Background: ATP synthase is a molecular turbine powered by the transmembrane proton flow.
Results: A molecular fusion of stator subunit a with the rotor subunit c incorporates into the ATP synthase and tethers the rotor to the stator.
Conclusion: Interaction of subunit a with monomeric subunit c is a key step in ATP synthase assembly.
Significance: The a/c fusion is a valid structural model of ATP synthase proton channel.
Keywords: ATP Synthase, Membrane Proteins, Membrane Transport, Protein Assembly, Protein Design
Abstract
Subunit a is the main part of the membrane stator of the ATP synthase molecular turbine. Subunit c is the building block of the membrane rotor. We have generated two molecular fusions of a and c subunits with different orientations of the helical hairpin of subunit c. The a/c fusion protein with correct orientation of transmembrane helices was inserted into the membrane, and co-incorporated into the F0 complex of ATP synthase with wild type subunit c. The fused c subunit was incorporated into the c-ring tethering the ATP synthase rotor to the stator. The a/c fusion with incorrect orientation of the c-helices required wild type subunit c for insertion into the membrane. In this case, the fused c subunit remained on the periphery of the c-ring and did not interfere with rotor movement. Wild type subunit a inserted into the membrane equally well with wild type subunit c and c-ring assembly mutants that remained monomeric in the membrane. These results show that interaction with monomeric subunit c triggers insertion of subunit a into the membrane, and initiates formation of the a-c complex, the ion-translocating module of the ATP synthase. Correct assembly of the ATP synthase incorporating topologically correct fusion of subunits a and c validates using this model protein for high resolution structural studies of the ATP synthase proton channel.
Introduction
ATP synthase catalyzes formation of ATP from ADP and inorganic phosphate at the expense of energy stored in the transmembrane gradient of protons or Na+ ions. In Escherichia coli and many other bacteria, the reaction is reversible under physiological conditions and can build up transmembrane ion gradients at the expense of ATP hydrolysis. ATP synthase is composed of the cytoplasmic F1 complex and the F0 complex, which is integrated into the cell membrane (1). ATP synthesis or hydrolysis takes place in active sites located in the F1 complex. This process is coupled to transmembrane ion translocation through the F0 complex.
In E. coli, the F0 complex consists of three types of subunits present in an ab2c10 ratio. ATP synthase functions as a molecular turbine. The transmembrane flow of protons through the F0 complex drives rotation of the subunit c oligomer (2), which is transmitted inside the core of the F1 complex by an elongated shaft built of F1 subunits γ and ϵ. The tip of subunit γ rotating inside the molecular bearing created by three pairs of symmetrically arranged α- and β-subunits causes cyclical conformational changes in the three nucleotide binding sites on the β subunits, which lead first to tight binding of ADP and phosphate and then to the release of the newly synthesized ATP molecules (3).
The complete structure of the F0 complex is still unknown, but x-ray structures of the rotor components of the F0 from Ilyobacter tartaricus (4), Spirulina platensis (5), and Bacillus firmus (6) show multiple copies of the hairpin-shaped subunit c arranged in a cylinder, with the N-terminal α-helix of each subunit located in the inner ring, and the C-terminal helix, in the outer ring. This architecture is consistent with models of the subunit c oligomer (7) and the a-c complex (8) of E. coli ATP synthase built on the basis of the NMR structure of monomeric subunit c (9) and intersubunit Cys-Cys cross-linking (10, 11). Subunit a is predicted to form five membrane-spanning α-helices (12) with C-terminal helices IV and V located at the contact surface between subunit a and the subunit c oligomer (10, 13). Although the three-dimensional structure of subunit a is unknown, the location of transmembrane segments in the primary sequence and packing of the helices has been deduced from a large body of biochemical and genetic data (14–17). The proton channel of ATP synthase is believed to lie at the interface of subunits a and c (13, 18, 19).
Insertion of the F0 subunits into the membrane has been investigated previously, but little is known about the assembly process of the ab2c10 complex. Insertion of subunit c into the membrane is independent of the Sec pathway and requires only the YidC chaperone protein (20). Insertion of subunits a and b requires participation of the Sec system and the signal recognition particle (21). The YidC chaperone is also involved in insertion of subunit a (22). In E. coli, subunits b and c were shown to insert into the membrane independently of the other two subunits of the F0 complex, but both b and c were required for incorporation of subunit a into the membrane (23). This is in contrast to earlier reports that found that subunit a inserts independently of b and c (24). Subunit c overproduced in the absence of the other two F0 subunits may spontaneously assemble into stable ring structures (25). Formation of the c-ring in the membrane, was reported to be assisted by the atpI gene product, a small hydrophobic protein, which is not a stoichiometric component of the mature ATP synthase complex (26, 27), although this protein is not strictly required for the assembly of functional ATP synthase in E. coli cells (28). In mitochondria, assembly of the c-ring, and its association with F1 is believed to precede attachment of subunit a (subunit 6) to the rotor (29). To determine the sequence of events in formation of the F0 complex and develop an experimental model for high-resolution structure determination of the ATP synthase proton channel, we have investigated assembly of ATP synthase incorporating fusion proteins consisting of subunits a and c.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
Plasmid pBWU13 (30) encoding the complete atp operon with the atpB gene (subunit a) modified to include an N-terminal His6 tag (31) was used to generate molecular fusions between subunits a and c (Fig. 1A) and for subsequent protein expression. In the first fusion protein, named a-c, the N terminus of subunit c (gene atpE) is connected to the C terminus of subunit a (gene atpB) by a linker peptide, GTASASNGASA, which is based on the sequence of linker peptides used previously to generate functional dimers of subunit c (32). To generate the a-c protein, a derivative of the pBWU13 plasmid containing atpIBEFHA genes only was amplified by around-the-plasmid PCR with 5′-phosphorylated primers. The first primer was designed to anneal to the 3′ terminus of the coding strand of atpB and included a noncomplementary 5′-sequence corresponding to the N-terminal half of the linker peptide, which also contained a KpnI site for the next cloning step. The second primer annealed to the 3′ terminus of the noncoding strand of the atpE gene and included a noncomplementary 5′ sequence corresponding to the C-terminal half of the linker peptide. Ligation of the purified PCR product resulted in formation of a plasmid containing an engineered gene encoding the a-c fusion protein instead of the separate atpB and atpE genes.
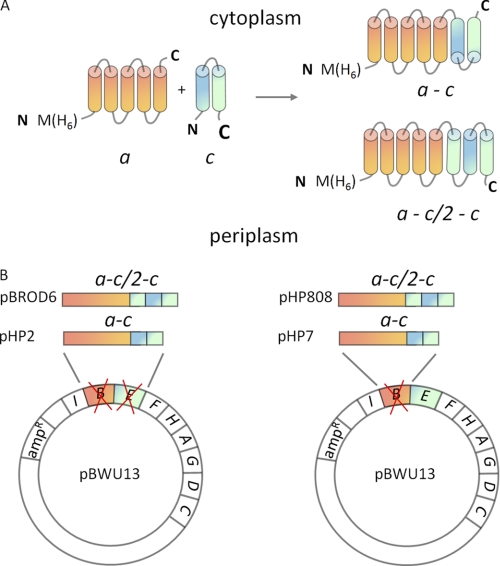
FIGURE 1.
Design of the molecular fusions of subunits a and c and plasmid construction. A, membrane topology of subunits a (orange) and c (helix I, blue; helix II, green) in the wild type ATP synthase (left) and predicted topology of the a-c and a-c/2-c fusion proteins (right). B, the engineered genes coding for the a-c/2-c and a-c fusion proteins were cloned into pBWU13 to replace either both atpB and atpE genes (plasmids pBROD6 and pHP2, respectively), or atpB only (plasmids pHP808 and pHP7, respectively).
In the second fusion protein, named a-c/2-c, a GT dipeptide followed by the sequence corresponding to the polar loop and the C-terminal transmembrane helix of subunit c (Gly38-Ala79) was inserted between the C terminus of subunit a and linker peptide GTASASNGASA of the a-c protein. This gene fusion was generated by amplifying the corresponding segment of the atpE gene by PCR and cloning it into the KpnI site introduced during the construction of the a-c gene at the previous step. The genes coding for the a-c and a-c/2-c fusion proteins were then cloned into the pBWU13 plasmid (Fig. 1B).
In the first set of plasmids, either a-c or a-c/2-c gene fusions replaced both atpB and atpE genes. A fragment of the atp operon containing the respective gene fusion was cloned into HindIII-SphI sites of pBWU13 to produce plasmids containing the a-c (pHP2) and a-c/2-c fusions (pBROD6) in the full operon context. In the second set of plasmids, genes coding for the fusion proteins only replaced the atpB gene, whereas wild type atpE was present on the plasmid in addition to the gene fusion. To generate these plasmids, pBWU13 was amplified by around-the-plasmid PCR with 5′-phosphorylated primers. The forward primer was designed to anneal at the 3′ end of the atpB gene and contained an NheI site immediately after the atpB stop codon. The reverse primer annealed 40 bases downstream of the unique PflMI site in atpB. The linear PCR product, which contained the full plasmid sequence except for the larger part of the atpB gene, was digested with NheI and PflMI and ligated with NheI-PflMI fragments of the a-c and a-c/2-c fusion genes yielding plasmids pHP7 and pHP808, respectively. The plasmids were then transformed into a recA− derivative of strain OM202 carrying complete deletion of the atp operon on the chromosome.
Membrane Preparation, Protein Purification, and Analytical Methods
The E. coli cell cultures were grown on a minimal medium to the late exponential phase (33). Cells were processed in a cell disruptor (Constant Systems Inc., Kennesaw, GA) at 35,000 p.s.i., and the cell membranes were isolated from the homogenate by ultracentrifugation (34). Oxidative phosphorylation activity was tested by growing cells on minimal agar plates containing 0.6% succinate as the sole source of carbon. ATP-dependent proton translocation was monitored by 9-amino-6-chloro-4-methoxyacridine fluorescence quenching (35) in a buffer containing 20 mm Tricine2-NaOH, pH 8.0, 10 mm MgCl2, and 300 mm KCl, with excitation at 410 nm and emission at 490 nm. Following SDS-gel electrophoresis (36), subunit a and the a/c fusion proteins were detected by Western blotting using monoclonal antibodies directed against the pentahistidine epitope (Qiagen). The F0 complex of ATP synthase was purified as described previously by Schneider and Altendorf (37). ATPase activity was measured by liberation of inorganic phosphate (38).
Modeling the a-c/2-c Protein Fold
Modeling was performed with the CNS software package (39, 40) using simulated annealing by restrained molecular dynamics in Cartesian space. The α-helices were defined by imposing dihedral angle and hydrogen-bond constraints based on secondary structure prediction (41) and NMR data for subunit a (42) and high-resolution structures (4, 9) for subunit c. Long range distance restraints between the helices were derived from the Cys-Cys cross-linking studies in the cell membrane (10, 13, 15, 32) as described previously (7), from several second site suppressor mutations in subunit a (7, 12, 43–45), and from NMR paramagnetic relaxation enhancement data (14).
RESULTS
Fusion of Subunits a and c with Correct Topology Inserts into the Membrane
Consistent with previous work (data not shown), we found expression of subunit a without subunit c toxic for the cells. Subunit a requires subunit c for stable insertion into the membrane, whereas subunit c inserts in the absence of the other two F0 subunits (23). In the absence of the partner subunits, subunit a is rapidly degraded by membrane protease FtsH (46, 47). Is interaction with the monomeric c subunit sufficient, or is an assembled c-ring required for membrane insertion of subunit a? To address this question we investigated insertion of fusion proteins constructed of a and c polypeptides, expressed in the context of the complete atp operon, with and without wild type subunit c.
Orientation of ATP synthase subunit c in the membrane is known, with both the N and C termini of the helical hairpin located in the periplasm. Subunit a is believed to have five transmembrane helices with the C terminus located in the cytoplasm (12), although this topology remains to be confirmed by high resolution structure determination. We have generated two polypeptide fusions linking subunit c to subunit a in a head-to-tail fashion. The two fusion proteins have a different number of transmembrane helices to account for the two possible orientations of the C terminus of subunit a relative to the N terminus of subunit c in the cell membrane. In the a-c fusion protein, the N terminus of subunit c is connected to the C terminus of subunit a through a flexible linker, which has a high content of polar amino acids and is expected to partition into the aqueous phase. Consequently, the N and C termini of the a-c fusion protein, which is predicted to contain seven transmembrane helices, should be located on the opposite sides of the cell membrane. This is different from the transmembrane topology predicted for subunits a and c in the native ATP synthase, where both the N terminus of subunit a and the C terminus of subunit c are located in the periplasm. The a-c/2-c fusion protein contains an additional segment corresponding to the polar loop and transmembrane helix II of subunit c inserted between the C terminus of subunit a and the N terminus of the full-length copy of subunit c. This construct is predicted to have eight transmembrane helices and should assume the same transmembrane topology as subunits a and c in the native ATP synthase (Fig. 1A).
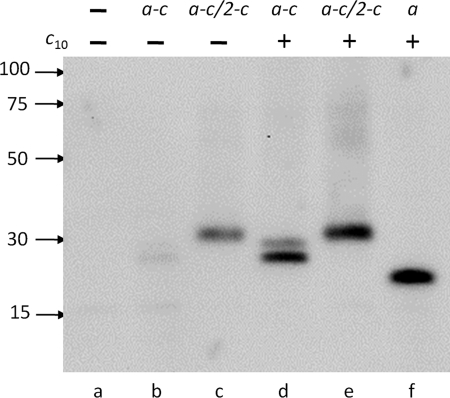
If interaction with monomeric subunit c is sufficient for subunit a insertion, the a-c/2-c fusion should insert into the membrane without wild type subunit c. Indeed, in a-c/2-c, the fusion protein was found in membranes of the strain containing plasmid pBROD6, which lacked the wild type atpE (subunit c) gene (Fig. 2, lane c), although the amount of protein in the membrane was somewhat reduced compared with the strain expressing the wild type atp operon (no a and c fusions, Fig. 2, lane f). In contrast, the a-c fusion protein, which is predicted to have incorrect membrane topology, was not found in the membrane without the wild type subunit c (plasmid pHP2, Fig. 2, lane b). Remarkably, both fusion proteins were readily incorporated into the cell membrane when the respective fusion proteins were coexpressed with wild type subunit c (plasmids pHP7 and pHP808, Fig. 2, lanes d and e). The amounts of fusion proteins found in the membrane in this case were similar to that of wild type subunit a expressed from the parent full operon plasmid pBWU13 (Fig. 2, lane f) as estimated from band intensity on Western blots. These results indicated that monomeric subunit c can chaperone subunit a insertion into the membrane, and an assembled ring is not required for this process, but the c subunit has to assume correct membrane topology. Although the incorrectly attached c subunit was not able to guide subunit a insertion in the case of a-c fusion, it did not interfere with the chaperone function of the wild type subunit c.
FIGURE 2.
Incorporation of a and c subunit fusions into the cell membrane. Western blot of the cell membranes from OM202recA− strains containing no plasmid (a), expressing the a-c fusion protein from plasmids pHP2 (b) or pHP7 (d), the a-c/2-c fusion protein from plasmids pBROD6 (c) or pHP808 (e), or wild type subunit a from plasmid pBWU13 (f) with (d-f) or without (a-c) coexpression of wild type subunit c. Antibody directed against the pentahistidine epitope was used for detection. Each lane contained 10 μg of total membrane protein. Positions of molecular mass markers and their masses in kDa are shown on the left.
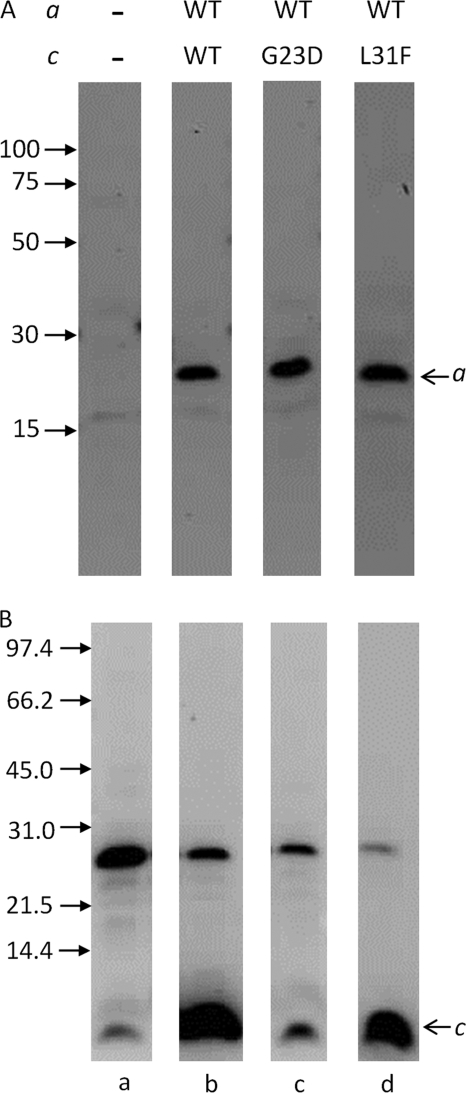
We then investigated insertion of wild type subunit a in the presence of subunit c mutants G23D and L31F, which insert into the membrane but do not assemble into the oligomeric rings (48, 49). Consistent with our previous conclusion, wild type subunit a was present in similar amounts in the membranes from strains expressing wild type subunit c, and cG23D or cL31F mutants (Fig. 3A), confirming that subunit c oligomerization is not required for subunit a insertion. In our experiments, the G23D mutation affected not only the c-ring assembly, but also insertion of subunit c into the membrane, as the amount of G23D subunit c in the membrane was much lower than of the wild type. The L31F mutant, on the other hand, was present in the membranes at wild type levels (Fig. 3B). As expected, both cG23D and cL31F mutants had little or no ATP-driven H+ translocation activity (supplemental Fig. S1, B and C). Importantly, even the comparatively low amount of G23D mutant present in the membrane was sufficient for normal insertion of subunit a, further supporting the notion that monomeric subunit c rather than the c10-oligomer is required for subunit a insertion.
FIGURE 3.
The c10-ring is not required for subunit a insertion into the membrane. Western blot of cell membranes from strains expressing no F0 subunits (a), wild type (wt) subunit a (b–d) with the wild type subunit c (b), G23D mutant of subunit c (c), or L31F mutant of subunit c (d) probed with antibody directed against the pentahistidine tag (A) or anti-subunit c antibody (B). The band in panel B at ∼25 kDa is an artifact commonly observed with this antibody.
ATP Synthase Incorporating the Fused a and c Subunits Is Assembled Correctly
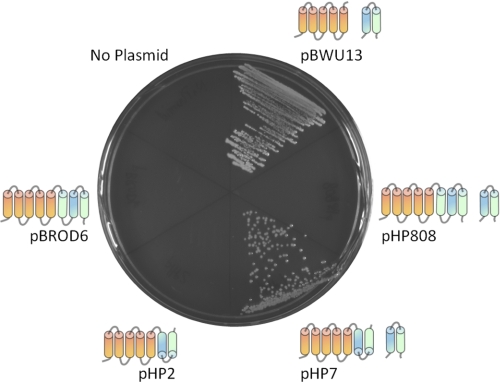
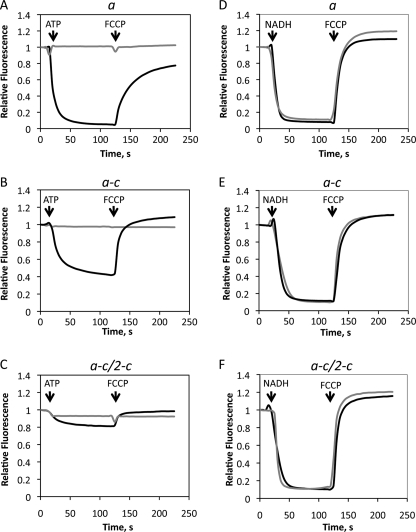
The OM202recA− cells, which have complete atp operon deletion on the chromosome, were able to grow by oxidative phosphorylation when the a-c, but not the a-c/2-c fusion protein was co-expressed with wild type subunit c from plasmids pHP7 and pHP808, respectively. As expected, the OM202recA−/pBROD6 and OM202recA−/pHP2 strains, which lack the wild type atpE gene, and therefore do not produce the c10-oligomer, were unable to grow by oxidative phosphorylation (Fig. 4), although growth on glucose minimal medium was normal (not shown). The a-c protein supported proton translocation driven by ATP hydrolysis, which was inhibited by dicyclohexylcarbodiimide (DCCD), a specific inhibitor of proton transport in ATP synthase, although the rate of H+-transport was lower than in the wild type (Fig. 5, A and B). In contrast, the rate of ATP-dependent H+-translocation with the a-c/2-c fusion protein was negligible (Fig. 5C). Lack of H+ accumulation in vesicles with the a-c/2-c protein reflects a drastic decrease in the rate of proton pumping coupled to ATP hydrolysis, and not increased proton leak across the membrane, because NADH-dependent H+ transport rates with both fusion constructs were similar to the wild type (Fig. 5, D–F).
FIGURE 4.
The a-c fusion protein supports growth by oxidative phosphorylation, but the a-c/2-c does not. The OM202recA− strains expressing wild type subunits a and c (pBWU13), or the a-c and a-c/2-c fusion proteins with (pHP7, pHP808) or without (pHP2, pBROD6) wild type subunit c were plated on succinate agar plates. Growth after 24 h at 37 °C is shown. The a (orange) and c (helix I, blue; helix II, green) polypeptides expressed in the respective strains are shown next to the corresponding sectors of the plate.
FIGURE 5.
The a-c fusion protein supports ATP-dependent proton translocation, but the a-c/2-c does not. ATP-driven (A–C) and NADH-driven (D–F) proton translocation was measured by 9-amino-6-chloro-4-methoxyacridine fluorescence quenching in the membrane vesicles (50 μg/ml of total membrane protein) with wild type subunit a from strain OM202recA−/pBWU13 (A and D), with the a-c fusion from strain OM202recA−/pHP7 (B and E), and with the a-c/2-c fusion from strain OM202recA−/pHP808 (C and F). Additions of 2.0 mm ATP or 0.2 mm NADH and 2.0 μm proton uncoupler carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) are indicated by the arrows. Gray traces were recorded with membranes pretreated with 80 μm DCCD.
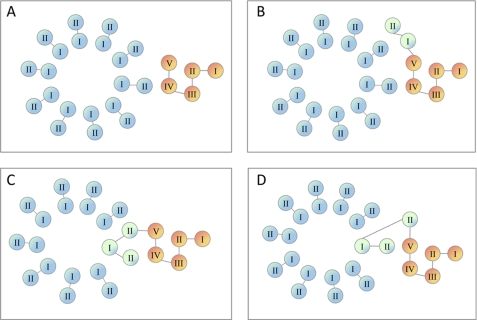
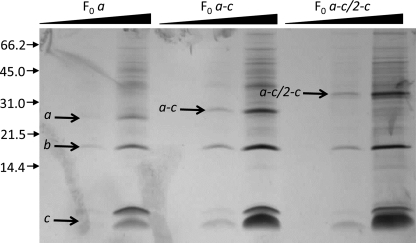
In the a-c protein, the c-polypeptide is fused to subunit a in the incorrect orientation, and therefore cannot be incorporated into the rotor. Because ATP synthase with the a-c fusion is active, the fused c-polypeptide must be displaced to the periphery of the ring, so that it does not interfere with movement of the rotor (Fig. 6, A and B). In the a-c/2-c protein the transmembrane helices of subunit c are in the correct orientation, and can be incorporated into the rotor together with wild type c-subunits. This would tether the rotor to the stator and produce an inactive enzyme (Fig. 6, C or D), explaining the lack of ATP-dependent H+-transport observed with the a-c/2-c protein. To further test this hypothesis, we purified the F0 complex from the membranes of strains expressing wild type subunit a, the a-c, and a-c/2-c fusion proteins as described by Schneider and Altendorf (37). Both fusion proteins were found in F0 complex preparations (Fig. 7), and the yield and ratio of the F0 subunits in these preparations were similar to the wild type.
FIGURE 6.
Models of a/c fusion proteins incorporation into the F0 complex. Transmembrane helices of wild type subunit c (blue), subunit a (orange), and the fused copy of subunit c (green) are shown for wild type subunit a (A), the a-c fusion (B), and the a-c/2-c fusion (C and D).
FIGURE 7.
The a-c and a-c/2-c fusion proteins are incorporated into the F0 complex. The F0 complex preparations from the membranes of OM202recA− strains transformed with pHP7 (a-c fusion), pHP808 (a-c/2-c fusion), or pBWU13 (wild type) were separated by SDS-PAGE and silver-stained. Three samples of each preparation containing 0.2, 1.0, or 5.0 μg of protein as indicated by the triangles are shown. Positions of molecular mass markers and their masses in kDa are shown on the left. Similar to wild type subunits a and c (37), the fusion proteins display anomalous electrophoretic mobility that does not correspond to their actual molecular weights, a common property of many highly hydrophobic proteins. The calculated and apparent molecular masses for subunit a are 30 and 26 kDa, respectively, for the a-c fusion protein, 38 and 28 kDa, and for the a-c/2-c fusion, 42 and 33 kDa.
ATP synthase maintains precise coupling between ATP synthesis, or hydrolysis, in the F1 complex and proton translocation through the F0 complex. For this reason, inhibition of rotor movement by the bulky adduct of the DCCD reaction with Asp61 of subunit c results in nearly complete loss of F0F1-ATPase activity. As a test of proper enzyme assembly with the a-c/2-c protein, we investigated if the F1 complex was attached to the F0 portion and if the coupling mechanism was intact. Because the rotor is immobilized in this construct, in the coupled enzyme the F1 complex should be attached to the F0 but have little ATPase activity. ATPase activity of the F1 should be restored upon F1 release from the membrane, which uncouples F1 from F0. First, we analyzed the amount of F1 bound to the cell membrane in the wild type and a-c/2-c containing ATP synthase. The F1 was released from the membrane by EDTA treatment, and analyzed by gel densitometry following separation by SDS-PAGE. The α and β subunits can be easily identified on electrophoregrams by comparison to background strain OM202 recA− which does not produce ATP synthase (supplemental Fig. S2). The content of α and β subunits in the a-c/2-c membranes was about 60% of the wild type. However, ATPase activity of the a-c/2-c membranes was less than 20% of the wild type (Table 1). The F1-ATPase activity was restored to about 50% of the wild type by removal of F1 from the membrane. In contrast, membranes from E. coli strains expressing truncated variants of subunit a that did not incorporate into the ATP synthase had ATPase activity equal to 70–90% of the wild type (23), indicating that F1 was associated with the membrane, but coupling between F0 and F1 was disrupted. Taken together these data indicate that although the amount of ATP synthase in the strain expressing the a-c/2-c fusion protein was about 40% less than in the wild type, most of ATP synthase present in the membrane was assembled correctly and F1 was coupled to the F0.
TABLE 1.
F1-ATPase activity of the ATP synthase with the a-c/2-c fusion protein
| ATPase activity of the membranes | DCCD inhibition | Released F1 activity |
||
|---|---|---|---|---|
| Units/mg membrane proteina | % wild type | |||
| units/mg | % | |||
| Wild type (pBWU13) | 2.09 ± 0.10 | 62 | 0.42 ± 0.02 | 100 |
| a-c/2-c (pHP808) | 0.40 ± 0.02 | 15 | 0.20 ± 0.01 | 50 |
| No ATP synthase (no plasmid) | 0.02 ± 0.002 | NDb | <0.01 | <0.5 |
a Membranes were suspended in 1 mm Tris-HCl, pH 7.5, 0.5 mm EDTA, and 10% glycerol at 5.0 mg of protein/ml and incubated for 2 h at room temperature. Membranes were removed by ultracentrifugation at 120,000 × g for 1.5 h. The ATPase activity of the supernatant containing soluble F1 was measured after addition of 5 mm MgCl2. Decrease in specific activity of F1 after release from the membrane results from the intrinsic inhibition by the ϵ–subunit (56).
b ND, not determined.
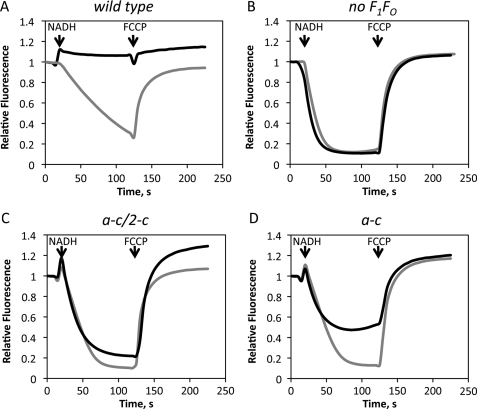
Removal of the F1 from the wild type E. coli ATP synthase by EDTA treatment of the membranes converts F0 into a passive proton pore and creates a proton leak in the membrane that can be detected by a decrease in the transmembrane pH difference generated by an active proton transporter, such as NADH dehydrogenase (Fig. 5, D–F). Indeed, proton accumulation inside EDTA-treated membrane vesicles from strain pBWU13/OM202recA−, which expresses wild type ATP synthase, was dramatically reduced, compared with background strain OM202recA−, which has no ATP synthase (Fig. 8, A and B). In contrast, the F0 complex with the a-c/2-c construct showed very low proton permeability compared with the wild type (Fig. 8C). The residual proton leak in this case may result from limited rotor oscillations within the range allowed by the length of the peptide linker between a and c. Finally, membranes containing F0 with the a-c fusion protein showed a decrease in proton conductance (Fig. 8D) compared with the wild type, consistent with the lower rates of ATP-dependent proton translocation catalyzed by F0F1-ATPase with this fusion construct (Fig. 5B). Proton conductance both with the wild type F0 and the a-c fusion was inhibited by DCCD.
FIGURE 8.
Proton permeability of the F0 complexes with the a-c and a-c/2-c fusion proteins. NADH-driven proton translocation was measured by 9-amino-6-chloro-4-methoxyacridine fluorescence quenching in membrane vesicles (50 μg/ml of total membrane protein) with wild type subunit a from strain OM202recA−/pBWU13 (A), control membranes containing no F0 from strain OM202recA− (B), with the a-c/2-c fusion from strain OM202recA−/pHP808 (C), and with the a-c fusion from strain OM202recA−/pHP7 (D). Additions of 0.2 mm NADH and 2.0 μm proton uncoupler carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) are indicated by arrows. Gray traces were recorded with the membranes pretreated with 80 μm dicyclohexylcarbodiimide.
DISCUSSION
Consistent with previous observations (50–52), we found that overexpression of subunit a without other subunits of ATP synthase is highly toxic to cells. In earlier work (52), overexpression of subunit a was shown to dissipate electric potential difference across the cell membrane, likely due to an increase in nonspecific ion permeability of the latter. This effect may result from spontaneous insertion of misfolded subunit a into the cell membrane at high expression levels. At the same time, subunit c can be overexpressed without other ATP synthase subunits and inserted into the cell membrane without any toxic effects. Interaction with subunit c may both facilitate insertion of subunit a into the membrane, and protect properly folded subunit a from degradation by membrane protease FtsH (46, 47).
We have shown that subunit c fused to subunit a through a peptide linker can chaperone insertion of subunit a into the membrane, if the relative orientation of a and c transmembrane helices in the fusion construct is the same as in the native enzyme. This clearly demonstrates that an assembled c-ring is not required for subunit a insertion. Notably, the G23D and L31F mutants of subunit c, which have low propensity for oligomerization and do not form rings in the membrane, can chaperone insertion of subunit a just as effectively as the wild type. This fact suggests that interaction with a single monomer of c subunit is sufficient to insert subunit a into the membrane. However, we cannot exclude the possibility that two copies of c subunit normally interact with subunit a during the insertion process because the a-c/2-c construct contains two copies of subunit c helix II, and modeling shows that subunit a may be in contact with two copies of the c subunit in the native F0.
Correct transmembrane topology of subunit c appears to be critical for proper insertion of subunit a into the membrane, because, unlike the a-c/2-c protein, the a-c fusion, which has inverted orientation of the helical hairpin of subunit c does not insert into the membrane in the absence of wild type subunit c. Interestingly, regardless of the orientation of the fused c-polypeptide, the a-component assembles properly into the F0 complex. Thus, consistent with the prediction of transmembrane topology of these fusion proteins (41), the a-polypeptide determines transmembrane topology of the fusion protein. Somewhat paradoxically, if the c-subunit is attached to a in the incorrect topology, ATP synthase is functional. In this case, the inverted helical c-hairpin is displaced to the periphery of the cylindrical oligomer formed by the wild type c-subunits, and does not interfere with rotor movement. In contrast, the c-subunit fused in the correct transmembrane topology is readily incorporated into the c-ring together with wild type c-subunits, and the assembled ATP synthase is inactive, because the rotor is tethered to the stator by the linker peptide. Several additional observations are consistent with this explanation. The a-c/2-c fusion protein was co-purified with subunits b and c in the standard procedure used for purification of the wild type F0 complex. The ratio of subunits in this preparation was very similar to the wild type. The F1 complex was attached to the membrane with the a-c/2-c construct but, the DCCD-sensitive ATPase activity was very low compared with the wild type, indicating that the coupling mechanism was intact, but the enzyme was inactive due to the jammed rotor. ATPase activity was restored when F1 was released from the membrane by EDTA treatment. Taken together, these data suggest that the fold of the a-c/2-c protein is similar to subunit a and the neighboring c monomers in the native F0 complex.
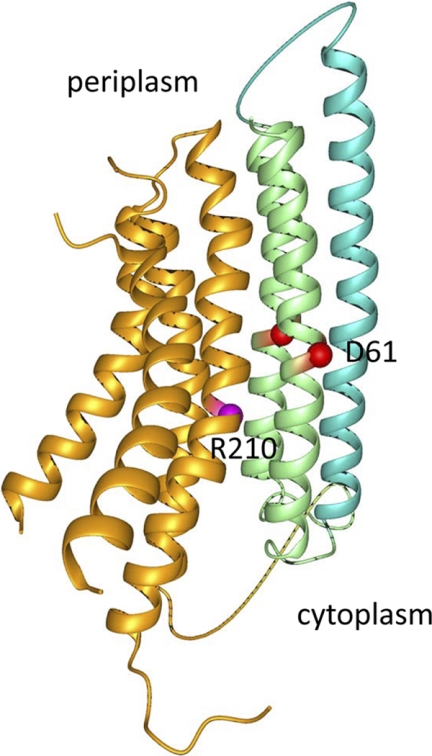
It is interesting to speculate about the location of transmembrane helix VI of the a-c/2-c fusion, which corresponds to the orphan helix II of subunit c, in the assembled F0. Structure modeling shows that changing the number of subunit c copies in the oligomeric complex by one or two does not significantly affect the interhelical contacts (53). It is possible then that both copies of subunit c helix II present in the fusion protein incorporate into the outer ring of helices of the c-rotor with some rearrangement of the inner ring to account for the missing helix I copy (Fig. 6C). This conformation seems energetically most favorable, because it would preserve the inter-helical contacts found between wild type a and c subunits in the F0 complex. A structural model of a-c/2-c fusion calculated from the Cys-Cys cross-linking studies within the F0 complex, secondary site suppressors in subunit a, and high resolution structure of subunit c oligomer, illustrates the arrangement of transmembrane helices in this protein (Fig. 9). In the model, helices IV and V of subunit a are in close contact with two copies of helix II of subunit c, similar to the arrangement predicted for the a-c interface in the native F0 complex (10, 13) with the Arg210 of subunit a in close proximity of Asp61 of subunit c. Although less likely, we cannot exclude the possibility that helix VI of the fusion protein is displaced to the periphery of the ring (Fig. 6D), similar to displacement of incorrectly oriented fused subunit c in the a-c protein (Fig. 6B), whereas the complete fused copy of subunit c (helices VII and VIII of the fusion protein) assembles into the ring normally.
FIGURE 9.
Model of the a-c/2-c fusion protein fold. Helices corresponding to subunit a are orange, the two copies of subunit c helix II are green, and helix I of subunit c is turquoise. The Arg210 of subunit a (magenta) and Asp61 of subunit c (red) are shown as spheres.
Efficient incorporation of the a-c/2-c fusion into ATP synthase suggests a complex of subunit a with one or two copies of monomeric subunit c as an important intermediate in the assembly of the F0. Formation of an ac or ac2 complex may prime assembly of the ac10 complex, which is followed by attachment of the b2-F1 intermediate complex. This is consistent with measurements showing that interactions between subunit b and F1 contribute the most to the free energy change of F1 binding to the F0, whereas contribution of subunit a is insignificant in free energy terms (54). Our data are also consistent with participation of more complex intermediates in the assembly process, such as a complex of b2, F1, and several copies of subunit c. In fact, subunit c was found in the purified preparation of F0F1-ATPase from a thermophilic Bacillus species assembled without subunit a (55). Subunit c was likely present in substoichiometric amounts in this preparation, as reconstitution with extraneous subunit a led to a very limited restoration of ATP-dependent proton transport. Although isolated E. coli subunit c may form ring structures spontaneously under certain conditions (25), in the course of F0 assembly in the membrane, subunit a is unlikely to attach to the preassembled c-ring because in that case the c-component of the a-c/2-c fusion protein would remain on the periphery of the ring and would not interfere with rotor movement, as observed with the a-c construct.
Analysis of ATP synthase assembly intermediates in yeast mitochondria suggested that in eukaryotes formation of the c-ring and its association with F1 precedes attachment of subunit 6 (atp6p), which is homologous to E. coli subunit a (29). This difference may reflect a more complex ATP synthase assembly process in eukaryotes involving additional chaperone proteins and minor subunits of the enzyme that have no homolog in bacterial ATP synthases. It is noteworthy that ring assembly monitored in this work appeared to be a slow process with newly synthesized monomeric subunit c (subunit 9 or atp9p in mitochondria) predominating in the mitochondrial membrane on a scale of minutes before being incorporated into the oligomeric complexes. In the context of a rapidly dividing bacterial cell, this would indicate low propensity for ring self-assembly in the membrane. Rather, an intermediate including subunit a and a subunit c monomer may prime assembly of the F0 complex in bacteria.
Despite remarkable success in solving the structure of F1 and deciphering the mechanism of ATP synthesis, the molecular mechanism of ion-driven rotation of the ATP synthase turbine remains hypothetical in the absence of a high resolution structure of subunit a and the a-c interface, where the transmembrane ion channel is believed to be located. Attempts to solve the structure of the complete ATP synthase or its membrane component (F0) have so far been unsuccessful. One of the reasons for the slow progress in this area is the difficulty of obtaining protein with defined subunit stoichiometry in a single conformational state. A possible solution for this problem is designing a simpler model protein, which preserves the structural features of the ion channel, but is more amenable to high resolution structural studies. With this goal in mind, we designed the a-c/2-c fusion protein built of subunits a and c connected in correct transmembrane topology. We have shown that this protein is inserted into the membrane and correctly incorporated in the ATP synthase complex. This is a strong indication that the a-c/2-c construct preserves the native fold of the a and c subunits and retains the rotor-stator interface structure that exists in the complete ATP synthase. Thus our work provides validation for using this construct for high resolution structure determination of ATP synthase ion channel.
Supplementary Material
Acknowledgments
We are grateful to Bob Fillingame for useful discussions and a generous gift of antibody against subunit c and Benjamin Rempel for constructing some of the plasmids used in this work.
This work was supported by a Saskatchewan Health Research Foundation New Investigator grant and a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant (to O. Y. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- DCCD
- N,N′-dicyclohexylcarbodiimide.
REFERENCES
- 1. Senior A. E. (1988) Physiol. Rev. 68, 177–231 [DOI] [PubMed] [Google Scholar]
- 2. Yoshida M., Muneyuki E., Hisabori T. (2001) Nat. Rev. Mol. Cell Biol. 2, 669–677 [DOI] [PubMed] [Google Scholar]
- 3. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 4. Meier T., Polzer P., Diederichs K., Welte W., Dimroth P. (2005) Science 308, 659–662 [DOI] [PubMed] [Google Scholar]
- 5. Pogoryelov D., Yildiz O., Faraldo-Gómez J. D., Meier T. (2009) Nat. Struct. Mol. Biol. 16, 1068–1073 [DOI] [PubMed] [Google Scholar]
- 6. Preiss L., Yildiz O., Hicks D. B., Krulwich T. A., Meier T. (2010) PLoS Biol. 8, e1000443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dmitriev O. Y., Jones P. C., Fillingame R. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7785–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rastogi V. K., Girvin M. E. (1999) Nature 402, 263–268 [DOI] [PubMed] [Google Scholar]
- 9. Girvin M. E., Rastogi V. K., Abildgaard F., Markley J. L., Fillingame R. H. (1998) Biochemistry 37, 8817–8824 [DOI] [PubMed] [Google Scholar]
- 10. Jiang W., Fillingame R. H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6607–6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jones P. C., Jiang W., Fillingame R. H. (1998) J. Biol. Chem. 273, 17178–17185 [DOI] [PubMed] [Google Scholar]
- 12. Valiyaveetil F. I., Fillingame R. H. (1998) J. Biol. Chem. 273, 16241–16247 [DOI] [PubMed] [Google Scholar]
- 13. Moore K. J., Fillingame R. H. (2008) J. Biol. Chem. 283, 31726–31735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dmitriev O. Y., Freedman K. H., Hermolin J., Fillingame R. H. (2008) Biochim. Biophys. Acta 1777, 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwem B. E., Fillingame R. H. (2006) J. Biol. Chem. 281, 37861–37867 [DOI] [PubMed] [Google Scholar]
- 16. Vik S. B., Long J. C., Wada T., Zhang D. (2000) Biochim. Biophys. Acta 1458, 457–466 [DOI] [PubMed] [Google Scholar]
- 17. Vik S. B., Ishmukhametov R. R. (2005) J. Bioenerg. Biomembr. 37, 445–449 [DOI] [PubMed] [Google Scholar]
- 18. Fillingame R. H., Angevine C. M., Dmitriev O. Y. (2003) FEBS Lett. 555, 29–34 [DOI] [PubMed] [Google Scholar]
- 19. Steed P. R., Fillingame R. H. (2008) J. Biol. Chem. 283, 12365–12372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van der Laan M., Bechtluft P., Kol S., Nouwen N., Driessen A. J. (2004) J. Cell Biol. 165, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yi L., Celebi N., Chen M., Dalbey R. E. (2004) J. Biol. Chem. 279, 39260–39267 [DOI] [PubMed] [Google Scholar]
- 22. Yi L., Jiang F., Chen M., Cain B., Bolhuis A., Dalbey R. E. (2003) Biochemistry 42, 10537–10544 [DOI] [PubMed] [Google Scholar]
- 23. Hermolin J., Fillingame R. H. (1995) J. Biol. Chem. 270, 2815–2817 [DOI] [PubMed] [Google Scholar]
- 24. Klionsky D. J., Brusilow W. S., Simoni R. D. (1983) J. Biol. Chem. 258, 10136–10143 [PubMed] [Google Scholar]
- 25. Arechaga I., Butler P. J., Walker J. E. (2002) FEBS Lett. 515, 189–193 [DOI] [PubMed] [Google Scholar]
- 26. Ozaki Y., Suzuki T., Kuruma Y., Ueda T., Yoshida M. (2008) Biochem. Biophys. Res. Commun. 367, 663–666 [DOI] [PubMed] [Google Scholar]
- 27. Suzuki T., Ozaki Y., Sone N., Feniouk B. A., Yoshida M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20776–20781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gay N. J. (1984) J. Bacteriol. 158, 820–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rak M., Gokova S., Tzagoloff A. (2011) EMBO J. 30, 920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iwamoto A., Omote H., Hanada H., Tomioka N., Itai A., Maeda M., Futai M. (1991) J. Biol. Chem. 266, 16350–16355 [PubMed] [Google Scholar]
- 31. Stalz W. D., Greie J. C., Deckers-Hebestreit G., Altendorf K. (2003) J. Biol. Chem. 278, 27068–27071 [DOI] [PubMed] [Google Scholar]
- 32. Jones P. C., Fillingame R. H. (1998) J. Biol. Chem. 273, 29701–29705 [DOI] [PubMed] [Google Scholar]
- 33. Dmitriev O. Y., Altendorf K., Fillingame R. H. (2004) FEBS Lett. 556, 35–38 [DOI] [PubMed] [Google Scholar]
- 34. Mosher M. E., White L. K., Hermolin J., Fillingame R. H. (1985) J. Biol. Chem. 260, 4807–4814 [PubMed] [Google Scholar]
- 35. Rottenberg H., Moreno-Sanchez R. (1993) Biochim. Biophys. Acta 1183, 161–170 [DOI] [PubMed] [Google Scholar]
- 36. Schägger H., von Jagow G. (1987) Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 37. Schneider E., Altendorf K. (1986) Methods Enzymol. 126, 569–578 [DOI] [PubMed] [Google Scholar]
- 38. Ekman P., Jäger O. (1993) Anal. Biochem. 214, 138–141 [DOI] [PubMed] [Google Scholar]
- 39. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 40. Brunger A. T. (2007) Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 41. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 42. Dmitriev O. Y., Abildgaard F., Markley J. L., Fillingame R. H. (2004) J. Biomol. NMR 29, 439–440 [DOI] [PubMed] [Google Scholar]
- 43. Hartzog P. E., Cain B. D. (1994) J. Biol. Chem. 269, 32313–32317 [PubMed] [Google Scholar]
- 44. Hatch L. P., Cox G. B., Howitt S. M. (1995) J. Biol. Chem. 270, 29407–29412 [DOI] [PubMed] [Google Scholar]
- 45. Vik S. B., Antonio B. J. (1994) J. Biol. Chem. 269, 30364–30369 [PubMed] [Google Scholar]
- 46. Akiyama Y., Kihara A., Ito K. (1996) FEBS Lett. 399, 26–28 [DOI] [PubMed] [Google Scholar]
- 47. Akiyama Y. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8066–8071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jans D. A., Fimmel A. L., Langman L., James L. B., Downie J. A., Senior A. E., Ash G. R., Gibson F., Cox G. B. (1983) Biochem. J. 211, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kol S., Majczak W., Heerlien R., van der Berg J. P., Nouwen N., Driessen A. J. (2009) J. Mol. Biol. 390, 893–901 [DOI] [PubMed] [Google Scholar]
- 50. Arechaga I., Miroux B., Runswick M. J., Walker J. E. (2003) FEBS Lett. 547, 97–100 [DOI] [PubMed] [Google Scholar]
- 51. Kanazawa H., Kiyasu T., Noumi T., Futai M. (1984) J. Bacteriol. 158, 300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. von Meyenburg K., Jørgensen B. B., Michelsen O., Sørensen L., McCarthy J. E. (1985) EMBO J. 4, 2357–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fillingame R. H., Dmitriev O. Y. (2002) Biochim. Biophys. Acta 1565, 232–245 [DOI] [PubMed] [Google Scholar]
- 54. Krebstakies T., Zimmermann B., Gräber P., Altendorf K., Börsch M., Greie J. C. (2005) J. Biol. Chem. 280, 33338–33345 [DOI] [PubMed] [Google Scholar]
- 55. Ono S., Sone N., Yoshida M., Suzuki T. (2004) J. Biol. Chem. 279, 33409–33412 [DOI] [PubMed] [Google Scholar]
- 56. Feniouk B. A., Suzuki T., Yoshida M. (2006) Biochim. Biophys. Acta 1757, 326–338 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.