Background: Yeast cells respond to high environmental iron by altering gene transcription.
Results: The high iron-sensing transcription factor Yap5 induces transcription of the iron-sulfur cluster-containing enzyme Tyw1.
Conclusion: The sequestration of iron by incorporation into protein-bound iron-sulfur clusters protects cells from iron toxicity.
Significance: Decreased free cytosolic iron is central for protection of iron toxicity.
Keywords: Iron, Metalloenzymes, Transcription, Transcription Factors, Yeast, Iron-Sulfur
Abstract
The budding yeast Saccharomyces cerevisiae responds to high cytosolic iron by inducing Yap5-mediated transcription. We identified genes regulated by Yap5 in response to iron and show that one of the genes induced is TYW1, which encodes an iron-sulfur cluster enzyme that participates in the synthesis of wybutosine-modified tRNA. Strains deleted for TYW1 do not show a phenotype in standard yeast medium. In contrast, overexpression of TYW1 results in decreased cell growth and induction of the iron regulon, leading to increased expression of the high affinity iron transporters. We identified a minimal domain of S. cerevisiae Tyw1 that is sufficient to induce the iron regulon. CCC1, a vacuolar iron importer, is a Yap5-regulated gene, and deletion of either CCC1 or YAP5 resulted in high iron sensitivity. Deletion of TYW1 in a Δccc1 strain led to increased iron sensitivity. The increased iron sensitivity of Δccc1Δtyw1 could be suppressed by overexpression of iron-sulfur cluster enzymes. We conclude that the Yap5-mediated induction of TYW1 provides protection from high iron toxicity by the consumption of free cytosolic iron through the formation of protein-bound iron-sulfur clusters.
Introduction
The budding yeast Saccharomyces cerevisiae responds to changes in the concentration of the redox-active metals copper and iron by altering the levels of transcripts for genes involved in metal transport and utilization. Alterations in gene expression can be ascribed primarily to metal-sensing transcription factors (for review, see Ref. 1). Low copper activates the transcription factor Mac1, which induces expression of CTR1, the gene encoding the high affinity copper transporter. Under high copper conditions, Ace1 induces the expression of metallothioneins, copper sequestration proteins. Yeast cells respond to low cytosolic iron by inducing a suite of ∼20 genes referred to as the iron regulon (2). These genes encode elemental iron transporters, siderophore iron transporters, and proteins involved in iron utilization. The genes in the iron regulon are activated by the transcription factor Aft1 and, to a lesser extent, by its paralog, Aft2. Aft1 and Aft2 reside in the cytosol and, under low iron conditions, translocate to the nucleus, where they then activate genes of the iron regulon (3). Yeast can also respond to high iron through activation of Yap5 (4). Yap5 is a member of the β-loop helix family, a family of eight homologous transcription factors (5). Yap5 was shown to induce transcription of CCC1, which encodes a vacuolar iron transporter that regulates the level of cytosolic iron by importing iron into the vacuole. In contrast to Aft1/Aft2, Yap5 is constitutively expressed in the nucleus and resides on the promoter of CCC1 (4). We demonstrate here that, in response to high iron, Yap5 also induces the expression of TYW1, which encodes a cytosolic iron-sulfur cluster-containing enzyme involved in the synthesis of the modified nucleoside wybutosine. We show that overexpression of Tyw1 and truncated versions of Tyw1 can induce the iron regulon. We further show that a potential function of the Yap5-induced expression of Tyw1 is to act as an iron sink, reducing the concentration of free cytosolic iron through formation of protein-bound Fe-S clusters.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Plasmids
All yeast strains for this study were from the W303 background (Table 1). Δccc1 and Δyap5 were deleted in wild-type DY150 (MATa ade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc)) as described previously (4). Δtyw1 and Δtyw2 were generated in DY150 using the gene-specific KanMX deletion cassette amplified from the homozygous deletion collection (Thermo Fisher Scientific). Deletion of CCC1 in Δyap5, Δtyw1, or Δtyw2 was generated by homologous recombination using PCR-amplified HIS3 as the marker for the double deletion strains as described previously (4). The Δaft1 and Δaft2 strains were the generous gifts of Dr. Dennis R. Winge (University of Utah). Mutant strains of nfs1 and isu1 were identified previously (6). The identification and behavior of strains carrying the erg25-2 allele were described previously (7).
TABLE 1.
Strains used in this study
| Yeast strain | Genotype | Source |
|---|---|---|
| DY150 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) | WT |
| Δccc1 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) Δccc1::HIS3 | Ref. 4 |
| Δyap5 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) Δyap5::KanMX | Ref. 4 |
| Δccc1Δyap5 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) Δyap5::KanMX Δccc1::HIS3 | Ref. 4 |
| Δtyw1 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) Δtyw1::KanMX | This study |
| Δccc1Δtyw1 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) Δtyw1::KanMX Δccc1::HIS3 | This study |
| Δtyw2 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) Δtyw2::KanMX | This study |
| Δccc1Δtyw2 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) Δtyw2::KanMX Δccc1::HIS3 | This study |
| Δaft1 | MATaleu2 his3 ura3 lys2 ade2 trp1 Δaft1::TRP1 | Ref. 24 |
| Δaft2 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) Δaft2::HIS3 | This study |
| OCY582 (nfs1 mutant) | MATα ade6 ura3-52 leu2-3,112 trp1-1 his3-11 can1-100(oc) | Ref. 6 |
| OCY401 (isu1 mutant) | MATα ade6 ura3-52 leu2-3,112 trp1-1 his3-11 can1-100(oc) | Ref. 6 |
| erg2-2 | MATα ade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) | Ref. 7 |
Complete minimal medium (CMM)2 was composed of yeast nitrogen base and dextrose and lacked the required nutrients. To induce GAL1-regulated genes, galactose was substituted for glucose in the medium. Media were made iron-limited by the addition of 80 μm bathophenanthroline disulfonate, and the concentration of added iron (FeSO4) is specified in the figure legends.
The YAP5-HA, GFP-YAP5, and FET3-lacZ plasmids have been described previously (4). The TYW1-lacZ reporter construct was generated by the insertion of a 600-bp upstream promoter region of the TYW1 gene into the lacZ reporter vector YEP354 at BamHI/HindIII sites using primers 5′-cgc gag tcc gat ggt gaa gag gtc atc tt-3′ and 5′-ccc aag ctt cat aat tta ctt act gtt aca g-3′. The Schizosaccharomyces pombe TYW1 gene was cloned into pYES2/CT (Invitrogen) with a His6 tag at the C terminus using primers 5′-ccc aag ctt atg ggg tat tct ttg ggg ta-3′ and 5′-ccg ctc gag cgc agc ggc ttc cga aat tg-3′. Full-length TYW1 (YPL207W) was cloned into the HindIII/XhoI sites of pYES2/CT with a His6 tag at the C terminus using primers P1 (5′-ccc aag ctt atg gat ggt ttt cgt gta gc-3′) and P2 (5′-ccg ctc gag agc agg aat agg agg gag ag-3′). The following truncations of TYW1 were cloned into pYES2/CT with a His6 tag at the C terminus using the indicated primers: t-TYW1(461–810) (Fe-S-containing S-adenosylmethionine (SAM) domain (SAM/Fe-S)), P3 (5′-ccc aag ctt atg tcg ctc ttc aat atc gcg tc-3′) and P2; t-TYW1(1–386), P1 and P4 (5′-ccg ctc gag acc ttc ttc ttc atc ttc ctc-3′); t-TYW1(200–386), P5 (5′-ccc aag ctt atg ctt ttg agt tca cag atc tat-3′) and P6 (5′-ccg ctc gag cgt ttc att cga aat tgt caa-3′); t-TYW1(1–199), P1 and P6; t-TYW1(9–199), P7 (5′-ccc aag ctt atg gcg tta acc gct gca tat tt-3′) and P6; t-TYW1(29–199), P8 (5′-ccc aag ctt atg gcg ctg gtc att ata gta gg-3′) and P7; t-TYW1(1–65), P9 (5′-ccg ctc gag gca aca tgg ctt ttt ctg ttg-3′) and P1; and t-TYW1(63–199), P10 (5′-ccc aag ctt atg cca tgt tgc agt gat aag aag-3′) and P6. Full-length TYW1 including the upstream promoter region (600 bp) was cloned with a 3′-FLAG tag into low copy pRS416. All constructs were verified by DNA sequencing.
Microarray and RT-PCR
Total RNAs were isolated using the Agilent RNA kit. Microarray was performed on an Affymetrix platform, and results were analyzed using GeneSifter software. For RT-PCR, cDNA was synthesized using SuperScript III (Invitrogen). Real-time PCR was performed using a LightCycler (Roche Applied Science) with TYW1 primers 5′-aac aaa tgg tgg cga aag ac-3′ and 5′-tgg atg aac atg cca aag aa-3′ and the following PCR conditions: denaturing at 95 °C for 20 s, annealing at 55 °C for 5 s, and elongation at 68 °C for 10 s for 40 cycles. The PCR quantification was normalized using primers to CMD1 as described previously (4).
Chromatin Immunoprecipitation
ChIP was performed as described (4). Sheared chromatin from cells grown in medium with or without 500 μm iron was immunoprecipitated for GFP-Yap5 using a rabbit anti-GFP antibody (Abcam, Cambridge, MA). The primers used for recognition of the Yap5-binding site of the TYW1 promoter were 5′-cag atg atg cca agg ctt tc-3′ and 5′-tac aca aat atc ggg cat cg-3′.
Mitochondrial Isolation and Metal Measurements
Mitochondrial fractions were isolated as described previously (8). Metal content was determined using a PerkinElmer inductively coupled plasma optical emission spectrometer. Protein concentrations of mitochondrial pellets were determined by bicinchoninic acid protein assay (Thermo Fisher Scientific), and the results were normalized to mitochondrial protein concentration.
Western Blotting and Immunofluorescence Microscopy
Cell lysates were generated by glass bead homogenization, and proteins were analyzed by Western blotting as described previously (7). The antibodies used included rabbit anti-His6 (1:3000; Covance, Princeton, NJ), mouse anti-CPY (1:5000; Invitrogen), and peroxidase-conjugated goat anti-rabbit IgG or peroxidase-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). Immunofluorescence was performed as described previously (9). Briefly, mid-log grown yeast cells were harvested and fixed in 4% formaldehyde for 1 h and in 4% paraformaldehyde overnight. Cell walls were removed with oxalyticase, and cells were permeabilized with 2% SDS. Samples were incubated overnight with rabbit anti-His6 (1:200) or mouse anti-Dpm1 (1:50; Invitrogen) antibody, washed, and then incubated with Alexa Fluor 594-conjugated goat anti-rabbit IgG or Alexa Fluor 488-conjugated goat anti-mouse IgG (1:750; Invitrogen). Fluorescent images were captured with an Olympus BX51 microscope.
Other Methods
β-Galactosidase activity was determined as described previously (6). Specific activity units were determined from a 10-min kinetic assay and calculated as nmol/min/mg of protein. Aconitase activity was measured as described (6).
RESULTS
TYW1 Is Transcriptionally Regulated by Yap5
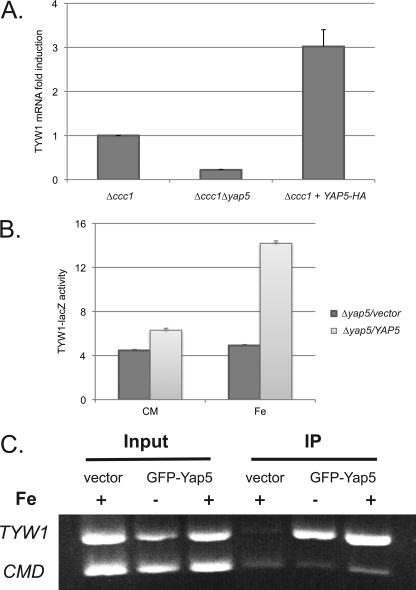
We utilized microarray analysis to identify Yap5-regulated genes. First, we examined changes in mRNA in Δccc1 cells expressing constitutively active Yap5 (Yap5-HA) (4). Second, we compared transcript levels of Δccc1 cells and Δccc1Δyap5 cells grown in high iron-containing medium. The reason for utilizing Δccc1 cells was that the activity of Ccc1 limits cytosolic iron and can reduce Yap5-induced gene expression (4). Yap5-dependent genes were examined for the presence of consensus Yap-binding sequences (TTACTAA or TTAGTAA) in their promoter regions. Those genes containing consensus Yap sequences that showed changes upon constitutive Yap5 expression and in Δccc1 cells but not in Δccc1Δyap5 cells were considered to be Yap5 targets. We identified a limited number of genes as Yap5 targets: GRX4, CUP1, YHB1, PHO3, GDH1, and TYW1 (Table 2). RT-PCR and/or S1 analysis confirmed that GRX4, TYW1, and CUP1 were increased under the above conditions. We focused on TYW1, which encodes an iron-sulfur cluster-containing enzyme involved in base modification in tRNA (10). Cells deleted for CCC1 and YAP5 had lower levels of the TYW1 transcript compared with Δccc1 cells (Fig. 1A). Expression of constitutively active Yap5-HA led to a 3-fold increase in TYW1 mRNA compared with control cells. Changes in mRNA levels could reflect either transcription or post-transcriptional regulation. To determine whether TYW1 is regulated transcriptionally, we generated a lacZ construct regulated by the predicted TYW1 promoter region. In the presence of iron, there was an increase in β-galactosidase activity in Yap5-expressing cells (Fig. 1B). No induction of β-galactosidase was seen in the absence of Yap5 or iron. We then performed ChIP to determine whether the Yap5-induced regulation of TYW1 is direct or indirect. Similar to the Yap5 regulation of CCC1 (4), Yap5 was bound to the TYW1 promoter in the presence or absence of iron (Fig. 1C). These results show that Yap5 occupies the TYW1 promoter and induces transcription in an iron-dependent manner.
TABLE 2.
Microarray and RT-PCR analysis of Yap5-induced gene
Microarray analysis was performed in duplicate, comparing Δccc1 cells expressing constitutively active Yap5 (Yap5-HA) with Δccc1 cells or comparing iron-loaded Δccc1 cells with Δccc1Δyap5 cells. Transcripts that were increased approximately 2-fold above the background level were then examined by RT-PCR or S1 analysis (n = 3). The promoter regions of genes were also examined for the presence of consensus Yap-binding sites (TTA(C/G)TAA).
| Gene name | Microarray |
TTA(C/G)TAA at promoter position | RNA level | |
|---|---|---|---|---|
| Yap5-HA | Δyap5 | |||
| -fold | ||||
| TYW1 | +3.18 | −3.72 | −250 bp | Verified |
| GRX4 | +2.20 | −6.18 | −250 bp | Verified |
| CUP1 | +2.56 | −1.89 | −460 bp | Verified |
| PHO3 | +3.64 | −1.80 | −150 bp | No |
| GDH1 | +1.90 | −1.62 | −750 bp | No |
| YHB1 | +2.37 | −1.30 | −400 bp | No |
FIGURE 1.
TYW1 transcription is regulated by Yap5. A, RT-PCR was performed on total RNA isolated from Δccc1 or Δccc1Δyap5 cells or from Δccc1 cells transformed with a constitutively active YAP5-HA plasmid. The level of expression of TYW1 mRNA was compared between Δccc1 cells and Δccc1 cells transformed with the YAP5-HA plasmid and between Δccc1 and Δccc1Δyap5 exposed to iron. B, Δyap5 cells containing a control or YAP5 plasmid were transformed with the TYW1-lacZ construct. Cells were grown overnight in CMM and then incubated with or without 500 μm ferrous iron for another 2 h. Cells were harvested, β-galactosidase activity was determined, and the results were normalized to protein concentration. C, Δccc1 cells transformed with a control vector or GFP-YAP5 were grown in iron-limited medium overnight and then incubated with 500 μm iron for 2 h before being processed for ChIP analysis. Calmodulin (CMD) was used a control, and all experiments were performed at least three times. Error bars represent S.E. IP, immunoprecipitation.
Overexpression of TYW1 Activates the Iron Regulon
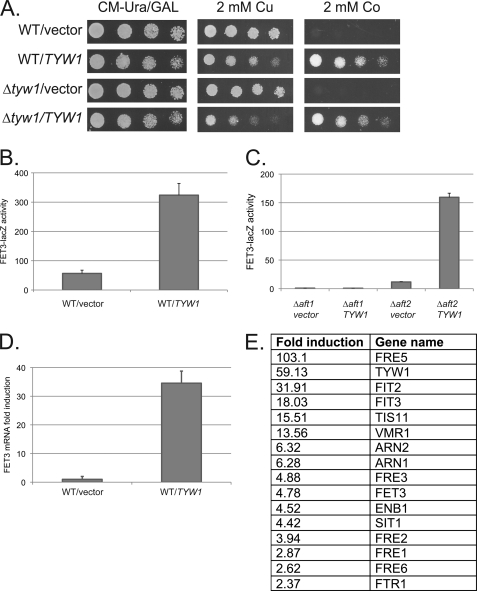
Overexpression of TYW1 was reported to be lethal (11). We transformed wild-type cells with a plasmid containing a GAL1-TYW1-His6 construct. We observed a slight growth deficit in TYW1-overexpressing cells (Fig. 2A). We also observed that TYW1-overexpressing cells were sensitive to 2 mm copper but were highly resistant to 2 mm cobalt. In contrast, deletion of TYW1 did not alter transition metal sensitivity. Copper sensitivity and cobalt resistance often result from induction of the iron regulon (12, 13). We determined that overexpression of TYW1 increased the activity of the FET3-lacZ reporter construct, FET3 being a member of the iron regulon (Fig. 2B). The effect of TYW1 on induction of FET3-lacZ expression was dependent on the low iron-sensing transcription factor Aft1, as deletion of AFT1 completely prevented FET3-lacZ expression (Fig. 2C). The effect of TYW1 on induction of the iron regulon was confirmed by RT-PCR (Fig. 2D) and microarray analysis (Fig. 2E). These results show that overexpression of TYW1 activates Aft1, leading to increased expression of the iron regulon.
FIGURE 2.
Overexpression of TYW1 induces the Aft1 iron regulon. A, wild-type and Δtyw1 cells were transformed with either a control vector or the GAL1-TYW1 plasmid. Cells were spotted in serial dilution on galactose medium with different concentrations of metals. Plates were incubated at 30 °C for 3 days and photographed. CM, complete minimal medium. B, wild-type cells with a control vector or the GAL1-TYW1 plasmid were transformed with the FET3-lacZ construct, and β-galactosidase activity and cell protein were determined. C, Δaft1 and Δaft2 cells were transformed with either a control vector or a TYW1 plasmid and a FET3-lacZ construct. Cells were processed for β-galactosidase activity and cell protein. D and E, total RNA was isolated from wild-type cells transformed with a control plasmid or a TYW1 plasmid and assayed by RT-PCR for FET3 mRNA (D) or genes in the iron regulon by microarray (E).
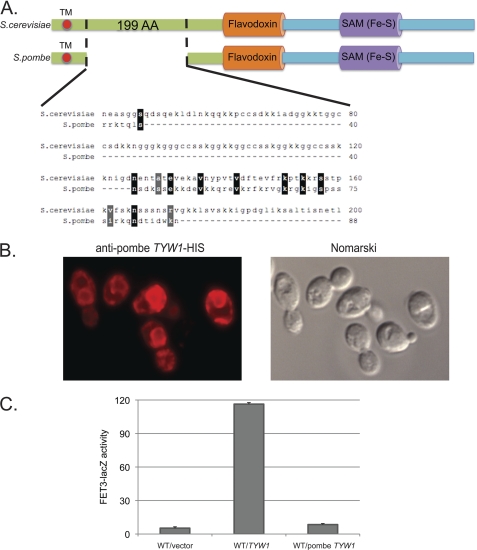
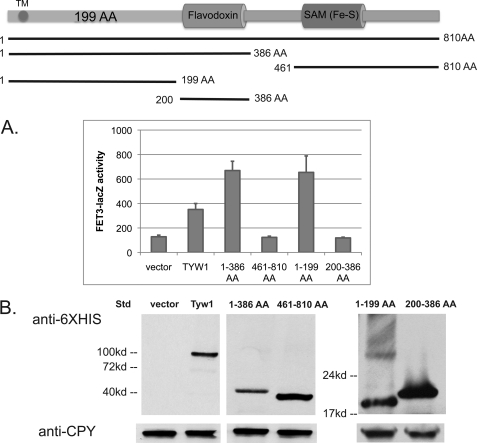
Tyw1 is an enzyme involved in the synthesis of wybutosine, a modified guanosine found in phenylalanine tRNA (10). The enzyme contains two domains: a flavodoxin-binding domain (FAD) and a [4Fe-4S]-containing radical SAM domain. Tyw1 has been localized to the cytosolic surface of the endoplasmic reticulum (ER) (14), and the protein sequence suggests that it has a single membrane-spanning domain. The protein is highly conserved throughout evolution, with the highest degree of conservation in the FAD and SAM domains. Comparison of the Tyw1 amino acid sequences of S. cerevisiae and S. pombe shows that both proteins are highly homologous (Fig. 3A). S. cerevisiae Tyw1 and its close relatives in the family Saccharomycetaceae contain a 199-amino acid region between the transmembrane domain and the FAD domain, which is not found in other species and is poorly conserved in S. pombe. S. pombe Tyw1 expressed in S. cerevisiae localized to the ER (Fig. 3B), but the expressed protein was unable to induce FET3-lacZ expression (Fig. 3C). This finding suggested that the region between the transmembrane and FAD domains might be responsible for Aft1 activation. We generated His6-tagged truncation constructs of TYW1 and expressed the truncated forms in wild-type cells. Expression of truncated Tyw1 lacking the Fe-S-containing SAM domain induced FET3-lacZ (Fig. 4A). In contrast, expression of truncated Tyw1 lacking the N-terminal portion and the FAD domain but including the Fe-S-containing SAM domain did not induce FET3-lacZ. Further truncations showed that the FAD domain was not necessary for induction of FET3-lacZ, whereas expression of the first 199 amino acids was sufficient. All constructs were expressed to similar levels (Fig. 4B).
FIGURE 3.
Overexpression of S. pombe TYW1 homolog does not induce the Aft1 regulon. A, the schematic compares the domains of S. cerevisiae TYW1 and S. pombe TYW1. Expanded is the 199-amino acid (AA) domain that is found in S. cerevisiae but that is poorly conserved in S. pombe. TM, transmembrane domain. B, wild-type cells with S. pombe TYW1-His6 were processed for immunofluorescence using an anti-His tag antibody, and images were captured. C, wild-type cells were transformed with either S. cerevisiae or S. pombe TYW1 plasmids and a FET3-lacZ plasmid. Cells were processed for β-galactosidase activity and cell protein.
FIGURE 4.
Identification of domains of Tyw1 that activate the iron regulon. A, wild-type cells were transformed with a control vector, full-length TYW1, or plasmids containing truncated regions as shown in the upper panel. All constructs contained a C-terminal His6 epitope. All cells were cotransformed with a FET3-lacZ construct. Cells were assayed for β-galactosidase activity and cell protein (lower panel). TM, transmembrane domain; AA, amino acids. B, samples were also assayed by Western blotting with anti-His6 or anti-CPY antibodies. Std, standard.
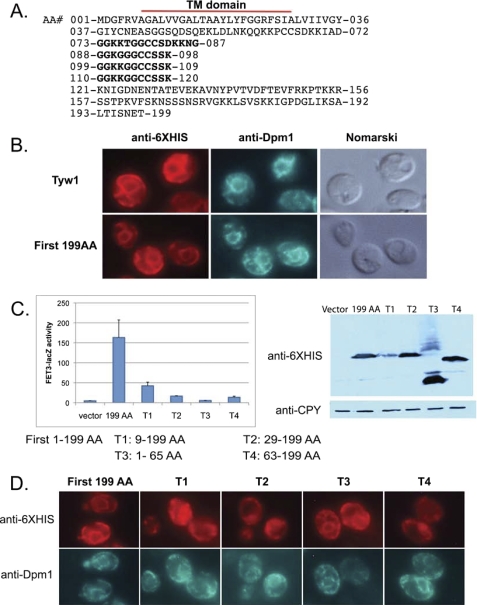
Examination of the first 199 amino acids revealed the presence of a hydrophobic region in the first 37 amino acids (Fig. 5A). Computer analysis of the protein sequence suggested that the hydrophobic domain encodes a transmembrane sequence, indicating that the bulk of the protein is localized in the cytosol. Wild-type Tyw1 and t-Tyw1(1–199) co-localized with the resident ER marker Dpm1, showing “classic” ER staining surrounding the nucleus with peripheral ER near the plasma membrane (Fig. 5B). t-Tyw1(1–199) was sufficient to induce FET3-lacZ (Fig. 5C). Deletion of the first nine amino acids (T1) resulted in an expressed protein that was not localized to the ER and that had a dramatically reduced ability to induce FET3-lacZ (Fig. 5, C and D). The T1 protein was expressed to a much lesser extent than the other constructs (Fig. 5C, right panel). Deletion of the transmembrane domain from the first 199 amino acids (T2) gave rise to a protein that localized to the cytosol and that did not activate FET3-lacZ. Expression of a construct that contained the first 65 amino acids (T3) resulted in a protein that was localized to the ER but did not induce FET3-lacZ. Similarly, deletion of the first 63 amino acids (T4) from the intact protein resulted in a widely diffused cytosolic protein that was unable to induce expression of FET3-lacZ. On the basis of these results, we conclude that localization to the ER and amino acids 63–199, which include the GGKK repeats, are important for inducing FET3-lacZ.
FIGURE 5.
Role of the membrane-tethering region of Tyw1 in the activation of the iron regulon. A, shown is the sequence of the first 199 amino acids (AA) of Tyw1. TM denotes the transmembrane region, and repetitive amino acids are shown in boldface. B, shown is the immunofluorescence of cells transfected with Tyw1-His6-containing constructs visualized using an anti-His6 antibody and an antibody to the ER marker Dpm1. C, wild-type cells were transformed with plasmids expressing amino acids 1–199, 9–199, 29–199, 1–65, or 63–199 and a FET3-lacZ construct. Cells were assayed for β-galactosidase activity and cell protein (left panel). The right panel shows a Western blot of protein levels in transformed cells. D, shown is the immunofluorescence of cells transformed with constructs expressing different truncations of Tyw1-His6 and stained for His6 and Dpm1.
Overexpression of Tyw1 Results in Increased Cellular Iron
There are two conditions known to induce the iron regulon: decreased cytosolic iron (15) or decreased mitochondrial Fe-S cluster synthesis or export (16). Reduced cytosolic iron can result from defects in the high affinity iron transport system composed of Fet3/Ftr1 or activation of the vacuolar iron transporter Ccc1, both leading to decreased cytosolic iron (13). To determine whether overexpression of Tyw1 results in defective iron transport activity, we assayed cellular iron content by inductively coupled plasma optical emission spectrometry. Overexpression of Tyw1 or t-Tyw1(1–199) resulted in an increase in whole cell iron (Fig. 6A), indicating that the high affinity iron transport system was induced and functional.
FIGURE 6.
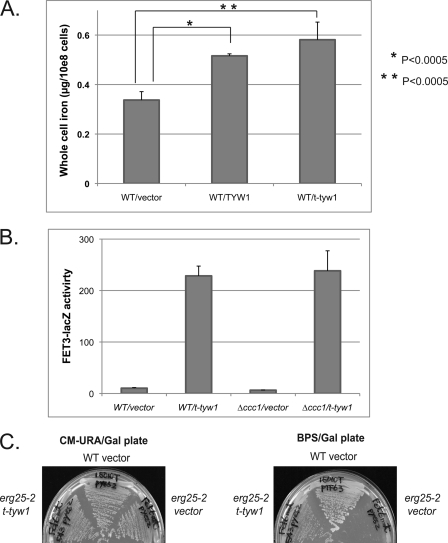
Overexpression of truncated TYW1 affects cellular iron accumulation. A, wild-type cells were transformed with a control vector, full-length TYW1, or t-TYW1 containing the first 199 amino acids. Cells were grown overnight in CMM, and iron levels were determined by inductively coupled plasma optical emission spectrometry. B, wild-type or Δccc1 cells were transformed with a control vector or t-TYW1(1–199) and a FET3-lacZ plasmid. Cells were incubated overnight, and β-galactosidase activity and cell protein were determined. C, wild-type and erg25-2 cells were transformed with either a control vector or a plasmid containing t-TYW1(1–199). Transformed cells were streaked on CMM−Ura/Gal (CM-URA/Gal) or iron-limited bathophenanthroline disulfonate (BPS)/Gal plates. Cells were grown for 3 days and photographed.
It may be possible that iron entering the cell is sequestered in an organelle or present in the cytosol in a form that is not bioavailable. Increased cytosolic iron may be transported into the vacuole. Previously, we showed that increased activity of the vacuolar transporter Ccc1 led to increased high affinity iron transport but decreased cytosolic iron as a result of increased vacuolar iron transport (17). We deleted CCC1 to determine whether vacuolar iron transport is involved in the Tyw1 induction of the iron regulon. Deletion of CCC1 did not affect the induction of FET3-lacZ in response to overexpressed t-Tyw1 (Fig. 6B).
To determine whether the cytosolic iron in Tyw1-overexpressing cells is bioavailable, we took advantage of the observation that a mutant allele of the methyl sterol oxidase Erg25 will not grow when cytosolic iron is low. Previously, we showed that an increase in cellular iron stores can rescue an erg25 mutant (erg25-2) growth defect under low iron conditions (7, 16). If expression of Tyw1 leads to an increase in cellular iron, then when cells are incubated in low iron medium, the increased cellular iron, if bioavailable, will maintain the activity of the erg25 mutant. We transformed t-TYW1 into erg25-2 mutant cells to assay for mobilizable iron stores. erg25-2 transformed with a control vector died on low iron plates, but cells overexpressing t-TYW1 suppressed the low iron growth defect (Fig. 6C). This result indicates that overexpression of t-TYW1 leads to increased cellular iron, which can be made bioavailable.
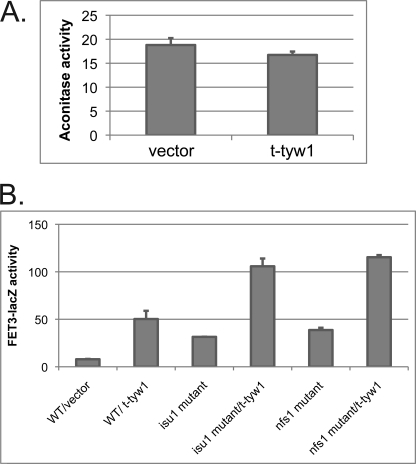
Decreases in mitochondrial Fe-S cluster synthesis are known to lead to increased expression of the iron regulon. Because disruption of Fe-S cluster formation leads to the activation of Aft1, we hypothesized that t-Tyw1 affects Aft1 activation through alterations in Fe-S cluster formation. Expression of t-TYW1 in wild-type cells did not affect the activity of the mitochondrial Fe-S cluster-containing aconitase (Fig. 7A). Cells with hypomorphic alleles of NFS1 or ISU1, two genes essential for Fe-S cluster formation, were transformed with t-TYW1 and FET3-lacZ. Overexpression of t-TYW1 in nfs1 or isu1 mutant cells showed additional increased FET3-lacZ activity compared with nfs1 or isu1 mutant cells alone (Fig. 7B). These results suggest that the effects of t-Tyw1 overexpression on Aft1 activation are additive with defects in the Fe-S cluster synthesis pathway.
FIGURE 7.
Overexpression of TYW1 does not affect Fe-S cluster synthesis. A, wild-type cells were transformed with a control vector or t-TYW1(1–199). Cells were assayed for aconitase activity and cell protein. B, wild-type, isu1 mutant, and nfs1 mutant cells were transformed with either s control vector or t-TYW1(1–199) and a FET3-lacZ plasmid. Cells were grown overnight, and β-galactosidase activity and cell protein were determined.
Role for TYW1 in Modulating High Iron Sensitivity
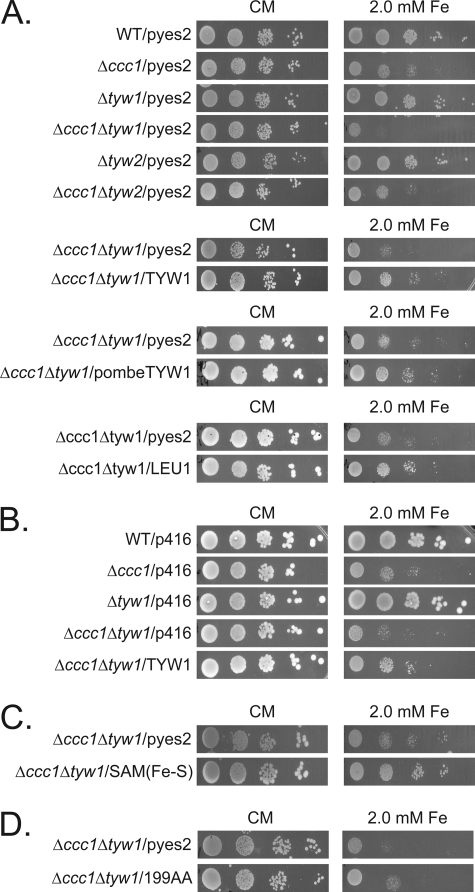
We next focused on investigating the reason for the Yap5-dependent induction of TYW1. We considered that wybutosine modification of tRNA is required for cell growth under high iron conditions. Deletion of TYW1 did not result in increased iron sensitivity. In contrast, a double deletion of TYW1 and CCC1 showed increased iron sensitivity relative to cells with a single deletion of either gene (Fig. 8A). The iron sensitivity was reduced by transforming TYW1 back into the double deletion strain (Fig. 8A). To determine whether the absence of wybutosine-modified tRNA is important in modulating iron sensitivity, we generated a strain that had deletions in CCC1 and TYW2, which encodes a tRNA methyltransferase whose substrate is the product of Tyw1 (10). Deletion of TYW2 in Δccc1 cells did not alter the iron sensitivity of Δccc1 cells (Fig. 8A). This result suggests that the lack of Tyw1 does not affect iron sensitivity through the absence of the wybutosine modification on tRNA but rather through the absence of the enzyme or the absence of the product of the enzyme. The sensitivity to high iron in Δccc1Δtyw1 cells could also be suppressed by S. pombe TYW1 (Fig. 8A), suggesting conservation in function. Overexpression of Leu1, an Fe-S cluster enzyme involved in leucine biosynthesis, also suppressed the increased iron toxicity of Δccc1Δtyw1 cells (Fig. 8A). Expression of TYW1 under the control of its own promoter, which is regulated by Yap5, could suppress the increased iron toxicity of Δccc1Δtyw1 cells (Fig. 8B). On the basis of these results, we hypothesized that it is the ability of Tyw1 to bind iron (through the formation of Fe-S clusters) that is responsible for the decrease in iron toxicity. We tested this possibility by expressing the Fe-S-containing SAM domain of Tyw1 in Δccc1Δtyw1 cells. Expression of this construct suppressed iron toxicity in Δccc1Δtyw1 cells (Fig. 8C). Overexpression of Tyw1(1–199) showed a slight reduction in high iron toxicity but not nearly to the extent as the full-length construct or the Fe-S-containing domain (Fig. 8D). These results indicate that formation of protein-bound Fe-S clusters can reduce free cytosolic iron, attenuating iron toxicity.
FIGURE 8.
Deletion of TYW1 in Δccc1 cells increases iron toxicity, and overexpression of Fe-S-containing proteins decreases that toxicity. A, cells transformed with an empty vector (pYES2), GAL1-TYW1, S. pombe GAL1-TYW1, or GAL1-LEU1 were spotted in serial dilution on galactose/CMM (CM) containing 2 mm iron. B, cells transformed with an empty vector (pRS416) or TYW1 were spotted in serial dilution on CMM containing 2 mm iron. C and D, Δccc1Δtyw1 cells transformed with either a control vector or a plasmid containing the t-TYW1(SAM/Fe-S) domain region (C) and Δccc1Δtyw1 cells transformed with either a control vector or a plasmid containing amino acids 1–199 (D) were spotted as described for A and photographed.
DISCUSSION
The ability of yeast to sense and respond to changes in iron levels is essential for cell viability. Under low iron conditions, the low iron-sensing transcription factor Aft1 affects the levels of ∼20 different genes. The only identified high iron-responsive transcription factor in S. cerevisiae is Yap5, which regulates the expression of the vacuolar iron transporter Ccc1. Here, we have shown that it also affects the levels of a limited number of other genes. The difference in gene transcription between low and high iron may reflect both the importance of Ccc1 in regulating cytosolic iron and perhaps the rarity of exposure of yeast to a high iron environment.
We focused on TYW1, as deletion of TYW1 increased the iron sensitivity of Δccc1 cell. Tyw1 is an Fe-S-containing enzyme involved in the synthesis of wybutosine, a modified base in tRNA. Our data indicate that the enzymatic product of Tyw1 does not play a role in the high iron response. Deletion of TYW1 did not affect growth in normal or iron-containing CMM. There was, however, increased iron sensitivity when TYW1 was deleted in Δccc1 cells. The finding that overexpression of the Fe-S cluster-containing protein Leu1 also protected Δccc1Δtyw1 cells from iron toxicity suggests that it is the sequestration of iron into the Fe-S clusters of the apoprotein that protects cells from high iron. These results support our previous finding that accumulation of free cytosolic iron leads to toxicity (18). Yap5 orchestrates the cellular response to high iron by decreasing the concentration of cytosolic free iron through sequestration of iron in vacuoles or through incorporation into protein-bound Fe-S clusters.
Overexpression of TYW1 was reported to result in lethality (11). Under our conditions, overexpression of TYW1 was not lethal and had only a modest effect on growth. A difference between the reported result and our data is that we did not use glutathione S-transferase-tagged versions. Glutathione S-transferase is known to dimerize, and it might be that dimerization of epitope-tagged Tyw1 leads to a loss of viability. We did identify, however, a phenotype associated with overexpression of Tyw1, which is induction of the iron regulon. Analysis of the mechanism of induction revealed several interesting features. First, induction of the iron regulon by overexpressed Tyw1 was ascribed not to the enzymatic domains of the protein but to the first 199 amino acids, which include the ER-tethering domain and a domain of the protein that is unique to a subgroup in Saccharomycetaceae, which is enriched in GGKKGGCCSSK repeats.
The second notable feature is that overexpression of Tyw1 affected the iron regulon in a unique way. To date, induction of the iron regulon has been ascribed to decreased free cytosolic iron and alterations in mitochondrial Fe-S cluster synthesis. Decreased cytosolic iron may result from mutations in genes that encode the high affinity iron transport system composed of Fet3/Ftr1 (19) or decreased assembly of copper-containing Fet3 (20, 21). These defects lead to decreased cellular iron accumulation. In contrast, in Tyw1-overexpressing cells, cellular iron transport and iron accumulation are increased. Induction of the iron regulon can also result from increased activity of the vacuolar iron transporter Ccc1 (13); however, deletion of CCC1 had no effect on the ability of overexpressed TYW1 to induce the iron regulon. Studies have shown that deletion of the monothiol glutaredoxins GRX3 and GRX4 induces the iron regulon (22). Mühlenhoff et al. (22) demonstrated that iron did not enter the mitochondria or vacuole, in Δgrx3Δgrx4 cells, leading to the lack of bioavailable iron. As shown here, iron trafficking to the vacuole or mitochondria occurs in Tyw1-overexpressing cells, indicating that intracellular iron movement is unaffected.
Induction of the iron regulon has also been seen in cells with deletions of genes that encode the mitochondrial Fe-S cluster-synthesizing proteins or mitochondrial Fe-S export machinery (1, 16). In these studies, the mutant cells showed decreased mitochondrial Fe-S enzyme activity, up-regulation of the iron regulon, and increased mitochondrial iron. Overexpressing Tyw1 in wild-type cells does not change iron levels in mitochondria or mitochondrial iron-sulfur cluster enzymes such as aconitase. Recently, it was suggested that increased activity of the iron regulon can occur in the absence of decreased mitochondrial Fe-S activity (23). How the membrane-spanning domain of S. cerevisiae Tyw1 affects the iron regulon is currently unknown. As suggested by a reviewer, it may be possible that the overexpressed protein titrates factors important for the regulation of Aft1 activity. The finding that overexpressed Tyw1 affects the iron regulon offers a new tool to dissect the induction of the iron regulon independent of iron or Fe-S cluster synthesis.
Acknowledgments
We thank our laboratory members for reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 030534 from NIDDK.
- CMM
- complete minimal medium
- t-TYW1/t-Tyw1
- truncated TYW1/Tyw1
- SAM
- S-adenosylmethionine
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Rutherford J. C., Ojeda L., Balk J., Mühlenhoff U., Lill R., Winge D. R. (2005) J. Biol. Chem. 280, 10135–10140 [DOI] [PubMed] [Google Scholar]
- 2. Philpott C. C., Protchenko O. (2008) Eukaryot. Cell 7, 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamaguchi-Iwai Y., Ueta R., Fukunaka A., Sasaki R. (2002) J. Biol. Chem. 277, 18914–18918 [DOI] [PubMed] [Google Scholar]
- 4. Li L., Bagley D., Ward D. M., Kaplan J. (2008) Mol. Cell. Biol. 28, 1326–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fernandes L., Rodrigues-Pousada C., Struhl K. (1997) Mol. Cell. Biol. 17, 6982–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kumánovics A., Chen O. S., Li L., Bagley D., Adkins E. M., Lin H., Dingra N. N., Outten C. E., Keller G., Winge D., Ward D. M., Kaplan J. (2008) J. Biol. Chem. 283, 10276–10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li L., Kaplan J. (1996) J. Biol. Chem. 271, 16927–16933 [DOI] [PubMed] [Google Scholar]
- 8. Li L., Kaplan J. (1997) J. Biol. Chem. 272, 28485–28493 [DOI] [PubMed] [Google Scholar]
- 9. Kumánovics A., Poruk K. E., Osborn K. A., Ward D. M., Kaplan J. (2006) J. Biol. Chem. 281, 22566–22574 [DOI] [PubMed] [Google Scholar]
- 10. Noma A., Kirino Y., Ikeuchi Y., Suzuki T. (2006) EMBO J. 25, 2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S. G., Cyert M., Hughes T. R., Boone C., Andrews B. (2006) Mol. Cell 21, 319–330 [DOI] [PubMed] [Google Scholar]
- 12. Li L., Kaplan J. (2004) J. Biol. Chem. 279, 33653–33661 [DOI] [PubMed] [Google Scholar]
- 13. Li L., Murdock G., Bagley D., Jia X., Ward D. M., Kaplan J. (2010) J. Biol. Chem. 285, 10232–10242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar A., Agarwal S., Heyman J. A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Cheung K. H., Miller P., Gerstein M., Roeder G. S., Snyder M. (2002) Genes Dev. 16, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamaguchi-Iwai Y., Stearman R., Dancis A., Klausner R. D. (1996) EMBO J. 15, 3377–3384 [PMC free article] [PubMed] [Google Scholar]
- 16. Chen O. S., Crisp R. J., Valachovic M., Bard M., Winge D. R., Kaplan J. (2004) J. Biol. Chem. 279, 29513–29518 [DOI] [PubMed] [Google Scholar]
- 17. Li L., Chen O. S., McVey Ward D., Kaplan J. (2001) J. Biol. Chem. 276, 29515–29519 [DOI] [PubMed] [Google Scholar]
- 18. Lin H., Li L., Jia X., Ward D. M., Kaplan J. (2011) J. Biol. Chem. 286, 3851–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Askwith C., Eide D., Van Ho A., Bernard P. S., Li L., Davis-Kaplan S., Sipe D. M., Kaplan J. (1994) Cell 76, 403–410 [DOI] [PubMed] [Google Scholar]
- 20. Yuan D. S., Stearman R., Dancis A., Dunn T., Beeler T., Klausner R. D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2632–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Radisky D. C., Snyder W. B., Emr S. D., Kaplan J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5662–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mühlenhoff U., Molik S., Godoy J. R., Uzarska M. A., Richter N., Seubert A., Zhang Y., Stubbe J., Pierrel F., Herrero E., Lillig C. H., Lill R. (2010) Cell Metab. 12, 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moreno-Cermeño A., Obis E., Bellí G., Cabiscol E., Ros J., Tamarit J. (2010) J. Biol. Chem. 285, 41653–41664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yun C. W., Ferea T., Rashford J., Ardon O., Brown P. O., Botstein D., Kaplan J., Philpott C. C. (2000) J. Biol. Chem. 275, 10709–10715 [DOI] [PubMed] [Google Scholar]