was performed as described in Fig. 1. Error bars show SEM; n = 3. (F) CENP-C depletion does not significantly affect M18BP1 levels in extract. Mock- and CENP-C–depleted extracts were Western blotted for M18BP1 and Anillin as a loading control. M18BP1-1 and M18BP1-2 levels in CENP-C–depleted extracts are 77 ± 8 and 92 ± 11% of control extracts, respectively (n = 3; error = SEM). Bars, 10 µm.
In contrast to the loss of M18BP1 from metaphase centromeres in CENP-C–depleted extracts, M18BP1 prematurely localized to interphase centromeres in extracts lacking CENP-C (Fig. 4, D and E). In CENP-C–depleted extracts, M18BP1 associated with centromeres within 45 min of release from mitosis, but in mock-depleted extracts, M18BP1 did not accumulate at centromeres until 60–75 min after mitotic exit (Fig. 2 C and Fig. 4, D and E). CENP-C depletion led to increased M18BP1 at interphase centromeres (Fig. 4, D and E), resulting in approximately threefold more centromeric M18BP1 by 90 min after mitotic exit (Fig. 4 E). Both isoforms of M18BP1 localized to interphase centromeres in the CENP-C–depleted extract (Fig. S5 B), indicating that CENP-C is required for M18BP1-1 targeting to centromeres in metaphase extract only. M18BP1 depletion did not affect CENP-C levels at centromeres, showing that the localization of CENP-C does not depend on M18BP1 (Fig. S5, C and D). Collectively, our observations demonstrate that CENP-C plays a critical role in the centromere targeting of M18BP1 during metaphase but antagonizes the localization of M18BP1 during interphase.
CENP-C and M18BP1 promote centromere targeting of HJURP
HJURP is a CENP-A–specific chaperone that localizes to the centromere in early G1, in which it functions to assemble new CENP-A nucleosomes into chromatin. HJURP localization to the centromere depends on the Mis18 complex (Dunleavy et al., 2009; Foltz et al., 2009; Barnhart et al., 2011). We tested whether CENP-C depletion and the accompanying loss of M18BP1 from metaphase centromeres would disrupt HJURP localization in Xenopus extracts. Our HJURP antibody (Fig. S2 B) does not work well for immunofluorescence, so we assayed the centromeric localization of exogenously added xHJURP-FLAG after CENP-C depletion (Fig. 5, A and B). We found that CENP-C depletion reduced xHJURP-FLAG levels at centromeres by 57 ± 8% relative to control extracts, indicating that CENP-C promotes localization of HJURP to centromeres (Fig. 5, A and B). We next tested whether M18BP1 depletion affected HJURP-FLAG localization to centromeres using the same assay (Fig. 5, C and D). We found that centromeric levels of HJURP-FLAG were reduced by 71 ± 11% in M18BP1-depleted extracts relative to mock-depleted extracts (Fig. 5, C and D), indicating that M18BP1 promotes HJURP localization to centromeres in Xenopus egg extract as it does in human cells (Barnhart et al., 2011). The HJURP targeting defects we observed were not a result of variation in HJURP-FLAG protein levels in extracts (Fig. 5, E and F). Our results demonstrate that both CENP-C and M18BP1 promote the recruitment of HJURP to centromeres.
Figure 5.
CENP-C and M18BP1 promote the recruitment of HJURP to centromeres. (A) CENP-C depletion inhibited HJURP assembly at centromeres. Xenopus sperm chromatin was incubated in mock- or CENP-C–depleted extracts supplemented with HJURP-FLAG protein and stained for FLAG and CENP-A. The depletion conditions are listed to the left of the images, and immunolocalized proteins are listed above. (B) Quantification of HJURP-FLAG fluorescence intensity at centromeres (normalized to the levels in mock-depleted extracts) after CENP-C depletion. Quantification was performed as described in Fig. 1. (C) M18BP1 depletion inhibited HJURP assembly at centromeres. Xenopus sperm chromatin was incubated in mock- or M18BP1-depleted extracts supplemented with HJURP-FLAG and stained for FLAG and CENP-A. The depletion conditions are listed to the left of the images, and immunolocalized proteins are listed above. (D) Quantification of HJURP-FLAG fluorescence intensity at centromeres (normalized to the levels in mock-depleted extracts) after M18BP1 depletion. Quantification was performed as described in Fig. 1. (E) Representative Western blot from HJURP-targeting assay described in A showing that equal amounts of the HJURP-FLAG protein were added to mock- and CENP-C–depleted extracts. Tubulin is shown as a loading control. (F) Representative Western blot from HJURP-targeting assay described in C showing that equal amounts of the HJURP-FLAG protein were added to mock- and M18BP1-depleted extracts. Tubulin is shown as a loading control. Error bars show SEM; n = 3. Bars, 10 µm.
CENP-C is required for CENP-A assembly in Xenopus egg extract
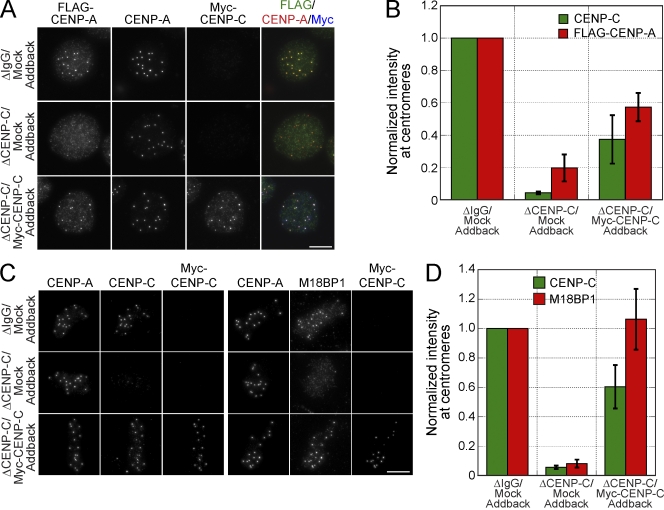
We determined whether CENP-C depletion and the resulting defects in M18BP1 and HJURP targeting caused a defect in CENP-A assembly. Depletion of CENP-C reduced the levels of centromeric FLAG–CENP-A by 80 ± 8.4% compared with mock-depleted extracts (Fig. 6, A and B). Complementation of the extract with myc–CENP-C restored CENP-C levels at centromeres to 40–60% of their original levels (Fig. 6, A–D; Milks et al., 2009; Carroll et al., 2010) and rescued FLAG–CENP-A levels to 57 ± 9% of that of mock-depleted extracts (Fig. 6, A and B). Furthermore, complementation of CENP-C–depleted extracts with myc–CENP-C fully rescued the M18BP1 localization defect (106 ± 21%; Fig. 6, C and D). Our ability to rescue CENP-A assembly and M18BP1 localization by complementation of CENP-C–depleted extracts with myc–CENP-C indicates that these defects are specific to the loss of CENP-C and not other factors. Collectively, these data show that CENP-C plays a critical role in new CENP-A assembly in Xenopus egg extract by promoting the centromeric localization of M18BP1 and/or HJURP.
Figure 6.
CENP-C depletion inhibits M18BP1 targeting to metaphase centromeres and xHJURP-dependent CENP-A assembly. (A) Depletion of CENP-C inhibits xHJURP-dependent CENP-A assembly. Representative images from CENP-A assembly assays were performed using mock- or CENP-C–depleted extracts supplemented with mock or myc–CENP-C IVT protein. The depletion and addback conditions are indicated to the left of the images, and immunolocalized proteins are listed above. (B) Quantification of myc–CENP-A fluorescence intensity at centromeres for CENP-A assembly reactions described in A, which were normalized to the levels in mock-depleted extracts. Quantification was performed as described in Fig. 1. (C) Addback of myc–CENP-C to CENP-C–depleted metaphase extract restores M18BP1 localization to centromeres. Representative images of sperm chromatin incubated in mock- or CENP-C–depleted metaphase extracts supplemented with mock or the myc–CENP-C IVT protein. The depletion and addback conditions are indicated to the left of the images, and immunolocalized proteins are listed above. (D) Quantification of CENP-C and M18BP1 levels in mock- or CENP-C–depleted metaphase extracts that were supplemented with mock or myc–CENP-C protein. Quantification was performed as described in Fig. 1. Error bars show SEM; n = 3. Bars, 10 µm.
CENP-C directly interacts with M18BP1
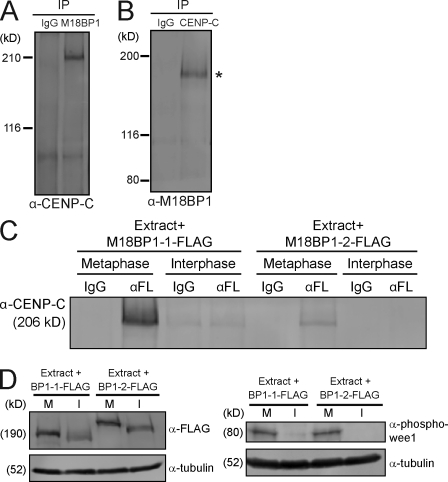
To better understand how CENP-C targets M18BP1 to metaphase centromeres, we tested whether CENP-C and M18BP1 are associated in metaphase extracts. Immunoprecipitation of M18BP1 from egg extracts coprecipitated CENP-C, and immunoprecipitation of CENP-C coprecipitated isoform 1 of M18BP1 (Fig. 7, A and B). We confirmed that the interaction was isoform specific by translating FLAG-tagged versions of xM18BP1-1 and xM18BP1-2 in Xenopus egg extracts, immunoprecipitating the tagged xM18BP1 from the extract, and then Western blotting for CENP-C. We found that M18BP1-1 precipitated CENP-C and that this association was specific to metaphase (Fig. 7, C and D). We also detected coprecipitation of M18BP1-2 and CENP-C from metaphase extracts, but this interaction appeared to be less robust (Fig. 7, C and D) than the interaction between M18BP1-1 and CENP-C given that equal amounts of FLAG-tagged M18BP1 isoforms were present in the extracts (Fig. 7 D). Together, this suggests that CENP-C’s association with M18BP1 in metaphase may recruit M18BP1 to mitotic centromeres.
Figure 7.
CENP-C associates with M18BP1-1 in metaphase extract. (A) M18BP1 coprecipitates CENP-C from Xenopus egg extract. Western blot for CENP-C after immunoprecipitation (IP) of metaphase Xenopus extract with IgG or α-M18BP1. (B) CENP-C coprecipitates M18BP1 from the Xenopus egg extract. Western blot for M18BP1 after immunoprecipitation of metaphase Xenopus extract with IgG or α–CENP-C. We only detected a single M18BP1 isoform (asterisk) coprecipitating with CENP-C, which migrates at the molecular mass of isoform 1. (C) M18BP1-1 associates with CENP-C in metaphase extract. FLAG epitope–tagged M18BP1 IVT proteins were added to metaphase or interphase extracts, reactions were split in half, and M18BP1-FLAG proteins were immunoprecipitated with control (IgG) or α-FLAG (αFL) antibodies. A Western blot of the immunoprecipitates for CENP-C is shown. (D) Representative Western blots from immunoprecipitation experiments described in C. Samples were taken after incubation of M18BP1-FLAG proteins in extract. (left) α-FLAG Western blot; (right) phospho-wee1 Western blot to assess the cell cycle state of the extract. M and I indicate metaphase and interphase extract, respectively. The molecular masses of protein standards and individual proteins in kilodaltons are listed on the left of each blot.
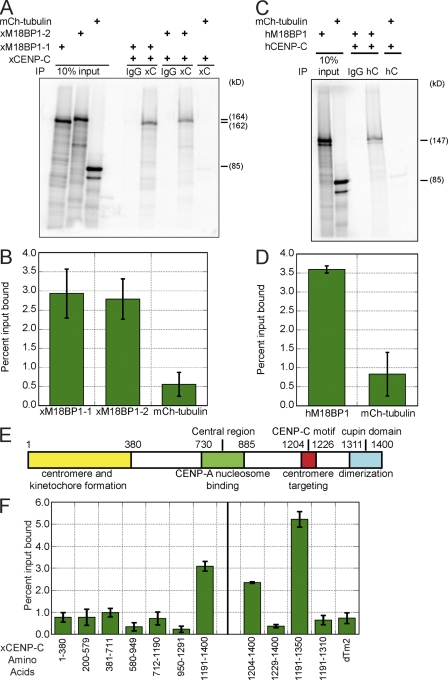
We determined whether CENP-C and M18BP1 bind one another by testing the coprecipitation of the two proteins translated in rabbit reticulocyte extracts in the presence of [35S]methionine. The two isoforms of M18BP1 or mCherry-tubulin as a control were mixed with unlabeled CENP-C and immunoprecipitated with either CENP-C antibody or IgG. We found that both M18BP1-1 and M18BP1-2 specifically associated with CENP-C, indicating that they directly interact (Fig. 8, A and B). This interaction was conserved in humans, as human M18BP1 and CENP-C bind to one another in the same assay (Fig. 8, C and D). Using this same in vitro binding assay, we mapped the region of CENP-C required for interaction with M18BP1-1 (Fig. 8, E and F). We found that a fragment of CENP-C spanning amino acids 1,191–1,350 was sufficient to bind M18BP1-1 (Fig. 8 F). This fragment contains the conserved CENP-C signature motif (residues 1,204–1,226) and the N-terminal half of the conserved cupin domain (residues 1,311–1,397), which mediates dimerization of CENP-C (Fig. 8 E). We found that removal of either the CENP-C motif or the cupin domain prevented the binding of CENP-C to M18BP1-1 (Fig. 8 F). Despite extensive mutagenesis of conserved amino acids throughout the 1,191–1,350 region of CENP-C, we have been unable to isolate a point mutant that disrupts the interaction between M18BP1-1 and CENP-C and yet still localizes normally to centromeres (unpublished data).
Figure 8.
CENP-C directly interacts with M18BP1 through two conserved domains in CENP-C. (A) xCENP-C binds directly to both isoforms of xM18BP1. A representative gel from an xCENP-C–xM18BP1 in vitro binding assay is shown. [35S]methionine-labeled in vitro translated xM18BP1-1 or xM18BP1-2 was mixed with unlabeled in vitro translated xCENP-C and immunoprecipitated with control (IgG) or α–xCENP-C antibodies. Samples were run on an SDS-PAGE gel, and bound xM18BP1 protein was detected by autoradiography. mCherry (mCh)-tubulin was used as a negative control. The addition of each component is shown at the top of the gel. (B) Quantification of the CENP-C–M18BP1 in vitro binding assay represented in A. (C) hCENP-C (hC) binds directly to hM18BP1. Representative gel from an hCENP-C–hM18BP1 in vitro binding assay. [35S]methionine-labeled in vitro translated hM18BP1 was mixed with unlabeled in vitro translated hCENP-C and immunoprecipitated with IgG or α–hCENP-C antibodies. Samples were run on an SDS-PAGE gel, and bound hM18BP1 protein was detected by autoradiography. mCherry-tubulin was used as a negative control. (D) Quantification of hCENP-C–hM18BP1 in vitro binding assay represented in C. (E) Schematic of CENP-C domains. (F) M18BP1 binding to CENP-C requires the CENP-C motif (amino acids 1,204–1,226) and the N-terminal half of the cupin domain (amino acids 1,311–1,397). In vitro binding assays with CENP-C fragments were performed as described in A. Domain mapping of the entire protein is shown to the left of the black vertical line, and fine-scale mapping of the C terminus is shown to the right. Error bars show SEM; n = 3. IP, immunoprecipitation.
Discussion
How centromeres are maintained at the same locus through cell divisions and organismal generations is not well understood. In this study, we developed an in vitro assay for CENP-A chromatin assembly in Xenopus egg extracts and used it to study the mechanisms that recruit the factors required for new CENP-A nucleosome assembly to the centromere. Previous studies have shown that CENP-C binds to CENP-A nucleosomes and is required for CENP-A assembly (Goshima et al., 2007; Erhardt et al., 2008; Carroll et al., 2010). We found that CENP-C is also required for M18BP1 recruitment to metaphase centromeres, and the two proteins interact directly through two highly conserved domains within CENP-C, the CENP-C motif and the cupin domain. In the absence of CENP-C, M18BP1 fails to target to centromeric chromatin, centromeric levels of HJURP are reduced, and new CENP-A nucleosome assembly is inhibited. This suggests that a key function for CENP-C is to recruit factors required for new CENP-A assembly to the existing centromere.
In vitro CENP-A chromatin assembly
The in vitro CENP-A assembly assay described here recapitulates several key aspects of CENP-A assembly in somatic cells. Namely, assembly of epitope-tagged CENP-A in Xenopus extracts is dependent on the CENP-A assembly factors HJURP and M18BP1, requires the exit from mitosis, and is independent of DNA replication (Shelby et al., 2000; Fujita et al., 2007; Jansen et al., 2007; Maddox et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009; Bernad et al., 2011). Our observations on CENP-A assembly using this assay are similar to a recent study (Bernad et al., 2011) with some noteworthy differences. In contrast to the previous study, we observed no effect on myc–CENP-A assembly when we supplemented egg extracts with hHJURP, but we observed a ∼30-fold stimulation of myc–CENP-A assembly when we supplemented the extract with xHJURP. We also found that the assembly of exogenously added CENP-A continued throughout interphase rather than being restricted to a narrow window before replication. The differences we observe may be because there is a limited pool of CENP-A in the extract and augmenting that pool with additional CENP-A allows for CENP-A assembly at any time during interphase.
CENP-C recruits proteins required for new CENP-A assembly to the centromere
We find that CENP-C is required for M18BP1 targeting to metaphase centromeres and that this is likely through direct interaction between the two proteins. M18BP1 depletion does not affect the centromeric localization of CENP-C, indicating that CENP-C functions upstream of M18BP1 in Xenopus egg extract. This result contrasts with observations in C. elegans, in which KNL-2 depletion prevented localization of CeCENP-C to centromeres. However, KNL-2 depletion in C. elegans embryos also caused CeCENP-A loss from centromeric chromatin, which may have in turn prevented CeCENP-C targeting (Maddox et al., 2007). We also find that CENP-C depletion reduces centromeric HJURP levels. M18BP1 depletion leads to a comparable defect in HJURP targeting, and HJURP requires M18BP1 for its localization to centromeres in human cells; thus, it is likely that the role of CENP-C in HJURP localization is through M18BP1 recruitment. However, we cannot exclude the possibility that CENP-C and M18BP1 function in parallel pathways to promote HJURP recruitment to centromeres in Xenopus extract.
We have demonstrated a role for CENP-C in CENP-A assembly in Xenopus egg extracts. Complementation of CENP-C–depleted extracts to 40–60% of the levels in mock-depleted extracts fully restored M18BP1 targeting to centromeres, whereas CENP-A assembly was only rescued to 60% of the levels in control extracts. This suggests that other factors that are dependent on CENP-C may act in concert with M18BP1 to promote CENP-A assembly.
CENP-C interacts directly with M18BP1 in a cell cycle–dependent manner
Our observations have identified a second important function for CENP-C in recruiting M18BP1 to centromeres to promote CENP-A assembly. CENP-C binds directly to M18BP1 in vitro, and this interaction requires the highly conserved CENP-C motif and the N-terminal half of the CENP-C cupin domain. The CENP-C box is an evolutionarily conserved motif found in all CENP-C homologues that is required for CENP-C targeting to centromeres (Meluh and Koshland, 1995; Fukagawa et al., 2001; Heeger et al., 2005; Milks et al., 2009), and the C-terminal cupin domain mediates dimerization of CENP-C (Sugimoto et al., 1997; Cohen et al., 2008). It will be interesting to determine whether CENP-C dimerization is required for the recruitment of M18BP1 to CENP-A chromatin and how the CENP-C box and cupin domain influence CENP-C binding to both CENP-A nucleosomes and M18BP1.
Both isoforms of M18BP1 bind to CENP-C in vitro, but in egg extract, we primarily detect an interaction with M18BP1-1 that is specific to metaphase. This suggests that the interaction between M18BP1-2 and CENP-C is inhibited in extracts and that the interaction between M18BP1-1 and CENP-C is cell cycle regulated. Currently, we do not know whether the interaction between M18BP1 and CENP-C is regulated through CENP-C, M18BP1, or both. Previous studies have demonstrated that CENP-C is regulated differently in mitosis and interphase. In human cells, CENP-C stably associates with mitotic centromeres but is dynamic in interphase (Hemmerich et al., 2008), and in chicken DT40 cells, CENP-C targeting to centromeres depends on CENP-H/I in interphase but not metaphase (Kwon et al., 2007). Understanding the cell cycle–dependent mechanisms that regulate the interaction between M18BP1 and CENP-C is likely to provide important insight into the process of new CENP-A nucleosome assembly.
We have shown that human M18BP1 and CENP-C directly interact, but unlike xM18BP-1, human M18BP1 does not localize to metaphase centromeres. In contrast to xM18BP1-1, purification of M18BP1 from asynchronous HeLa cell extracts did not identify CENP-C as an M18BP1-interacting protein (Fujita et al., 2007; Lagana et al., 2010). The interaction between M18BP1 and CENP-C in human cells may have been below the detection limit in these experiments because the interaction is mitosis specific, and only a fraction of the total M18BP1 in extract is associated with CENP-C. It will be important to understand whether CENP-C plays a role in M18BP1 targeting to human centromeres, whether the human CENP-C–M18BP1 interaction is regulated by the cell cycle, and how this interaction relates to the isoform specific interactions we have described in Xenopus.
Regulation of M18BP1 localization in interphase
In the absence of CENP-C, M18BP1 accumulates at interphase centromeres precociously and to higher levels. This demonstrates a role for CENP-C in regulating the interphase localization of M18BP1 and indicates that there exists a second, CENP-C–independent mechanism for targeting M18BP1 to centromeres. We have previously shown that CENP-K does not localize to centromeres in CENP-C–depleted extract (Milks et al., 2009). Moreover, in human cells, removal of CENP-C causes the loss of multiple CCAN proteins from the centromere, including CENP-H, -I, -K, and -T (Carroll et al., 2010; Gascoigne et al., 2011). Thus, one possibility is that CENP-A chromatin itself recruits M18BP1 to interphase centromeres.
M18BP1 is required for CENP-A assembly during the exit from mitosis, but it is possible that it performs an additional function during interphase. The GTPase-activating protein MgcRacGAP was recently identified as an M18BP1-associated protein that is required for the maintenance of newly assembled CENP-A nucleosomes at interphase centromeres (Lagana et al., 2010). It will be interesting to test whether M18BP1 might function in interphase to recruit MgcRacGAP to stabilize newly assembled CENP-A.
Collectively, our observations demonstrate that CENP-C plays an essential role in CENP-A assembly by localizing M18BP1 and HJURP to centromeres. By directly linking CENP-A nucleosomes with M18BP1, CENP-C may provide a mechanism to reinforce new CENP-A assembly at the site of preexisting centromeres and, thereby, ensure accurate centromere propagation. An important future goal will be to understand how the factors required for new CENP-A assembly function in concert to transfer soluble CENP-A into chromatin. The ability to assemble centromeric chromatin in vitro will provide a valuable tool for dissecting the biochemical and cell cycle regulatory mechanisms that control new CENP-A assembly and ensure faithful centromere propagation.
Materials and methods
Cloning and antibody generation
xHJURP (available from GenBank/EMBL/DDBJ under accession no. HQ148662) and M18BP1 were identified through BLAST (Basic Local Alignment Search Tool) analysis and were amplified from a Xenopus ovary cDNA library by PCR. We identified two separate, but closely related, isoforms of xM18BP1, which we have designated xM18BP1-1 (GenBank accession no. HQ148660) and xM18BP1-2 (GenBank accession no. HQ148661).
For in vitro production of RNA or proteins, genes were cloned into the AscI and PacI sites of modified pCS2+ vectors containing insertions of epitope tag sequences adjacent to the multiple cloning site (MCS). For expression of hM18BP1, xCENP-C, xHJURP, and hHJURP in rabbit reticulocyte lysate, the xCENP-C, xHJURP, and hHJURP sequences were codon optimized for Escherichia coli and rabbit reticulocyte expression by gene synthesis (DNA 2.0). Plasmids used in this study are listed in Table I.
Table I.
Plasmids used in this study
| Plasmid name (ASP) | Construct |
| 451 | pGEX-6P-1 (GE Healthcare) containing a modified MCS for bacterial expression of GST-tagged proteins |
| 1,633 | GST–xM18BP1-2161–415 |
| 1,980 | GST–xM18BP1-1161–375 |
| 1,639 | GST-xHJURP |
| 1,938 | 6His-xHJURP42–194 |
| 447 | pCS2+ containing a modified MCS for IVT of the protein in rabbit reticulocyte lysate or in vitro transcription of RNA |
| 459 | pCS2+ containing 6×myc sequence upstream of the MCS for IVT of the protein in rabbit reticulocyte lysate or in vitro transcription of RNA |
| 1,730 | pCS2+ containing 3×FLAG sequence upstream of the MCS for IVT of the protein in rabbit reticulocyte lysate or in vitro transcription of RNA |
| 1,853 | pCS2+ containing 3×FLAG sequence downstream of the MCS for IVT of protein in rabbit reticulocyte lysate or in vitro transcription of RNA |
| 1,592 | p447-xHJURP for production of xHJURP RNA |
| 1,815 | p447-xHJURP (codon optimized) for production of the xHJURP IVT protein |
| 1,970 | p447-hHJURP (codon optimized) |
| 1,968 | p1853-xHJURP (codon optimized) |
| 1,969 | p1853-hHJURP (codon optimized) |
| 559 | p459–xCENP-A |
| 1,819 | p1730–xCENP-A |
| 1,870 | p1853–xM18BP1-1 |
| 1,871 | p1853–xM18BP1-2 |
| 867 | p459–xCENP-C (codon optimized) |
| 1,330 | p459–xCENP-C amino acids 1–380 (codon optimized) |
| 1,925 | p459–xCENP-C amino acids 200–579 (codon optimized) |
| 1,928 | p459–xCENP-C amino acids 381–711 (codon optimized) |
| 1,926 | p459–xCENP-C amino acids 580–949 (codon optimized) |
| 1,920 | p459–xCENP-C amino acids 712–1,190 (codon optimized) |
| 1,927 | p459–xCENP-C amino acids 950–1,291 (codon optimized) |
| 1,150 | p459–xCENP-C amino acids 1,191–1,400 (codon optimized) |
| 1,943 | p459–xCENP-C amino acids 1,204–1,400 (codon optimized) |
| 1,935 | p459–xCENP-C amino acids 1,229–1,400 (codon optimized) |
| 1,921 | p459–xCENP-C amino acids 1,191–1,350 (codon optimized) |
| 1,936 | p459–xCENP-C amino acids 1,191–1,310 (codon optimized) |
| 866 | p447–xCENP-C (codon optimized) |
| 1,350 | p447–xM18BP1-1 |
| 1,351 | p447–xM18BP1-2 |
| 844 | p447–hCENP-C |
| 1,828 | p447-hM18BP1 (codon optimized) |
| 572 | p447–mCherry-tubulin |
| 1,981 | p459-dTm2 (Drosophila melanogaster tropomyosin 2) |
ASP, A.F. Straight plasmid.
Antibodies to M18BP1 and HJURP were produced in rabbits (Cocalico Biologicals). For xM18BP1 antibody production, a fragment of xM18BP1-2 spanning amino acids 161–415 was expressed as a GST fusion in E. coli and affinity purified on glutathione agarose (Sigma-Aldrich) according to the manufacturer’s instructions. To generate an affinity column for antibody purification, a fragment of xM18BP1-1 spanning amino acids 161–375 was expressed as a GST fusion in E. coli and purified using glutathione agarose. Rhinovirus 3C protease was used to cleave xM18BP1-1161–375 from the GST tag after fusion protein binding to the glutathione agarose column. xM18BP1-1161–375 was further purified by chromatography on S-Sepharose by using a linear gradient from 25 mM to 1 M NaCl in 20 mM MES, pH 6.0, 10% glycerol, and 1 mM DTT. The protein was then coupled to N-hydroxy succinimide–activated Sepharose 4 Fast Flow (GE Healthcare). For xHJURP antibody production, xHJURP was expressed as a GST fusion protein and purified using glutathione agarose. To generate an affinity column for antibody purification, a fragment of xHJURP spanning amino acids 42–194 was expressed with an N-terminal 6His tag in E. coli, affinity purified on nickel–nitrilotriacetic acid agarose (QIAGEN) according to the manufacturer’s instructions, and coupled to N-hydroxy succinimide–activated Sepharose 4 Fast Flow. Other antibodies used in this study that are not commercially available are α–xCENP-A (Maddox et al., 2003), α–xCENP-C (Milks et al., 2009), α-xAnillin (Straight et al., 2005), and α-myc (Milks et al., 2009). The supernatant of 9E10 mouse hybridoma cells was used directly for myc immunofluorescence. The phospho-wee1 antibody was provided by J.E. Ferrell (Stanford University, Stanford, CA). Antibodies used in this study are listed in Table II.
Table II.
Antibodies used in this study
| Antibody | Species | Catalog number | Company | Dilution |
| α-xM18BP1 | Rabbit | A.F. Straight | 1.5 µg/ml (IF); 5 µg/ml O/N (WB) | |
| α-xHJURP | Rabbit | A.F. Straight | 2 µg/ml (WB) | |
| α-myc | Mouse | A.F. Straight | 1 µg/ml (WB) | |
| α–xCENP-A | Rabbit | A.F. Straight | 2 µg/ml (WB); 1 µg/ml (IF) | |
| 594-α–xCENP-A | Rabbit | A.F. Straight | 2 µg/ml (IF) | |
| 488-α–xCENP-A | Rabbit | A.F. Straight | 2 µg/ml (IF) | |
| α-xAnillin | Rabbit | A.F. Straight | 1 µg/ml (WB) | |
| α–xCENP-C | Rabbit | A.F. Straight | 1 µg/ml (IF); 1.5 µg/ml (WB) | |
| α–phospho-wee1 | J.E. Ferrell | 1:1,000 O/N (WB) | ||
| α-tubulin | Mouse | T9026 | Sigma-Aldrich | 0.25 µg/ml (WB) |
| α-BrdU | Mouse | 555627 | BD | 10 µg/ml (IF) |
| α–histone H4 | Rabbit | ab7311 | Abcam | 2 µg/ml (WB) |
| α–histone H2A | Rabbit | ab13923 | Abcam | 2 µg/ml (WB) |
| α-FLAG | Mouse | F1804 | Sigma-Aldrich | 2 µg/ml (IF); 1 µg/ml (WB) |
| 647-anti-myc | Mouse | 2233 | Cell Signaling Technology | 20 µg/ml (IF) |
IF, immunofluorescence; O/N, overnight; WB, Western blot.
Xenopus extracts and tissue culture
Xenopus cytostatic factor (CSF) and interphase extracts and demembranated sperm were prepared as previously described (Desai et al., 1999). In brief, Xenopus eggs were washed in MMR buffer (5 mM Na-Hepes, pH 7.8, 0.1 mM EDTA, 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 2 mM CaCl2) and then subsequently dejellied in MMR + 2% l-cysteine. After dejellying, eggs were washed in CSF-XB buffer (100 mM KCl, 50 mM sucrose, 2 mM MgCl2, 0.1 mM CaCl2, 10 mM K-Hepes, pH 7.7, and 5 mM K-EGTA, pH 7.7) followed by washing in CSF-XB + protease inhibitor buffer (CSF-XB + 10 µg/ml LPC [leupeptin/pepstatin A/chymostatin]). Eggs were then placed in a 13 × 51–mm ultraclear tube (Beckman Coulter) and packed by low-speed spin in a table top clinical centrifuge for 45 s. After removal of excess buffer, eggs were centrifuged in a rotor (SW50.1; Beckman Coulter) for 15 min at 10,000 rpm. The soluble cytoplasmic material was removed from the centrifuge tube and supplemented with energy mix (7.5 mM creatine phosphate, 1 mM ATP, and 1 mM MgCl2), 50 mM sucrose, 10 µg/ml LPC, and 10 µg/ml cytochalasin D. Cycloheximide (Sigma-Aldrich) was added to extracts to 100 µg/ml, BrdU (Sigma-Aldrich) was added to extracts to 40 µM, and aphidicolin (Sigma-Aldrich) was added to extracts to 50 µg/ml. Xenopus tissue-culture (S3) cells were grown in 70% Leibovitz’s L-15 media (Invitrogen) supplemented with 15% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 0.1 mg/ml streptomycin.
CENP-A assembly assay in Xenopus extract
The TnT Sp6 Coupled Rabbit Reticulocyte System (Promega) was used for in vitro transcription/translation (IVT) of plasmid DNA according to the manufacturer’s protocol, except twice the recommended amount of DNA was added. RNA was produced using the either the mMESSAGE mMACHINE SP6 kit (Invitrogen) or the RiboMAX SP6 Large Scale RNA Production System and Ribo m7G cap analogue (Promega), and RNA was added to the extract to a final concentration of 2 µg RNA per 100 µl extract.
For standard CENP-A assembly assays, 2 µl HJURP IVT protein was added per 20 µl assembly reaction. HJURP IVT protein and myc–xCENP-A RNA were added to CSF extract, reactions were incubated for 30 min at 16–20°C to allow for translation of myc–xCENP-A, and then cycloheximide was added to block further translation. Subsequently, demembranated sperm (final concentration of ∼3 × 105 sperm per 100 µl extract) and CaCl2 (final concentration of 0.75 mM) were added, and the reaction was incubated for an additional 60–75 min to allow for release into interphase. Reactions were then processed for immunofluorescence as described under the Immunofluorescence subheading. For experiments to test replication dependence of CENP-A assembly, DMSO or aphidicolin was added concurrent with xHJURP IVT protein and myc–CENP-A RNA. For CENP-A assembly assays using M18BP1-depleted extracts (Fig. 3, C and D), 1 µl HJURP IVT protein was added per 20 µl assembly reaction. For CENP-A assembly assays using CENP-C–depleted extracts (Fig. 6, A and B), HJURP RNA was used instead of the HJURP IVT protein. Protocol adjustments were made so that the volume of IVT lysate added was never >10% of the total reaction volume.
Immunofluorescence
Xenopus S3 cells grown on coverslips were washed with 70% PBS and then fixed for 5 min in 2% formaldehyde/0.5% Triton X-100. Coverslips were then blocked in antibody dilution buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 2% BSA, and 0.1% sodium azide) and stained with α-xM18BP1 and α–CENP-A antibody directly conjugated to the Alexa Fluor 594 fluorophore.
The localization of M18BP1 and CENP-A on demembranated sperm nuclei was assayed by adhering sperm to poly-l-lysine–coated acid-washed coverslips without fixative, treating sperm with 5 µM recombinant Xenopus Nap1 in sperm dilution buffer (10 mM K+ Hepes, pH 7.7, 1 mM MgCl2, 100 mM KCl, and 150 mM sucrose) for 30 min, and then fixing sperm for 5 min using 2% formaldehyde. Sperm chromatin from egg extract experiments were washed in dilution buffer (BRB-80 [80 mM K-Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA], 30% glycerol, 0.5% Triton X-100, and 150 mM KCl) for 5 min, fixed for 5 min using 2% formaldehyde, and spun through 40% glycerol–BRB-80 cushions onto coverslips. For experiments to test for stable incorporation of myc–CENP-A into chromatin, reactions were washed for 5 min in dilution buffer with increasing concentrations of KCl (0 mM–1 M) before fixation. Sperm immunostained for BrdU were postfixed with cold methanol, treated with 2 N HCl/0.5% Triton X-100 to denature DNA, and then neutralized with 0.1 M borate, pH 8.5, before immunostaining. After fixation, coverslips were blocked in antibody dilution buffer and then immunostained with the antibodies indicated in Fig. S1. Alexa Fluor–conjugated secondary antibodies were used according to the manufacturer’s specifications (Invitrogen). When required, coverslips were blocked using 1 mg/ml whole-rabbit IgG or whole-mouse IgG (Jackson ImmunoResearch Laboratories, Inc.).
Immunodepletions
Depletion experiments were performed using affinity-purified antibodies bound to protein A beads (Dynabeads; Invitrogen). For 100 µl extract, 2.5 µg α-xM18BP1 antibody or 0.6 µg α–CENP-C antibody was bound to 33 µl of beads in 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Triton X-100 for 1 h at 4°C. An equivalent amount of whole-rabbit IgG was used for control depletions. The beads were then washed and resuspended in extract for 1 h at 4°C. Beads were removed from the extract by two 5-min rounds of exposure to a magnet.
Complementation experiments in Xenopus extract
For CENP-C addback experiments, 1 µl (Fig. 6, C and D) or 2 µl (Fig. 6, A and B) myc–CENP-C IVT protein was added per 20 µl CENP-C–depleted extract. For M18BP1 addback experiments, 0.5 µl each of M18BP1-1–FLAG and M18BP1-2–FLAG IVT proteins was added per 20 µl M18BP1-depleted extract. For CENP-A assembly assays using depleted extracts, myc–CENP-C and M18BP1-FLAG IVT proteins were added to CSF extract concurrent with the addition of HJURP IVT protein and CENP-A RNA.
Immunoblotting
Samples were separated by SDS-PAGE and transferred onto polyvinylidene fluoride membrane (Bio-Rad Laboratories). Samples were transferred in CAPS transfer buffer (10 mM 3-(cyclohexylamino)-1-propanesulfonic acid, pH 11.3, 0.1% SDS, and 20% methanol) for CENP-A, CENP-C, H2A, and H4 or Tris-glycine transfer buffer (20 mM Tris-HCl and 200 mM glycine) for xHJURP, hHJURP, and M18BP1. Alexa Fluor 488– or Alexa Fluor 647–conjugated goat anti–rabbit or anti–mouse antibodies (1:2,500; Invitrogen) were used for detection by fluorescence using a gel imager (Typhoon 9400 Variable Mode Imager; GE Healthcare). For detection of xM18BP1, a tertiary amplification step was performed using Dylight 649–conjugated bovine anti–goat (1:2,500; Jackson ImmunoResearch Laboratories, Inc.). Fluorescence of detected bands was background corrected and quantified using ImageJ (National Institutes of Health). Background correction was performed by measuring the integrated intensity of a region encompassing the detected band and then subtracting from that the intensity as measured in an identical region below the band in the same lane. For egg extract experiments, 2 µl extract was loaded into each lane.
Isolation of chromatin after CENP-A assembly reactions
CENP-A assembly reactions were prepared as described under the CENP-A assembly assay in Xenopus extract subheading, except reactions were prepared to a final volume of 500 µl. 60 min after calcium addition, assembly reactions were diluted into 2 ml dilution buffer (BRB-80 [80 mM K-Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA], 30% glycerol, 0.5% Triton X-100, and 300 mM KCl) and incubated for 5 min at room temperature. Reactions were then overlaid onto 5 ml of cushion (40% glycerol/80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EDTA) in a 15-ml conical tube and centrifuged in a rotor (JS4.2; Beckman Coulter) for 20 min at 3,500 rpm. After centrifugation, the supernatant was removed, and the chromatin pellets were resuspended in micrococcal nuclease (MNase) buffer (250 mM NaCl, 20 mM Hepes, pH 7.7, 2 mM EDTA, 1% NP-40, 1 mM DTT, 1 mM PMSF, 1 mM benzamidine HCl, and 10 µg/ml LPC). Samples were then centrifuged for 5 min at 5,000 rpm in a microcentrifuge at 4°C. The supernatant was removed, and the chromatin pellets were resuspended in 50 µl MNase buffer supplemented with 4 mM CaCl2 and sonicated for 30 s in a cup sonicator. MNase (Worthington Biochemical Corporation) was then added to a final concentration of 1 U/µl and incubated at 37°C for 15 min. After 15 min, EDTA was added to 10 mM, and the samples were sonicated for 30 s. After sonication, the samples were diluted with 50 µl of 4× sample buffer (200 mM Tris, pH 6.8, 40 mM EDTA, 0.05% bromophenol blue, 10% SDS, and 50% glycerol) to a final volume of ∼100 µl. For Western blots, 30 µl of each sample was loaded per lane.
Microscopy
All coverslips were mounted onto glass slides with mounting media (0.5% p-phenylenediamine, 20 mM Tris, pH 8.8, and 90% glycerol) and sealed with clear nail polish. Immunofluorescence microscopy images were acquired at room temperature using a 60× 1.4 NA Plan Apochromat VC oil immersion lens (Nikon), a Sedat quad filter set (Chroma Technology Corp.), and a charge-coupled device camera (CoolSNAP HQ; Photometrics) mounted on a microscope (Eclipse 80i; Nikon). Wavelength selection was performed with a Lambda 10–3 controller and 25-mm high-speed filter wheels (Sutter Instrument). Z sections were acquired with a z-axis drive (MFC2000; Applied Scientific Instrumentation) at 0.2-µm intervals. Microscope instrumentation was controlled via Metamorph imaging software (Molecular Devices). Immunofluorescence microscopy images were also acquired at room temperature using a 60× 1.4 NA Plan Apochromat oil immersion lens (Olympus), a Sedat quad filter set (Semrock, Inc.), and a charge-coupled device camera (CoolSNAP HQ) mounted on a microscope (IX70; Olympus) outfitted with a Deltavision Core system (Applied Precision). Microscope instrumentation was controlled via softWoRx 4.1.0 software (Applied Precision). Maximum intensity projections were generated from the z sections and then analyzed with custom-written software.
Images of Xenopus S3 cells were deconvolved using softWoRx 4.1.0. All images were postprocessed in Photoshop (Adobe) after data analysis for display purposes. Images were cropped and contrast adjusted. No changes to image γ were performed.
Quantification of myc–CENP-A loading percentage
To quantify the proportion of centromeres that had loaded myc–CENP-A, we fit a Gaussian distribution to a histogram of the background subtracted per pixel intensity values at every centromere in the no-HJURP condition (best fit parameters: mean = 21.8, standard deviation = 52.39, and R2 = 0.99). We then chose a cutoff for myc–CENP-A loading as the background-subtracted intensity value, in which the cumulative distribution function of the fitted Gaussian was equal to 0.992 (corresponding to a false positive rate of 1 in 125). For each condition and each experiment, we totaled the number of centromeres whose intensity was above this cutoff and divided by the total number of centromeres counted in that condition and experiment to determine the proportion of centromeres loading myc–CENP-A. At least 250 centromeres were counted in each condition in each experiment.
Image analysis
An automated image analysis was performed using custom-written software in C++. Source code and documentation for all programs used in this study are available under the Mozilla Public License as previously described (Fuller and Straight, 2010). The analysis code consists of four steps: image normalization, centromere finding, cell clustering, and image quantification.
In brief, images were normalized by median filtering the image and then dividing the original image by the filtered intensity value. Centromeres were found using Otsu’s thresholding method (Otsu, 1979) modified to recursively divide regions larger than centromeres (Xiong et al., 2006) and then size filtered to remove objects much larger or smaller than centromeres. Centromeres were grouped into cells using Gaussian mixture model clustering (McLachlan and Basford, 1988). The initial guess for clustering the image was made by first heavily Gaussian blurring the image, thresholding the image for nonzero pixel intensities, and using the resulting regions as a first cluster approximation. These initial clusters were then refined by randomly subdividing the image and then iteratively Gaussian mixture model clustering until no new successful subdivisions were made. Centromere intensities were then quantified in the original image, and the mean over the entire image was calculated. The images were manually examined to exclude misidentified centromeres, but the automated analysis was accurate enough that the manual examination did not measurably change the results. The validity of the segmentation method was verified by manual analysis of a subset of images (in which centromeres were identified by drawing circles around them). No significant difference between the two methods could be detected.
Online supplemental material
Fig. S1 characterizes the properties of xHJURP-mediated CENP-A assembly in the Xenopus egg extract. Fig. S2 shows the localization of xHJURP to interphase centromeres and its ability to stimulate myc–CENP-A assembly. Fig. S3 shows xHJURP’s ability to promote the assembly of the endogenous CENP-A present in extract. Fig. S4 characterizes the cell cycle requirements for CENP-A assembly. Fig. S5 provides additional characterization of M18BP1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106079/DC1.
Acknowledgments
The authors would like to thank the Straight Laboratory members for support and helpful comments and James E. Ferrell for reagents.
B. Moree was supported by T32GM007276, C.B. Meyer was supported by grants from the National Science Foundation and the National Defense Science and Engineering Graduate Fellowship Program, and C.J. Fuller was supported by a Stanford Graduate Fellowship. This work was supported by National Institutes of Health R01GM074728 to A.F. Straight.
Footnotes
Abbreviations used in this paper:
- CCAN
- constitutive centromere-associated network
- CSF
- cytostatic factor
- hHJURP
- human HJURP
- IVT
- in vitro transcription/translation
- MCS
- multiple cloning site
- MNase
- micrococcal nuclease
- xHJURP
- Xenopus HJURP
References
- Allshire R.C., Karpen G.H. 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9:923–937 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M., Suzuki A., Hori T., Backer C., Okawa K., Cheeseman I.M., Fukagawa T. 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 186:173–182 10.1083/jcb.200903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart M.C., Kuich P.H., Stellfox M.E., Ward J.A., Bassett E.A., Black B.E., Foltz D.R. 2011. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194:229–243 10.1083/jcb.201012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry A.E., Howman E.V., Cancilla M.R., Saffery R., Choo K.H. 1999. Sequence analysis of an 80 kb human neocentromere. Hum. Mol. Genet. 8:217–227 10.1093/hmg/8.2.217 [DOI] [PubMed] [Google Scholar]
- Bernad R., Sánchez P., Rivera T., Rodríguez-Corsino M., Boyarchuk E., Vassias I., Ray-Gallet D., Arnaoutov A., Dasso M., Almouzni G., Losada A. 2011. Xenopus HJURP and condensin II are required for CENP-A assembly. J. Cell Biol. 192:569–582 10.1083/jcb.201005136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R., Li B., Florens L., Swanson S.K., Washburn M.P., Gerton J.L. 2007. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 26:853–865 10.1016/j.molcel.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Carroll C.W., Straight A.F. 2006. Centromere formation: from epigenetics to self-assembly. Trends Cell Biol. 16:70–78 10.1016/j.tcb.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C., Godek K.M., Jansen L.E., Straight A.F. 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11:896–902 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., Straight A.F. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Baker R.E., Keith K.C., Harris K., Stoler S., Fitzgerald-Hayes M. 2000. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 20:7037–7048 10.1128/MCB.20.18.7037-7048.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Mao Y., Sullivan K.F. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421 10.1016/S0092-8674(03)00115-6 [DOI] [PubMed] [Google Scholar]
- Cohen R.L., Espelin C.W., De Wulf P., Sorger P.K., Harrison S.C., Simons K.T. 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 19:4480–4491 10.1091/mbc.E08-03-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Deacon H.W., Walczak C.E., Mitchison T.J. 1997. A method that allows the assembly of kinetochore components onto chromosomes condensed in clarified Xenopus egg extracts. Proc. Natl. Acad. Sci. USA. 94:12378–12383 10.1073/pnas.94.23.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T.J., Walczak C.E. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412 10.1016/S0091-679X(08)61991-3 [DOI] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 137:485–497 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Erhardt S., Mellone B.G., Betts C.M., Zhang W., Karpen G.H., Straight A.F. 2008. Genome-wide analysis reveals a cell cycle–dependent mechanism controlling centromere propagation. J. Cell Biol. 183:805–818 10.1083/jcb.200806038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., III, Cleveland D.W. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R., III, Bassett E.A., Wood S., Black B.E., Cleveland D.W. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 137:472–484 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. 2007. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 12:17–30 10.1016/j.devcel.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Regnier V., Ikemura T. 2001. Creation and characterization of temperature-sensitive CENP-C mutants in vertebrate cells. Nucleic Acids Res. 29:3796–3803 10.1093/nar/29.18.3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller C.J., Straight A.F. 2010. Image analysis benchmarking methods for high-content screen design. J. Microsc. 238:145–161 10.1111/j.1365-2818.2009.03337.x [DOI] [PubMed] [Google Scholar]
- Furuyama T., Dalal Y., Henikoff S. 2006. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl. Acad. Sci. USA. 103:6172–6177 10.1073/pnas.0601686103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., Cheeseman I.M. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., Stuurman N. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi C., Ono Y., Doi N., Kitamura F., Tagami M., Mineki R., Arai T., Taguchi H., Yanagida M., Hirner S., et al. 2008. Multiple molecular interactions implicate the connectin/titin N2A region as a modulating scaffold for p94/calpain 3 activity in skeletal muscle. J. Biol. Chem. 283:14801–14814 10.1074/jbc.M708262200 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 118:715–729 10.1016/j.cell.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Heeger S., Leismann O., Schittenhelm R., Schraidt O., Heidmann S., Lehner C.F. 2005. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 19:2041–2053 10.1101/gad.347805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., Diekmann S. 2008. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180:1101–1114 10.1083/jcb.200710052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Jansen L.E., Black B.E., Foltz D.R., Cleveland D.W. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., Fukagawa T. 2007. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 18:2155–2168 10.1091/mbc.E07-01-0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana A., Dorn J.F., De Rop V., Ladouceur A.M., Maddox A.S., Maddox P.S. 2010. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat. Cell Biol. 12:1186–1193 10.1038/ncb2129 [DOI] [PubMed] [Google Scholar]
- Maddox P., Straight A., Coughlin P., Mitchison T.J., Salmon E.D. 2003. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162:377–382 10.1083/jcb.200301088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Hyndman F., Monen J., Oegema K., Desai A. 2007. Functional genomics identifies a Myb domain–containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176:757–763 10.1083/jcb.200701065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S.E., Borusu S., Amaro A.C., Winter J.R., Belwal M., McAinsh A.D., Meraldi P. 2007. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 26:5033–5047 10.1038/sj.emboj.7601927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan G.J., Basford K.E. 1988. Mixture Models: Inference and Applications to Clustering. Dekker M., New York: 253 pp [Google Scholar]
- Meluh P.B., Koshland D. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 6:793–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks K.J., Moree B., Straight A.F. 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell. 20:4246–4255 10.1091/mbc.E09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Xiao H., Wisniewski J., Smith M.M., Wu C. 2007. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 129:1153–1164 10.1016/j.cell.2007.04.026 [DOI] [PubMed] [Google Scholar]
- Otsu N. 1979. Threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9:62–66 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Palmer D.K., O’Day K., Wener M.H., Andrews B.S., Margolis R.L. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805–815 10.1083/jcb.104.4.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D.K., O’Day K., Trong H.L., Charbonneau H., Margolis R.L. 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA. 88:3734–3738 10.1073/pnas.88.9.3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L., Choi E.S., Abbott J.K., Liu X., Kagansky A., Castillo A.G., Hamilton G.L., Richardson W., Rappsilber J., He X., Allshire R.C. 2009. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell. 33:299–311 10.1016/j.molcel.2009.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. 1998. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8:310–318 10.1016/S0962-8924(98)01299-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L., Pidoux A.L., Ponting C.P., Allshire R.C. 2009. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 137:1173–1174 10.1016/j.cell.2009.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., Lehner C.F., Heidmann S. 2007. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 17:237–243 10.1016/j.cub.2006.11.051 [DOI] [PubMed] [Google Scholar]
- Shelby R.D., Monier K., Sullivan K.F. 2000. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151:1113–1118 10.1083/jcb.151.5.1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.C., Jansen L.E. 2009. At the right place at the right time: novel CENP-A binding proteins shed light on centromere assembly. Chromosoma. 118:567–574 10.1007/s00412-009-0227-3 [DOI] [PubMed] [Google Scholar]
- Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., Baker R.E. 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. USA. 104:10571–10576 10.1073/pnas.0703178104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Field C.M., Mitchison T.J. 2005. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol. Biol. Cell. 16:193–201 10.1091/mbc.E04-08-0758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Kuriyama K., Shibata A., Himeno M. 1997. Characterization of internal DNA-binding and C-terminal dimerization domains of human centromere/kinetochore autoantigen CENP-C in vitro: role of DNA-binding and self-associating activities in kinetochore organization. Chromosome Res. 5:132–141 10.1023/A:1018422325569 [DOI] [PubMed] [Google Scholar]
- Sullivan K.F., Hechenberger M., Masri K. 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127:581–592 10.1083/jcb.127.3.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voullaire L.E., Slater H.R., Petrovic V., Choo K.H. 1993. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am. J. Hum. Genet. 52:1153–1163 [PMC free article] [PubMed] [Google Scholar]
- Williams J.S., Hayashi T., Yanagida M., Russell P. 2009. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell. 33:287–298 10.1016/j.molcel.2009.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G., Zhou X., Ji L., Bradley P., Perrimon N., Wong S. 2006. Segmentation of Drosophila RNAi fluorescence images using level sets. IEEE International Conference on Image Processing. 13: 73–76 [Google Scholar]