Background: The role of long proline-rich segments, as they are abundantly present in the spliceosome, is elusive.
Results: Cell biological and biophysical data show the significance of multiple motifs for tandem-WW domain recognition.
Conclusion: The dynamic assembly of the pre-spliceosome is enabled by transient multivalent interactions.
Significance: Our results have general implications for the recognition of proline-rich sequence hubs in modular protein assemblies.
Keywords: Isothermal Titration Calorimetry, Mass Spectrometry (MS), NMR, Protein Domains, Spliceosome, Spin-label, WW Domain, Multivalent
Abstract
The high abundance of repetitive but nonidentical proline-rich sequences in spliceosomal proteins raises the question of how these known interaction motifs recruit their interacting protein domains. Whereas complex formation of these adaptors with individual motifs has been studied in great detail, little is known about the binding mode of domains arranged in tandem repeats and long proline-rich sequences including multiple motifs. Here we studied the interaction of the two adjacent WW domains of spliceosomal protein FBP21 with several ligands of different lengths and composition to elucidate the hallmarks of multivalent binding for this class of recognition domains. First, we show that many of the proteins that define the cellular proteome interacting with FBP21-WW1-WW2 contain multiple proline-rich motifs. Among these is the newly identified binding partner SF3B4. Fluorescence resonance energy transfer (FRET) analysis reveals the tandem-WW domains of FBP21 to interact with splicing factor 3B4 (SF3B4) in nuclear speckles where splicing takes place. Isothermal titration calorimetry and NMR shows that the tandem arrangement of WW domains and the multivalency of the proline-rich ligands both contribute to affinity enhancement. However, ligand exchange remains fast compared with the NMR time scale. Surprisingly, a N-terminal spin label attached to a bivalent ligand induces NMR line broadening of signals corresponding to both WW domains of the FBP21-WW1-WW2 protein. This suggests that distinct orientations of the ligand contribute to a delocalized and semispecific binding mode that should facilitate search processes within the spliceosome.
Introduction
Protein interactions mediated by proline-rich sequences (PRS)3 are characterized by low affinity and promiscuous binding of adaptor domains (1–5). These types of interactions seem to be a hallmark of molecular assemblies that undergo rapid rearrangements. One such example of a highly dynamic molecular machine is the spliceosome where several subunits, also denoted as small nuclear ribonuclear particles (snRNP), are steered into a final complex that is responsible for the excision of introns. Proline-rich sequence hubs have been identified in several snRNP proteins and shown to play a crucial role in the assembly of the early spliceosome (6, 7). WW and GYF domain containing proteins are attracted by these sequences and thereby allow for molecular crowding at the long and repetitive PRS of essential splicing proteins.
Formin-binding protein 21 (FBP21) was initially found as a binding partner of the cytoskeletal protein formin (8). Later investigations, however, clearly put the protein into the context of splicing and transcription (9). It binds to the core splicing protein SmB/B′ and was recently shown to be important for splicing in a cellular context (10). FBP21 contains two WW domains that both have been characterized to interact with PRS ligands of group Rb (11). In these ligands proline residues are flanked by arginine and are indeed abundantly present in several splicing proteins. The linker connecting the two WW domains is 10–12 amino acids in length and shows enhanced flexibility compared with the individual WW domains. Upon ligand encounter, this linker is thought to adopt a more defined conformation to allow for cooperative binding of both WW domains (10). However, the exact mode of interaction between the tandem-WW domains of FBP21 and elongated ligands is still not well understood and the cellular proteome specifically targeted to the tandem-WW domains is also elusive. We therefore set out to profile cellular interaction partners of FBP21-tandem-WW by SILAC/MS analysis. Several candidate proteins were identified that match the requirement of bivalent binding with some targets displaying 3 or more PRS motifs. We show in vitro that the strength of the interaction of FBP21 with SmB and SF3B4 peptides depends on the valency of the ligand. Affinity enhancement due to the tandem arrangement of the WW domains is moderate for bivalent ligands but increases significantly for the tetravalent SmB binding partner. Moreover, the tandem action of both WW domains seems to be more relevant for ligands containing more than two motifs. Variation of the length and composition of the linker between the two WW domains does not compromise the interaction with different SmB ligands. Finally, we show data that are consistent with two perpendicular binding orientations, supporting a model of a dynamic positional and orientational equilibrium of ligands interacting with the FBP21-WW domains.
EXPERIMENTAL PROCEDURES
Constructs
For cloning of the human FBP21-WW constructs, a fragment encoding residues 122–196 (tandem-WW wt) was amplified by PCR and cloned into pET-28a via NdeI and XhoI restriction sites. To disrupt the PRS-binding site, relevant Trp residues 151 and 191 (position 29 and 70 in the tandem-WW construct) were replaced by Ala. Constructs with various linker length between the individual WW domains were designed by deleting residues 156–162 (35–41, “Δ35–41”) or 154–165 (33–44, “Δ33–44”). Additionally we replaced residues 156–163 (35–42, “glycine linker”) by a GGGGSGGG stretch.
Cloning of the GST fusion construct of FBP21-tandem-WW was achieved by inserting FBP21 residues 122–196 into pGEX4T1 via BamHI/XhoI restriction sites. For the CFP- and YFP-tagged constructs, DNA fragments encoding FBP21 residues 122–196, full-length SmB, and full-length SF3B4 were amplified by PCR and cloned into a pECFP/EYFP-N1 vector (Invitrogen) using EcoRI/AgeI restriction sites for insertion of SmB and BamHI/AgeI of the FBP21 and SF3B4 variants. A Kozak sequence was included in the 5′ primers for efficient expression.
HEK293FT Cells
HEK293FT cells (Invitrogen) were grown in DMEM with 4.5 g liter−1 of glucose and stable glutamine (PAA Laboratories) containing 10% fetal bovine serum at 37 °C with 5% CO2. Transfection using PromoFectin (PromoKine) was performed with 1.5 μg of plasmid DNA according to the manufacturer's instructions. Cells were cultured on 60-mm cell culture dishes coated with gelatin 24 h prior to transfection.
FRET Measurements
Images were obtained using a Leica SP2 confocal microscope. The FRET signal and the YFP reference channel were recorded with photomultiplier 3 between 530 and 600 nm. The CFP emission was detected with photomultiplier 2 between 470 and 510 nm. 458 nm excitation was used to record CFP and FRET emission, 514 nm was used for excitation of YFP. For FRET measurements the double dichroic mirror DD458/514 was used. The scan speed was 400 Hz, the image resolution 1024 × 1024 pixels, the pinhole diameter 100 μm. For each set of transformation, images from more than 20 cells were obtained, and each experiment was repeated independently (12). The correction factors α (0.88) and β (0.64) were determined with cells solely expressing YFP and CFP, respectively. The apparent FRET efficiency E was defined as,
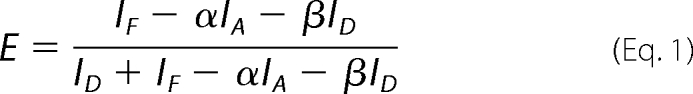 |
Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and Pulldown
Experiments were performed as described (7). SILAC medium was prepared by adding stable isotope-labeled l-[13C6]/[15N2]lysine and l-[13C6]/[15N4]arginine and l-[2H4]lysine and l-[13C6]arginine (Cambridge Isotope Laboratories). For competitive inhibition, the peptide SmB-2 (GTPMGMPPPGMRPPPPGMRGLL) was added to a final concentration of 3 mm. GST-tandem-WW of FBP21 alone was incubated with lysate containing l-[13C6]/[15N2]lysine and l-[13C6]/[15N4]arginine in the first pulldown and with l-[2H4]lysine and l-[13C6]arginine in the second pulldown. Inversely, GST-tandem-WW of FBP21 containing inhibitory SmB-2 peptide was incubated with lysates supplied with l-[2H4]lysine and l-[13C6]arginine in the first pulldown and with l-[13C6]/[15N2]lysine and l-[13C6]/[15N4]arginine in the second pulldown experiment. This inverse labeling strategy prevents the overestimation of enrichment factors from proteins that are differentially expressed in either of the two labeling media.
Mass Spectrometry
For MS analysis, proteins were separated using a Tris glycine gradient 4–20% gel (Invitrogen) and Coomassie-stained gel bands were cut into 40 slices of equal size.
Tryptic in-gel digestion of proteins and nano-LC-MS/MS experiments were performed as described previously (13). In brief, tryptic peptides were separated by a reversed-phase capillary liquid chromatography system (Eksigent 2D nanoflow LC, Axel Semrau GmbH) connected to an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific). Mass spectra were acquired in a data-dependent mode with one MS survey scan (with a resolution of 60,000) in the Orbitrap followed by MS/MS scans of the five most intense precursor ions in the LTQ. The MS survey range was m/z 350–1500. The dynamic exclusion time (for precursor ions) was set to 120 s and automatic gain control was set to 3 × 106 and 20,000 for Orbitrap-MS and LTQ-MS/MS scans, respectively. Identification and quantification of proteins was carried out with version 1.0.12.31 of the MaxQuant software package (14). Generated peak lists (msm files) were submitted to a MASCOT search engine (version 2.2, Matrix Science Ltd.) and searched against an IPI human protein data base (version 3.52). The mass tolerance of precursor and sequence ions was set to 7 ppm and 0.35 Da, respectively. Methionine oxidation and the acrylamide modification of cysteine were used as variable modifications. False discovery rates were <1% based on matches to reversed sequences in the concatenated target-decoy data base. Proteins were considered for further analysis if at least two sequenced peptides could be quantified.
Isothermal Titration Calorimetry
ITC experiments have been performed at 281 K using the VP-ITC device (Originlab). Proteins were dialyzed against PBS, 1 mm EDTA, pH 7.4, peptides were dissolved in dialysis buffer and solutions were corrected to pH 7.4. Protein concentrations in the measurement cell ranged between 20 and 350 μm and were titrated against increasing amounts of 0.25–4 mm peptide stock solutions. To rule out other unspecific effects, peptide was also titrated against buffer and buffer against protein. To obtain the stoichiometry (N), association constant (KA), and the change of enthalpy (ΔH) and entropy (ΔS) of the reaction, peak areas were integrated and reaction heats were plotted against the molar ratio by fitting to a “One Set of Sites” model until χ-square reached a minimum.
Protein Preparation
Proteins were expressed as His- or GST-tagged fusion constructs in Escherichia coli BL21(DE3). Proteins were purified by the corresponding affinity matrices, thrombin cleaved in case of the His-tagged proteins, and applied to a gel filtration column (Superdex-75) at 50 mm phosphate, 150 mm NaCl, 1 mm EDTA, pH 6.5, or 1× PBS, 1 mm EDTA, pH 7.4. Proteins were concentrated by Centrifugal Filter Units (regenerated cellulose, 3 kDa cutoff, Millipore) if necessary. In the case of NMR samples, proteins were expressed in M9 minimal medium supplemented with [15N]NH4Cl and/or [13C]glucose and had a final concentration of 0.1–0.9 mm.
Peptide SPOT Experiments
SPOT analyses were performed as described (15). Membranes were incubated with either 7.5 μg/ml of GST-WW1/GST-WW2 or with 10 μg/ml of GST-tandem-WW.
NMR Spectroscopy
Measurements were performed at 300 K on a 750 MHz AV and a 600 MHz DRX spectrometer (Bruker) equipped with triple resonance cryoprobes, respectively. Protein samples were buffered in 50 mm phosphate, 150 mm NaCl, 1 mm EDTA, 10% D2O at pH 6.5. NMR data were processed using Topspin (Bruker) and spectra evaluation was performed using CCPNMR Analysis software (16).
Assignment of Backbone NH Resonances
Resonance assignment of the unbound tandem-WW WT was performed via two-dimensional 15N-HSQC spectra in combination with standard triple resonance CBCA(CO)NNH/CBCANNH experiments. The resonance assignment of the ligand-bound WT was generated by a 15N-HSQC based titration experiment. Resonances of the tandem-WW domain knock out mutants (W29A/W70A) were assigned by standard triple resonance HNCA/HN(CO)CA-BEST experiments in a constant time mode (17).
Chemical Shift Mapping
The determination of relevant binding epitopes was realized by titrating unlabeled ligand (supplemental Table 1) to 15N-labeled tandem-WW WT (0.2 mm). The protein-ligand ratios were increased stepwise to a final ratio of 1:6 (SmB-4), 1:10 (SmB-2 and SF3B4-2), or 1:100 (SmB-1, SmB-1A, SmB-1B, SmB-1C, SmB-1D, and SF3B4-1). The weighted chemical shift changes observed in 15N-HSQC spectra of every titration end point were calculated based on Equation 2.
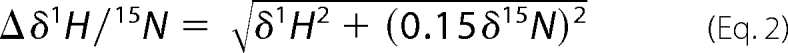 |
15N-T1/T2 Backbone Relaxation Experiments
R1 and R2 rates were calculated for 15N-labeled tandem-WW WT (0.2 mm) in the unbound state, in complex with SmB-2 (1:6) and SmB-4 (1:3) at 750 MHz. The extraction of R1/R2 rates for NH backbone resonances was based on a set of 15N-T1/T2-HSQC spectra (18) with incremented relaxation delays of 6, 10, 14, 18, 22, 26, 34, 50, 82, 162, and 242 ms for T2 and 12, 52, 102, 152, 202, 402, 802, 1602, and 3202 ms for T1. Peak heights were plotted against the corresponding relaxation times and fitted to an exponential decay function using CCPNMR Analysis software. Correlation times were calculated based on Equation 3 (19).
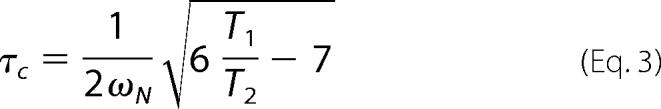 |
Paramagnetic Relaxation Enhancement (PRE) Experiments
To address the binding orientation of a bivalent ligand, the peptide X-DAPA-MPPPGMRPPPPGMRGLL, X represents the TEMPO moiety and DAPA the 2,3-diaminopropionic acid (TEMPO-SmB-2), was synthesized (EMC Microcollections). 15N-Labeled samples (0.2 mm) of tandem-WW WT and tandem-WW domain mutants were complexed with TEMPO-SmB-2 added in 6-fold excess. The TEMPO-based PRE effect for the two constructs was analyzed by a comparison of the peak heights of NH resonances in the case of active and inactive spin labels by 15N-HSQC-SOFAST experiments (20). The inactivation of the TEMPO group within the ligand was achieved by the addition of a 2-fold excess of ascorbic acid.
Homology Modeling of FBP21-Tandem-WW in Complex with a Bivalent SmB Ligand (SmB Amino Acid 213–227)
Each FBP21-WW domain bound to a ligand segment was modeled separately. As a starting point the crystal structure of the FE65 WW-domain in complex with a PRS peptide (Protein Data Bank code 2HO2) was used. Missing atoms were added using default internal coordinates of CHARMM22 (21, 22). Furthermore, several possible binding modes were considered. The criteria to select the appropriately bound ligand segments are: (i) maximal contact area between ligand and pocket, (ii) fitting of the prolines into the known pocket of WW-domains, (iii) favorable hydrophobic contacts of the ligand with the WW-domain, and (iv) formation of a conserved hydrogen bond between the pocket tryptophan Nϵ (in both WW domains) and the carbonyl group of the residue i+1 to the last proline in the canonical orientation or the carbonyl group of residue i−2 in the inverted orientation. Both ligand segments bound to the individual WW domains were connected to a linear structure followed by the introduction of the interdomain linker. The model was subsequently optimized using the CHARMM22 force field. The procedure requires a stepwise minimization by gradually relaxing the structural strain as described (23, 24). Finally global energy minimization without constraints was applied. Backbone root mean square deviations of WW1/WW2 compared with the first coordinate set of NMR structure PDB code 2JXW is 0.93 Å/1.53 Å for the canonical and 1.01 Å/1.72 Å for the inverted binding mode.
RESULTS
FBP21-Tandem-WW Domains Interact with RNA-processing Factors
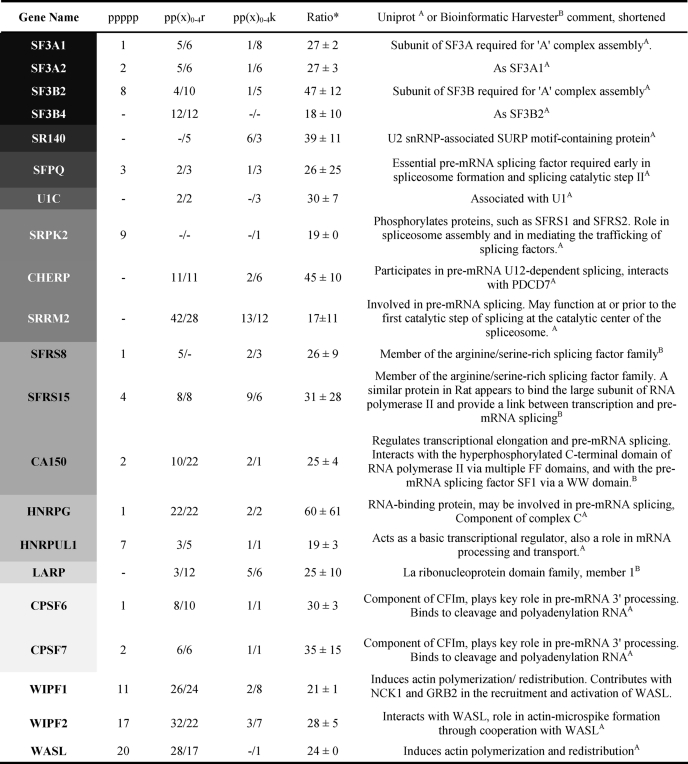
Because we were interested in the characteristic features of the FBP21-tandem-WW domains binding to PRS containing proteins we started out by a proteomic experiment based on SILAC/MS. We thereby utilized an epitope inhibition approach as it was previously described for GYF and UEV domains (7, 25) and that capitalizes on the specific isotope enrichment of proteins bound to a binding site blocked by an inhibitor in the second pulldown. We used the SmB-2 peptide (supplemental Table S1) from the known interaction partner SmB as an inhibitor and obtained a large number of highly enriched proteins. To increase the robustness of our results the second experiment was performed with inversely labeled cell lysates (see “Experimental Procedures”). Furthermore, we only considered proteins meeting stringent enrichment criteria as putative binders and the list of the hundred most highly enriched proteins is given under supplemental Table S2. Interestingly, 74% of the proteins in this list are associated either with mRNA processing or RNA binding in agreement with previous results obtained with the second WW domain of FBP21 (6). A prominent fraction of these targets is functionally associated with the spliceosome (supplemental Fig. S1) and only a few expected binding partners of FBP21 could not be detected in our pulldown (6), e.g. the known interactor SmB/B′, which runs close to the GST-WW fusion construct in SDS-PAGE. However, the presence of SmB/B′ in the co-precipitate was independently demonstrated by Western blot analysis (7) and binding to individual SmB motifs was ensured by peptide-walk SPOT analysis (supplemental Fig. S2). Peptide SPOT analysis also showed that the occurrence but not the exact position of a positive charge is important for binding (supplemental Fig. S3).
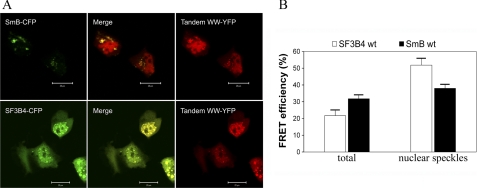
Closer inspection of the highly enriched spliceosomal proteins shows that many of them can be grouped into submodular entities of known function (supplemental Fig. S1). For example, the snRNP particles of the early spliceosome (U1/U2 snRNPs) were almost completely represented in the highly enriched fraction of the pulldown and this list further includes spliceosomal core and U5 snRNP proteins. We conclude that PRS binding by FBP21-tandem-WW is of sufficient strength and lysis conditions are sufficiently mild to allow co-elution of the entire subspliceosomal complexes. In addition to the U2 and U5 snRNP components 3′-RNA-processing proteins belonging to the CPSF or PABP family were also found within the cohort of highly enriched proteins. In contrary, RNA 5′-binding molecules were rare. Three additional classes of protein families are overrepresented as potential binding partners, namely proteins of the DEAD box RNA helicases family (DDX proteins), RNA-binding motif proteins, and Ser/Arg-rich pre-mRNA splicing factors (SFRS proteins). SFRS15 and SFRS8 both contain many PRS that may also serve as interaction sites for the tandem-WW domains of FBP21 (Table 1). Of particular interest was the finding that most proteins of the SF3 complex are among the highly enriched proteins (Table 1 and supplemental Fig. S1). The SF3 complex plays a pivotal role in maturation of the early spliceosome with the SF3B subunit enabling branch point recognition (26), whereas its dissociation commits the spliceosome to its first catalytic step (27). Several components of SF3 contain multiple PRS motifs and the SF3B subunit directly recognize the branch point region of the intron (26). Similar to FBP21, small molecule interference with SF3B leads to the modulation of VEGF receptor expression (28, 29). We therefore exemplarily investigated the colocalization of the newly identified interaction partner SF3B4 in living cells and compared its cellular interaction with the compartmentation of the known binding partner SmB/B′. Although full-length FBP21 fused to the N terminus of both CFP and YFP was toxic to HEK cells when overexpressed, we performed cellular experiments and subsequent in vitro biophysical experiments with a fragment of FBP21(122–196). As shown in Fig. 1A, this FBP21-tandem-WW domain construct shows nuclear accumulation, which is also observed for SmB and full-length SF3B4. Overexpression of the tandem-WW domain construct with either of the two target proteins expressed as corresponding CFP- or YFP-fusion constructs leads to significant FRET. The FRET efficiency resulting from energy transfer from SF3B4-CFP to tandem-WW-YFP was 21.7 ± 2.9% (mean ± S.E., n = 98) in total (Fig. 1B). Considering nuclear speckle localization alone a higher FRET efficiency of 51.9 ± 4.2% (n = 15) was observed. Coexpression of SmB-CFP and tandem-WW-YFP leads to overall FRET efficiency of 31.8 ± 3.0 (n = 48) and 38.0 ± 2.0% if only nuclear speckles were considered (Fig. 2B, n = 24).
TABLE 1.
Multiple PRMs found within the 100 most highly enriched proteins
The gray intensity of the gene name column correlates with the protein complex integration/molecular function (spliceosome/splicing) and is listed top-down according to common function or localization with FBP21. Number of motifs in front of the slash represents canonically oriented motifs (N- to C-terminal), whereas those behind the slash are noncanonical (C- to N-terminal). Means of enrichment (ratio) and S.D. results are from two independent experiments, whereby in the second replicate the labeling strategy was inverted (“sample” versus “control,” see “Experimental Procedures”).
FIGURE 1.
In vivo interaction of FBP21-tandem-WW domains with splicing factors. A, colocalization of SmB-YFP and FBP21-tandem-WW-YFP (upper panel) and SF3B4-CFP and FBP21-tandem-WW-YFP (lower panel) in nuclear speckles. HEK293FT cells were transfected with the respective constructs and analyzed on a Leica SP2 confocal microscope. The bar is 20 μm. B, FRET efficiencies for the energy transfer from SF3B4-CFP and SmB-CFP to FBP21 tandem-WW-YFP were determined as described under “Experimental Procedures.”
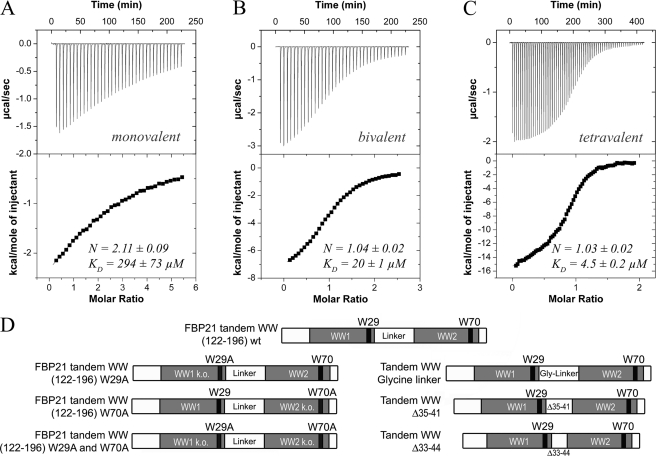
FIGURE 2.
Increase in affinity due to elevated ligand valency for the interaction of the FBP21-tandem- WW domains with SmB-derived peptides detected by isothermal titration calorimetry and scheme of constructs used in this study. Shown are the interactions detected by ITC of the WT tandem-WW domain of FBP21 with a supposedly monovalent (A), bivalent (B), and tetravalent (C) ligand. Upper panel, shows enthalpy changes upon injection of peptide into the measurement cell containing the FBP21-tandem-WW domains. Bottom panel, shows integrated power peaks fitted with a 1:1 model. Enthalpy and entropy changes are listed in Table 2. S.D. results from at least two experiments. Peptide sequences are listed under supplemental Table S1. D, scheme of constructs. Top construct, position 122–196 of FBP21-tandem-WW domains WT (WT). Second line left, the tryptophan at position 29 (corresponds to position 150 in the full-length protein) was substituted by alanine resulting in loss of binding of the N-terminal WW domain (W29A). Second line right, the tryptophan at position 70 (corresponds to position 191 in the full-length protein) was substituted by alanine and compromises the binding of the C-terminal WW domain (W70A). Third line left, both tryptophans at positions 29 and 70 were substituted by alanine resulting in a construct with two compromised WW domains (W29A/W70A). Third line right, the linker region regarding positions 35–42 (corresponding to 156–163 in the full-length protein) was replaced by a highly flexible GGGGSGGG stretch (glycine linker), enhancing the flexibility of the linker. Bottom line left, linker residues 35–41 (156–162 in the full-length protein, Δ35–41) were deleted. Bottom line right, the linker region at positions 33–44 (154–165 in the full-length protein, Δ33–44) was deleted.
Ligand Valency Strongly Affects Binding Affinities
To assess the ligand binding properties of SmB/B′ in regard to the WW domains of FBP21 we first fragmented the PRS tail into individual peptides. Four peptides each containing a potential binding motif were analyzed for their interaction with the tandem WW domains by NMR and isothermal titration calorimetry (supplemental Fig. S4). All of the peptides induced very similar chemical shift changes in both of WW domains of the tandem construct and therefore indicate a conserved recognition mode. This is in line with the very similar interaction profile of the individual WW domains with respect to longer overlapping PRS sequences (15–19 amino acids in length, see supplemental Fig. S2). The binding affinities for the individual peptides differ, with the best individual peptide still displaying a very high KD of 220 μm. We then assessed the importance of multivalency within the PRS ligand by measuring ITC curves for SmB ligands containing 1 (SmB-1), 2 (SmB-2), and 4 (SmB-4) binding motifs and for SF3B4 comprising 1 (SF3B4-1) or 2 (SF3B4-2) motifs. A large effect on affinities can be observed when comparing a bivalent and a monovalent PRS ligand. Here, reductions in affinity >10 are observed with KD values of 20 ± 1 μm for the interaction with a ligand containing two binding sites and 295 ± 73 μm for a monovalent ligand (Fig. 2, A and B). A similar difference in affinity was observed when comparing the SF3B4-derived peptides with either one (SF3B4-1) or two (SF3B4-2) binding sites (KD values of 570 ± 5 and 38 ± 2 μm, respectively, Table 2). The importance of a second proline-arginine motif in longer ligands is also evident by analyzing a truncation series of peptide spots from bivalent SmB peptides (supplemental Fig. S5). Here the intensity arising from bound tandem-WW domains is significantly reduced when relevant residues of either of the two motifs are deleted. To obtain information about the contribution of each single domain to the binding affinity we determined binding constants for tandem-WW constructs where binding-relevant tryptophans were substituted by alanine. NMR spectroscopy showed that the mutations did not hamper folding of the WW domains (data not shown), whereas binding to the respective PRS motifs was compromised by the mutation (10). Substitution of the binding relevant Trp-29 or Trp-70 by alanine resulted in KD values of ∼60 μm for the interaction with SmB-2 (Table 2). A moderate decrease in affinity compared with the WT construct was also observed for the interaction of these W29A and W70A mutants with SF3B4-2 (Table 2). When both Trp were replaced, no relevant binding between the tandem-WW construct (W29A/W70A) with SmB-2 and SF3B4-2 was observed (data not shown).
TABLE 2.
Thermodynamic constants determined by ITC
S.D. results from at least two experiments. The peptides used in this experiment are listed under supplemental Table S1.
| N | KD | ΔH | −TΔS | |
|---|---|---|---|---|
| μm | kJ/mol | |||
| Smb-1 | ||||
| WT | 2.1 ± 0.1 | 295 ± 70 | −21 ± 1 | 2 ± 1 |
| Smb-2 | ||||
| WT | 1.04 ± 0.02 | 20 ± 1 | −37 ± 2 | 10 ± 2 |
| W29A | 1.15 ± 0.20 | 62 ± 10 | −38 ± 3 | 16 ± 3 |
| W70A | 0.98 ± 0.06 | 60 ± 1 | −35 ± 1 | 12 ± 1 |
| Δ35–41 | 1.05 ± 0.02 | 26 ± 4 | −37 ± 2 | 13 ± 2 |
| Δ33–44 | 0.98 ± 0.02 | 33 ± 12 | −21 ± 5 | −3 ± 6 |
| Glycine linker | 0.99 ± 0.09 | 40 ± 9 | −46 ± 5 | 23 ± 6 |
| Smb-4 | ||||
| WT | 1.03 ± 0.02 | 4.5 ± 0.2 | −65 ± 4 | 37 ± 5 |
| W29A | 0.74 ± 0.02 | 50 ± 5 | −51 ± 1 | 28 ± 1 |
| W70A | 0.72 ± 0.03 | 35 ± 2 | −55 ± 4 | 31 ± 4 |
| Δ35–41 | 1.02 ± 0.01 | 2.7 ± 0.1 | −61 ± 1 | 32 ± 1 |
| Δ33–44 | 1.04 ± 0.02 | 1.8 ± 0. 3 | −53 ± 2 | 22 ± 2 |
| Glycine linker | 1.05 ± 0.02 | 4.7 ± 0. 2 | −62 ± 1 | 33 ± 1 |
| SF3B4–1 | ||||
| WT | 2.0 ± 0.1 | 570 ± 5 | −8 ± 1 | −9 ± 1 |
| SF3B4–2 | ||||
| WT | 1.02 ± 0.02 | 38 ± 2 | −30 ± 1 | 6 ± 1 |
| W29A | 1.04 ± 0.03 | 134 ± 32 | −27 ± 1 | 7 ± 1 |
| W70A | 1.12 ± 0.10 | 91 ± 11 | −19 ± 1 | −3 ± 1 |
| Δ35–41 | 1.03 ± 0.01 | 38 ± 4 | −34 ± 2 | 10 ± 2 |
| Δ33–44 | NDa | ND | ND | ND |
| Glycine linker | 1.11 ± 0.07 | 43 ± 5 | −35 ± 8 | 12 ± 8 |
a ND, not determined.
To further investigate the effect of multiple motif recognition we studied interaction of the WW domain constructs with a SmB peptide containing four PRS motifs in ITC experiments. An apparent KD of 4 μm was measured for the WT construct, indicating that multivalency of the ligand further enhances affinity (Fig. 2C). When the W29A and W70A mutant constructs were used, a ∼10-fold higher KD was observed, indicating that the WW domain tandem arrangement is more efficient in promoting affinity enhancement at higher ligand valencies. Affinity enhancement for the long SmB-4 ligand is driven by increased enthalpy (Table 2), whereas entropy compensation is observed in all cases. Furthermore, the stoichiometry for complex formation of the WW1-WW2 construct with monovalent ligands was 1:2 (Fig. 2), whereas it was 1:1 for all longer peptides, including SmB-4 (Table 2). NMR chemical shift perturbation showed that SmB-1 and SF3B4-1 affect both WW domains of FBP21, suggesting the binding of two peptides to a single tandem-WW fragment (supplemental Fig. S6). In contrast, when more than one motif was available for binding the tandem-WW domains was engaged in a bivalent complex. Interestingly, the binding stoichiometry to the tetravalent ligand, on average, determined by ITC was still 1:1 and this was confirmed by NMR relaxation measurements (supplemental Fig. S7) that showed very similar overall correlation times values for the tandem WW domains in complex with either bivalent (τc = 8 ± 0.3 ns) or tetravalent ligands (τc = 9 ± 0 ns), both in agreement with a complex of an approximate molecular mass of 13 kDa.
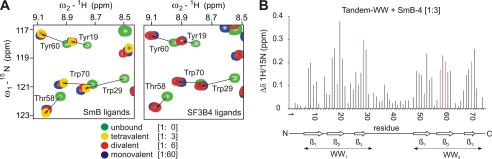
To investigate whether these bivalent complexes preserve the known binding pockets of WW domains, NMR chemical shift experiments were used to map the respective binding epitopes. The interactions are in fast exchange relative to the NMR time scale, independent of the length of the ligand. Clearly, the epitopes affected by the individual SmB and SF3B4 ligands are very similar and comprise the expected proline-binding pockets within both WW domains (supplemental Fig. S6) similar to those reported by Huang et al. (10). Interestingly we could observe a clear relationship between the saturation point of chemical shift changes and the number of presented PRS within the ligand. An increase of potential binding sites leads to a decrease in the required concentration based on an increased affinity. Consequently, whereas a 3-fold excess of the tetravalent ligand results in similar shifts as a 6-fold excess of the bivalent and a 60-fold excess of the monovalent peptide (Fig. 3). These findings are in line with our ITC data (Table 2) and show that affinity enhancement is correlated with the multivalency of the ligand.
FIGURE 3.
Chemical shift changes of the tandem-WW domains of FBP21 upon binding different PRS peptides. A shows an overlay of 1H-15N-HSQC spectra of the FBP21-tandem-WW domains unbound and after saturation with peptides containing a different number of PRS: tetravalent (1:3 ratio), bivalent (1:6), and monovalent (1:60). The sequence of the peptide ligands was deduced from the known interaction partner SmB (left overlay) and from the new binder SF3B4 (right overlay), respectively (supplemental Table S1). The NH backbone resonances of FBP21-tandem-WW affected by the respective binding event are very similar for all ligands as exemplified for Trp-29 and Trp-70 located in the central proline binding pocket of WW1 and WW2, respectively. B, comprehensive mapping of weighted 15N-1H chemical shifts for backbone NH resonances of FBP21-tandem-WW upon binding the tetravalent SmB ligand (Smb-4). Chemical shifts for residues shown in A are colored in red.
The Effect of Linking the Two WW Domains
To address the question, whether the binding of a bivalent ligand leads to an altered flexibility within the protein backbone, we compared R2 rates for backbone NH resonances of the unbound tandem-WW and the tandem-WW domains in complex with the bivalent SmB-2 ligand. The comparison was based on averaged and truncated R2 mean values for WW1 (amino acids 7–29), the linker region (amino acids 37–45), and WW2 (amino acids 48–70). We could observe a significant increase of the R2 value for the linker region (unbound: 7.59 ± 0.16 s−1, bound: 9.12 ± 0.1 s−1) and for WW2 (unbound, 10.92 ± 0.15 s−1; bound, 16.45 ± 0.14 s−1). The R2 value of the first WW domain was not affected (unbound, 17.25 ± 0.82 s−1; bound, 16.53 ± 0.14 s−1) by the binding event. We therefore asked the question of how far changes in linker length would influence coupling of the WW domains and whether correct spacing between the domains is critical for binding to bi- or multivalent ligands. To assess linkage phenomena, we used variants that either contained a more flexible linker of the composition GGGGSGGG or truncation constructs with either 7 or 12 amino acid deletions (Fig. 2D, a scheme of the constructs used here). Dissociation constants for these artificial linker constructs did not differ significantly from WT values (Table 2), indicating that linker composition and flexibility between the domains is not crucial for bivalent ligands with shortly spaced motifs.
Bidirectional Binding of PRS to Tandem-WW Domains
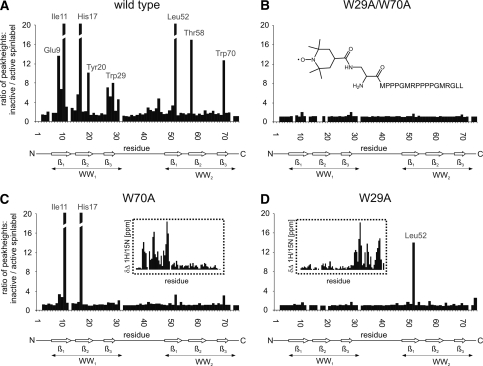
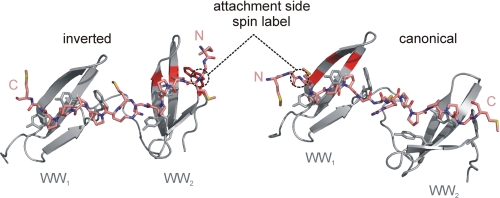
Ligand titration experiments of the tandem-WW domains indicate that bivalent ligands as well as the tetravalent ligand are in fast exchange relative to the NMR time scale, however, the binding mode for these longer ligands is incompletely understood. More specifically, it has been observed that individual motifs can bind in two orientations, due to the pseudosymmetry of the PPII helix. We therefore asked the question of whether such a bidirectional binding mode might be adopted by ligands of FBP21-tandem-WW domains. A TEMPO spin label was attached to the N terminus of the bivalent SmB peptide that should lead to PRE for residues in the vicinity of the unpaired electron. Relaxation enhancement was monitored by a comparison of the peak heights of NH backbone resonances before and after inactivation of the spin label. Fig. 4 shows the result of this experiment as it was performed either with the WT, the single domain and the double domain mutant. For the WT protein severe relaxation enhancement was observed for a distinct set of resonances close to the proline binding pockets in both domains. The NH resonances showing the largest relaxation enhancement in the individual domains of the WT protein were also affected in the respective single domain mutants. In the case of the double domain knock-out mutant no relevant relaxation enhancement takes place, indicating that the effects within the WT and single domain mutants are ligand-dependent. Most strikingly, the strong relaxation enhancement for NH resonances Glu-9, Ile-11, and His-17 in WW1 and for Leu-52, Thr-58, and Trp-70 in WW2 is not compatible with a single rigid binding mode. In the case of canonical binding, the N-terminal spin label would be in the vicinity of the backbone NH groups of the three most significantly affected WW1 residues (Fig. 5). The drastic PRE effect on the WW2 residues, however, could not be explained by this peptide orientation because the distance between the spin label and the NH group is larger than 15 Å, even when the mobility of the spin label was taken into account. However, assuming a second binding mode where the bivalent peptide binds in an inverted direction, the PRE effect on the three affected residues within the WW2 domain can be reasonably explained (Fig. 5). Vice versa, the position of the N-terminal spin label close to the WW2 domain residues cannot explain the effect on the NH groups of the WW1 domain residues in the case of an inverted peptide orientation. We therefore have to postulate a dynamic equilibrium of at least two orientations for the ligand as visualized by a conceivable homology model of FBP21-tandem-WW in complex with a bivalent SmB ligand (Fig. 5).
FIGURE 4.
Paramagnetic relaxation enhancement of 15N-1H resonances of the FBP21-tandem-WW domain upon binding of a bivalent PRS ligand, which comprises an N-terminal TEMPO label. The figure shows the ratio of peak heights for NH backbone resonances of FBP21-tandem-WW WT (A), the binding deficient “W29A/W70A” mutant (B), the single domain mutants W70A (C) and W29A (D) all titrated with a 6-fold excess of TEMPO-labeled bivalent SmB. The obtained ratios of peak heights were calculated by comparing signals from measurements with active and inactive TEMPO-labeled peptides. The structure of the TEMPO-labeled peptide is shown as an inlet in panel B. Chemical shift changes of NH backbone resonances are shown as an inlet (dotted lines) for the single domain mutants (C and D).
FIGURE 5.
Model of bidirectional binding of a bivalent SmB-derived ligand to the FBP21-tandem-WW domains confirmed by spin label experiments. The figure shows a model of the tandem-WW domains (gray) in complex with a bivalent peptide (SmB amino acids 213–227, pink). Side chains of hydrophobic residues (Trp, Tyr) involved in ligand binding are displayed. The ligand could bind either in a canonical (right) or inverted (left) orientation as confirmed by spin label experiments. The three most affected residues within each WW domain are highlighted in bright red. Their close spatial location to the spin label attachment side underlines the proposed bidirectional binding mode.
DISCUSSION
Affinity Enhancement by Multivalency
Proline-rich sequence recognition, with few exceptions, is characterized by recognition rules that allow for peptide degeneracy. Because individual PRS motifs in many cases occur as multiple variants within a single protein, it is of importance to understand the mechanism of multivalent binding at a molecular level. Also, it is not clear how far multiple low-affinity interactions of PRS with protein domains such as WW or GYF contribute to spliceosomal function. Although efficient experimental setups that unambiguously define the functional role of long PRS in proteins are missing we now provide evidence that multivalent binding partners are preferred targets for the tandem-WW domains containing protein FBP21. First, we show that proteins containing long PRS are highly enriched in the soluble cellular fraction when precipitated by FBP21-tandem-WW domains. Furthermore, the cellular interaction of functionally related proteins such as SF3B4 argues for a role of extended regions of PRS in early spliceosomal assembly. Interestingly, the affinity of the tandem-WW domains construct of FBP21 for single PRS motifs is very low, and this is conceivably true for several of the spliceosomal proteins identified here. For FBP21 binding is already significantly increased for a bivalent ligand (Table 2) and this increase is also observed for the W29A and W70A mutants, indicating the importance of ligand valency. However, whereas only a minor further increase in affinity is observed for the mutant constructs upon addition of the tetravalent ligand a 4–5-fold affinity enhancement is seen for the WT tandem-WW domains. Therefore, the effect of WW domain linkage is more prominent for a multi- versus a bivalent ligand and high-affinity binding in this system is seemingly optimized for efficient binding of the tandem-WW domains to multivalent PRS ligands. It has to be noted, however, that the definition of multivalency is not unambiguous for many of the proteins found in our pulldown, because motifs often overlap and because nonproline residues such as arginine are certainly of importance for the stabilization of the respective complexes.
Influence of the Linker Region
Our results indicate that a shorter linker between the WW domains does not interfere with the observed affinity enhancement upon binding to the SmB and SF3B4 ligands (Table 2). Although the structure of the tandem WW domains does not give any indication of direct domain-domain interactions (10) it has been found that the short ∼10 residue linker is dynamically more restricted in the complexed form. Although such motional coupling upon binding could come at an entropic cost, it will be more than counterbalanced by the experimentally observed negative enthalpies (Table 2). Furthermore, the observed overall affinity enhancement, at least in part, will be a consequence of avidity that is operative over a large range of distances between WW domains as well as the PRS motifs. In the case of the YAP and TAZ transcription factors the linker region between WW domains is significantly longer (∼28 residues) and a 6-fold higher affinity binding to a bi- versus monovalent ligand is observed, indicating that avidity, whereas still operative, decreases with increasing linker length (30). A somewhat different scenario has recently been described for the Smad ubiquitination regulatory factor, where the presence of an additional 26 residues between its WW2 and WW3 domain in the long isoform of the protein severely uncouples the binding to a composite ligand (31). However, in this case, the two WW domains directly contact each other and the small but common interface probably invokes true cooperativity. An interesting twist of domain-domain coupling has been observed in the Nedd4-like protein suppressor of deltex (Su(dx)), where an autoinhibitory interface between the third and fourth WW domain is released upon ligand binding (32). Similarly, the rigid helix connecting the two WW domains of splicing factor Prp40 leads to an overall structure of defined geometry, requiring a distinct topology of proline-rich ligands (33). However, even for Prp40 a set of different ligands can be bound by the tandem-WW domains arguing for promiscuity to be a general property of WW domain recognition (33).
Proline-rich Sequences in the Spliceosome
It is noteworthy that FBP21, as several other nuclear WW domain proteins, also contains an RNA binding zinc finger domain. RNA binding of these types of domains is thought to be of limited specificity and it is conceivable that such promiscuous interaction allows for movement of the domains along longer stretches of nucleotides. This has been investigated in more detail for DNA-binding domains and demonstrated that sparsely populated encounter complexes precede the formation of a specific DNA-binding complex (34). This process allows for inter- as well as intramolecular translocations and is steered by long-range electrostatic interactions. Charge attraction is found to be of importance for the tandem-WW domains binding of FBP21 to a bivalent ligand (supplemental Fig. S3) and we envisage that similar search processes are of importance for spliceosomal maturation (35). It is well conceivable that checkpoint decisions such as splice-site selection or poly(A) tail addition are complemented by highly dynamic protein recognition events. We suggest the PRS within spliceosomal proteins provide a scaffold for “delocalized” binding by protein adaptor domains such as WW and our data are in line with the existence of multiple complexes along long unstructured PRS. The observation that the long PRS sequences of core splicing proteins, such as SmB/B′ have not been assigned to measurable electron densities by EM (36) could be an indication that these sequences display intrinsic flexibility. Together with the observed degenerate binding of PRS recognition domains, a conformational space within several hundred Å of a certain intracellular locale would be accessible for the formation of encounter complexes within the spliceosome. It will be challenging to prove this hypothesis and to show the importance of semi-specific multivalent binding modes, because proline-rich sequences are generally not highly conserved during eukaryotic evolution. In many cases, certain proteins of the spliceosome have lost PRS recognition sites, whereas others acquired new ones. Despite this shuffling of PRS motifs within larger protein complexes more recent studies suggest that long PRS stretches are of physiological importance. For example, deletion of PRS in the transcription factor Ssdp1 leads to severe defects in mice neuronal development (37).
Ligand Orientation
Although it has been observed that individual WW domains can bind to PRS in two perpendicular orientations relative to the central binding pocket of the domain (38, 39), our study provides the first evidence that a bivalent peptide can bind in two different binding modes to WW domains arranged in tandem repeats. Bleaching of resonances in both WW domains is most easily explained by an inverted binding mode as suggested in Fig. 5 and the experiments with individually mutated WW1 or WW2 domains shows that unspecific binding of the spin-label to the proteins does not occur. However, we cannot rule out register shifts of the ligand or inter-domain flexibility to contribute to the observed line-broadening pattern. Although the exact binding mode needs to be investigated in future experiments, orientational equilibria can conceivably allow different encounter complexes to be formed in the early spliceosome and it remains to be seen in how far such flexibility is required for proper positioning of other RNA or protein recognition elements of the assembling spliceosome.
Supplementary Material
Acknowledgments
We thank M. Beyermann for synthesis of membrane-bound and of soluble peptides and Dr. Jana Sticht for carefully reading the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grants FG806 and SFB765.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1 and S2.
- PRS
- proline-rich sequence
- snRNP
- small nuclear ribonuclear particle
- FBP
- formin-binding protein
- SILAC
- stable isotope labeling by amino acids in cell culture
- ITC
- isothermal titration calorimetry
- PRE
- paramagnetic relaxation enhancement.
REFERENCES
- 1. Zarrinpar A., Bhattacharyya R. P., Lim W. A. (2003) Sci. STKE. 2003, RE8 [DOI] [PubMed] [Google Scholar]
- 2. Ball L. J., Kühne R., Schneider-Mergener J., Oschkinat H. (2005) Angew. Chem. Int. Ed. Engl. 44, 2852–2869 [DOI] [PubMed] [Google Scholar]
- 3. Kofler M. M., Freund C. (2006) FEBS J. 273, 245–256 [DOI] [PubMed] [Google Scholar]
- 4. Sudol M. (1996) Prog. Biophys. Mol. Biol. 65, 113–132 [DOI] [PubMed] [Google Scholar]
- 5. Mayer B. J. (2001) J. Cell Sci. 114, 1253–1263 [DOI] [PubMed] [Google Scholar]
- 6. Ingham R. J., Colwill K., Howard C., Dettwiler S., Lim C. S., Yu J., Hersi K., Raaijmakers J., Gish G., Mbamalu G., Taylor L., Yeung B., Vassilovski G., Amin M., Chen F., Matskova L., Winberg G., Ernberg I., Linding R., O'donnell P., Starostine A., Keller W., Metalnikov P., Stark C., Pawson T. (2005) Mol. Cell. Biol. 25, 7092–7106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kofler M., Schuemann M., Merz C., Kosslick D., Schlundt A., Tannert A., Schaefer M., Lührmann R., Krause E., Freund C. (2009) Mol. Cell. Proteomics 8, 2461–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan D. C., Bedford M. T., Leder P. (1996) EMBO J. 15, 1045–1054 [PMC free article] [PubMed] [Google Scholar]
- 9. Bedford M. T., Reed R., Leder P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10602–10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang X., Beullens M., Zhang J., Zhou Y., Nicolaescu E., Lesage B., Hu Q., Wu J., Bollen M., Shi Y. (2009) J. Biol. Chem. 284, 25375–25387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Otte L., Wiedemann U., Schlegel B., Pires J. R., Beyermann M., Schmieder P., Krause G., Volkmer-Engert R., Schneider-Mergener J., Oschkinat H. (2003) Protein Sci. 12, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seidel T., Golldack D., Dietz K. J. (2005) FEBS Lett. 579, 4374–4382 [DOI] [PubMed] [Google Scholar]
- 13. Lange S., Sylvester M., Schümann M., Freund C., Krause E. (2010) J. Proteome Res. 9, 4113–4122 [DOI] [PubMed] [Google Scholar]
- 14. Cox J., Mann M. (2008) Nat. Biotechnol 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 15. Kofler M., Heuer K., Zech T., Freund C. (2004) J. Biol. Chem. 279, 28292–28297 [DOI] [PubMed] [Google Scholar]
- 16. Fogh R., Ionides J., Ulrich E., Boucher W., Vranken W., Linge J. P., Habeck M., Rieping W., Bhat T. N., Westbrook J., Henrick K., Gilliland G., Berman H., Thornton J., Nilges M., Markley J., Laue E. (2002) Nat. Struct. Biol. 9, 416–418 [DOI] [PubMed] [Google Scholar]
- 17. Schanda P., Van Melckebeke H., Brutscher B. (2006) J. Am. Chem. Soc. 128, 9042–9043 [DOI] [PubMed] [Google Scholar]
- 18. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 19. Fushman D., Weisemann R., Thüring H., Ruterjans H. (1994) J. Biomol. NMR 4, 61–78 [DOI] [PubMed] [Google Scholar]
- 20. Schanda P., Brutscher B. (2005) J. Am. Chem. Soc. 127, 8014–8015 [DOI] [PubMed] [Google Scholar]
- 21. Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) J. Comput. Chem. 4, 187–217 [Google Scholar]
- 22. MacKerell A. D., Jr., Bashford D., Bellott M., Dunbrack R. L., Jr., Evanseck J., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., 3rd, Roux B., Schlenkrich M., Smith J., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 23. Popović D. M., Zarić S. D., Rabenstein B., Knapp E. W. (2001) J. Am. Chem. Soc. 123, 6040–6053 [DOI] [PubMed] [Google Scholar]
- 24. Gámiz-Hernández A. P., Kieseritzky G., Galstyan A. S., Demir-Kavuk O., Knapp E. W. (2010) ChemPhysChem 11, 1196–1206 [DOI] [PubMed] [Google Scholar]
- 25. Schlundt A., Sticht J., Piotukh K., Kosslick D., Jahnke N., Keller S., Schuemann M., Krause E., Freund C. (2009) Mol. Cell. Proteomics 8, 2474–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spadaccini R., Reidt U., Dybkov O., Will C., Frank R., Stier G., Corsini L., Wahl M. C., Lührmann R., Sattler M. (2006) RNA 12, 410–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lardelli R. M., Thompson J. X., Yates J. R., 3rd, Stevens S. W. (2010) RNA 16, 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ishiyama A., Iwatsuki M., Namatame M., Nishihara-Tsukashima A., Sunazuka T., Takahashi Y., Ōmura S., Otoguro K. (2011) J. Antibiot. 64, 381–384 [DOI] [PubMed] [Google Scholar]
- 29. Furumai R., Uchida K., Komi Y., Yoneyama M., Ishigami K., Watanabe H., Kojima S., Yoshida M. (2010) Cancer Sci. 101, 2483–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Webb C., Upadhyay A., Giuntini F., Eggleston I., Furutani-Seiki M., Ishima R., Bagby S. (2011) Biochemistry 50, 3300–3309 [DOI] [PubMed] [Google Scholar]
- 31. Chong P. A., Lin H., Wrana J. L., Forman-Kay J. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18404–18409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jennings M. D., Blankley R. T., Baron M., Golovanov A. P., Avis J. M. (2007) J. Biol. Chem. 282, 29032–29042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wiesner S., Stier G., Sattler M., Macias M. J. (2002) J. Mol. Biol. 324, 807–822 [DOI] [PubMed] [Google Scholar]
- 34. Iwahara J., Clore G. M. (2006) Nature 440, 1227–1230 [DOI] [PubMed] [Google Scholar]
- 35. Wahl M. C., Will C. L., Lührmann R. (2009) Cell 136, 701–718 [DOI] [PubMed] [Google Scholar]
- 36. Stark H., Dube P., Lührmann R., Kastner B. (2001) Nature 409, 539–542 [DOI] [PubMed] [Google Scholar]
- 37. Enkhmandakh B., Makeyev A. V., Bayarsaihan D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11631–11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang X., Poy F., Zhang R., Joachimiak A., Sudol M., Eck M. J. (2000) Nat. Struct. Biol. 7, 634–638 [DOI] [PubMed] [Google Scholar]
- 39. Verdecia M. A., Bowman M. E., Lu K. P., Hunter T., Noel J. P. (2000) Nat. Struct. Biol. 7, 639–643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.