Background: Glycans are synthesized in the Golgi by sequentially acting glycosyltransferases, but it is not known how their functions are coordinated in live cells.
Results: N- and O-glycosyltransferases form enzymatically active homo- and/or heteromeric complexes.
Conclusion: Glycosyltransferases function as physically distinct enzyme complexes rather than single enzymes.
Significance: The results help understand the overall functioning of the Golgi glycosylation pathways both in health and disease.
Keywords: Cancer, Fluorescence Resonance Energy Transfer (FRET), Glycosylation, Glycosyltransferases, Golgi pH Function, Complex Formation
Abstract
Glycosylation is one of the most common modifications of proteins and lipids and also a major source of biological diversity in eukaryotes. It is critical for many basic cellular functions and recognition events that range from protein folding to cell signaling, immunological defense, and the development of multicellular organisms. Glycosylation takes place mainly in the endoplasmic reticulum and Golgi apparatus and involves dozens of functionally distinct glycosidases and glycosyltransferases. How the functions of these enzymes, which act sequentially and often competitively, are coordinated to faithfully synthesize a vast array of different glycan structures is currently unclear. Here, we investigate the supramolecular organization of the Golgi N- and O-glycosylation pathways in live cells using a FRET flow cytometric quantification approach. We show that the enzymes form enzymatically active homo- and/or heteromeric complexes within each pathway. However, no complexes composed of enzymes that operate in different pathways, were detected, which suggests that the pathways are physically distinct. In addition, we show that complex formation is mediated almost exclusively by the catalytic domains of the interacting enzymes. Our data also suggest that the heteromeric complexes are functionally more important than enzyme homomers. Heteromeric complex formation was found to be dependent on Golgi acidity, markedly impaired in acidification-defective cancer cells, and required for the efficient synthesis of cell surface glycans. Collectively, the results emphasize that the Golgi glycosylation pathways are functionally organized into complexes that are important for glycan synthesis.
Introduction
A typical mammalian cell contains an estimated 1 billion individual proteins (1) of which about half are glycosylated (2, 3). Most of them bear N-linked and/or O-linked glycan chains that are synthesized co- or post-translationally in the endoplasmic reticulum and the Golgi apparatus by dozens of functionally distinct glycosidases and glycosyltransferases. These enzymes are thought to sequentially either remove or add single sugar residues from/to the growing oligosaccharide chains (4, 5). Their concerted functioning gives rise to a vast array of various glycan structures that often are cell-, tissue-, organism-, or species-specific (3).
Given the diversity of glycan structures produced and the competitive nature of many of the glycosyltransferases, it is remarkable that the cell can faithfully synthesize all the different glycan structures with such high precision and speed. Generally, cells can achieve such specificity by the spatial (or temporal) organization of a discrete subset of proteins into specific compartments and/or assemble them into functionally relevant protein complexes. These are already known to be utilized to coordinate many cellular activities such as intracellular or intercellular signaling networks, metabolic pathways, and protein folding processes (1). In addition to providing increased specificity and speed, the regulation of complex formation and/or disassembly provides additional means to modulate these processes in a cell.
Complex formation has been suggested to play a role in glycosylation processes as well (6–8). However, there is very little direct in vivo evidence for the formation of complexes by the Golgi glycosylation enzymes. This is due to the fact that complexes have been previously accessed mainly by using in vitro pulldown assays (for a review, see Ref. 6). Most of the existing in vivo data on N-glycosylation has also been obtained by methods that allow detection of the complexes only in the endoplasmic reticulum (9). Moreover, the functional relevance of the complexes in glycan synthesis has been confirmed only in a few cases by showing that the complexes are enzymatically active (10–12).
To investigate these issues, we have previously used the bimolecular fluorescence complementation approach (13) to screen N-glycosyltransferase complexes in the Golgi of live cells. We identified a novel N-glycosyltransferase complex consisting of GalT-I3 and ST6Gal-I. However, bimolecular fluorescence complementation does not allow detection of dynamic interactions and their potential regulation by the Golgi microenvironment. Therefore, we systematically screened for potential Golgi N- and O-glycosyltransferase interactions in live cells by using the recently developed FRET-flow cytometric quantification approach (14, 15). In contrast to microscopy-based FRET, which has been previously used to assess potential interactions between ganglioside-synthesizing glycosyltransferases (16), the flow cytometric quantification approach is well suited for systematic screening of multiple enzyme interactions in a large population of cells.
Here, we show that Golgi glycosyltransferases form enzymatically active homomeric and/or heteromeric complexes within both the N- and the O-glycosylation pathways. The fact that none of the N-glycosylation enzymes interacted with any of the O-glycosylation enzymes tested indicates that the two pathways are physically distinct. We also show that the interactions were mediated in nearly all cases by the catalytic domains of the interacting partners. Finally, we show that the formation and functioning of the heteromeric complexes are pH-dependent and markedly impaired in acidification-defective cancer cells. Taken together, the results provide novel insights into the overall functioning of the Golgi glycosylation pathways in higher eukaryotes.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
All glycosyltransferase expression plasmids were prepared using commercially available full-length cDNA clones (Imagenes GmbH, Berlin, Germany) and PCR amplification with specific primers (supplemental Table S1). For each enzyme, the PCR product was inserted into the pcDNA3 vector (Clontech Laboratories, Inc., Mountain View, CA), and the sequence was verified before use. The vector also contained sequences encoding a C-terminal five amino acid linker region (RSIAT) followed by either the monomeric Cerulean (mCer) (17), monomeric Venus (mVen) (18), HA (YPYDVPDYA), or FLAG (DYKDDDDK) tag. Non-tagged forms of the enzyme constructs were used for the enzyme activity measurements. These were prepared by replacing the five amino acid linker region with a stop codon.
Cell Cultivation and Transfections
COS7, HeLa, MCF7, and SW48 cells (all from ATCC, Manassas, VA) were cultivated in DMEM as described previously (19). One day after plating, the cells were transfected using 0.5 μg of each plasmid cDNA and the FuGENE 6TM transfection reagent (Roche Applied Sciences). After cultivation for 24 h at 37 or 30 °C, the cells were processed either for fluorescence microscopy, FRET, or enzyme activity measurements. Chloroquine (Sigma Aldrich) treatments were performed by adding the drug (40 μm) to cells either for 20 h (overnight treatment) or for the last 4 h before the measurements.
Fluorescence Microscopy
Cells were processed for indirect immunofluorescence as described previously (19). After fixation, the cells were permeabilized with 1 ml of 0.1% saponin in 1% BSA-PBS, pH 7.4, and stained with the anti-GM130 Golgi marker (610822, BD Biosciences) and secondary Alexa Fluor 594 -conjugated antibodies (Invitrogen). After staining, the cells were imaged using a Zeiss Observer. Z1 equipped with a LSM 700 confocal unit, 63× PlanApo oil immersion objective and appropriate filter sets for cyan fluorescent protein, YFP, and Alexa Fluor 594, and Zen2009 software (Carl Zeiss AG, Oberkochen, Germany). When appropriate, FRET images were taken and analyzed using the Youvan method.
Quantification of FRET Signal by Flow Cytometry
FRET measurements were done according to Banning et al. (14). Briefly, 1 day after plating, the cells were transfected with the FRET plasmid constructs that encoded selected enzymes tagged with either the mCer or the mVen. After 24 h, the cells were detached from the plates with 2× trypsin-EDTA (Sigma-Aldrich), diluted, and analyzed on a flow cytometer (CyFlow, Partec GmbH, Münster, Germany) equipped with appropriate filter sets for mCer (excitation, 405 nm; emission, 425–475 nm) and mVen (excitation, 488 nm; emission, 515–540 nm). In each measurement, 2–5 × 103 cells expressing both mCer and mVen were selected by gating (for setting and validation of the gates, see supplemental Fig. S1) and quantified in triplicate from three different experiments (as the mCer/mVen ratios). The FRET filter set (excitation, 405 nm; emission, 515–540 nm) and FloMAX software were used for quantification. The FRET signal is presented as the percentage of cells expressing both constructs that are FRET-positive (FRET+; mean ± S.D.). A value higher than 5% was considered relevant (14).
Determination of Cell Surface Sialic Acid Content and T-antigen Expression Levels
Cell surface sialic acid content was determined by using a fluorescence-based approach developed recently by Zeng et al. (20). Briefly, cells transfected with the indicated plasmids were subjected first to periodate oxidation (Sigma-Aldrich), then treated with aminooxy-biotin (Biotium, Inc. Hayward, CA), and finally, with FITC-conjugated streptavidin (Invitrogen). After washing, stained cells were either examined by fluorescence microscopy or analyzed by flow cytometry (supplemental Fig. S2). When appropriate, cells were treated with sialidase A (10 units/sample, ProZyme, Inc., Hayward, CA) for 90 min at 37 °C prior to staining. Cell surface T-antigen expression was quantified mainly as described previously (21). Briefly, cells grown on plates were treated first with peptide N-glycosidase F (20,000 units/sample, New England Biolabs) to remove cell surface N-glycans (22). After blocking with 1% BSA-PBS (pH 7.4) and washing, the cells were stained with Alexa Fluor 594-conjugated peanut agglutinin (3 μg/ml, Invitrogen). After washing, the cells were detached from the plates with EDTA-PBS, resuspended in fresh DMEM, and analyzed by flow cytometry.
In Vitro Enzymatic Activity Measurements
Activity measurements were performed with slight modifications as described by Hansske et al. (23) and Legaigneur et al. (24). In short, cells grown in 35-mm plates were solubilized in 500 μl of lysis buffer (1% Triton X-100, 300 mm NaCl, pH 7.0) supplemented with 1× CompleteTM EDTA-free protease inhibitor mixture (Roche Diagnostics). Enzyme activity measurements were performed using a reaction buffer, which contained 50 mm Tris-HCl, pH 7.0, 0.5% Triton X-100 and 2.5 mm ATP. For galactosyltransferase activity measurements, the reaction buffer was supplemented with 20 mm MnCl2 and 1 μCi of UDP-[3H]galactose (40 Ci/mmol, American Radiolabeled Chemicals, Inc., St. Louis, MO) and 60 μm non-labeled UDP-galactose. For sialyltransferase activity measurements, the reaction buffer was supplemented with 2 mm MnCl2, 1 μCi CMP-[14C]sialic acid (0.55 Ci/mmol, American Radiolabeled Chemicals) and 60 μm CMP-sialic acid. Ovalbumin (18 mg/ml) and asialofetuin (6 mg/ml, both from Sigma-Aldrich) were used as acceptor proteins for galactosyltransferase and sialyltransferase activities, respectively. Enzyme activity of the heteromeric GalT-I·ST6Gal-I complex was measured using 2 mm MnCl2 and both nucleotide sugar donors simultaneously. Scintillation counting was used for quantification. The distributions of enzyme proteins in the fractions were visualized after TCA precipitation, SDS-PAGE, and immunoblotting by using anti-HA and anti-FLAG antibodies.
Size-exclusion Chromatography
COS7 cells transfected with GalT-I-HA or ST6Gal-I-FLAG constructs were solubilized for 1 h on ice using 500 μl of lysis buffer (300 mm NaCl, 1% Triton X-100, in phosphate buffer, pH 7.0, 1× complete EDTA-free protease inhibitor mixture (Roche Diagnostics). Cell lysates were precleared by centrifugation (10,000 × g at 4 °C for 10 min) before loading onto a Bio-Gel A0.5m column (Bio-Rad). Proteins were fractionated using 0.1% Triton X-100, 300 mm NaCl in phosphate buffer, pH 7.0. Protein concentrations in the fractions (1 ml) were measured using a Bio-Rad Protein Assay. Every second fraction was precipitated by adding TCA to 10% (1 h, +4 °C) and washed twice with 5% TCA and once with acetone before subjecting to SDS-PAGE. The column was calibrated using blue dextran (void volume marker) and molecular weight markers β-amylase (200 kDa), alcohol dehydrogenase (120 kDa), and bovine serum albumin (60 kDa) (Sigma-Aldrich). In vitro activity of the remaining fractions was measured as described above.
Immunoblotting
SDS-PAGE and immunoblotting was performed as described previously (19). Briefly, cells transfected with the selected enzyme constructs were lysed in 2× SDS sample buffer and subjected to SDS-PAGE before transfer onto a nitrocellulose membrane. Monoclonal anti-HA, anti-FLAG, and anti-α-tubulin antibodies (Sigma-Aldrich), peroxidase-conjugated secondary antibodies (P.A.R.I.S., Compiègne, France), ECL substrates, and Chemidoc XRS+ (Bio-Rad) were used for the visualization of the bands. NIH ImageJ software (version 1.43) and Image Lab 3.0 software were used for quantification.
Identification of Interacting Domains
To identify the interacting domains for each enzyme, we utilized the domain swapping approach. We first constructed two general plasmid vehicles, one that encodes the first 120 amino acids (i.e. the transmembrane domain plus stem region (TMDS) of the ST3Gal-III), and another that encodes only the catalytic domain of the ST6Gal-I and lacks the first 120 amino acids of the full-length protein. These domains were selected for this purpose, as our preliminary tests (supplemental Fig. S3) showed that they are not required for the homomerization of either enzyme.
In the case of ST3Gal-III, we PCR amplified the required cDNA region using the full-length ST3Gal-III cDNA clone as a template. The PCR product was then inserted into the mCer- or mVen-encoding pcDNA3 plasmids using the HindIII/XhoI restriction sites. The cDNAs encoding the catalytic domains of the test enzymes were also PCR-amplified and inserted into the ST3Gal-IIITMDS-mCer/mVen vector using XhoI and XbaI restriction sites. To screen for the TMDS domain interactions, we PCR amplified the relevant cDNA region from ST6Gal-I and inserted it into the mCer or mVen pcDNA3 plasmids using the XhoI and XbaI restriction sites. The TMDS domains of the test enzymes were then PCR-amplified and inserted into the ST6Gal-Icat-mCer/mVen plasmid using the HindIII and XhoI restriction sites. Construction of the C2GlcNAcT-1 plasmids required the use of the BamHI/XhoI cleavage sites. All of the primers used are listed in supplemental Table S2.
Statistical Analyses
Statistical analyses were performed using the Student's t test for the comparison of small normally distributed samples.
RESULTS
Formation of N-Glycosyltransferase Complexes
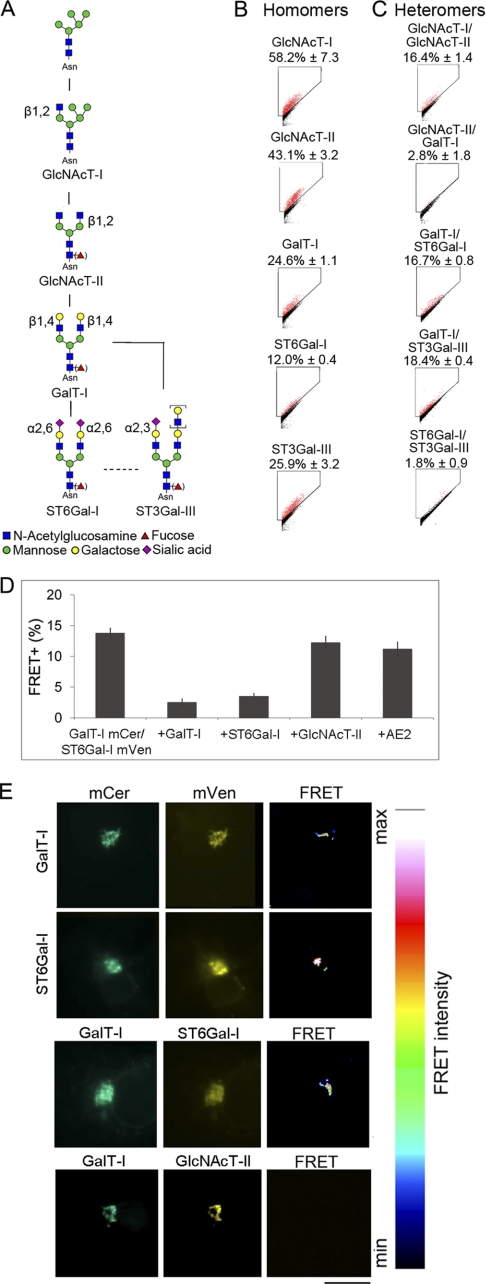
To systematically screen for potential interactions between the main Golgi N-glycosyltransferases, we selected known enzymes (Fig. 1A, for EC numbers and naming, see also supplemental Table S1), and in each case, first confirmed in COS7 cells that the constructs localized correctly in the Golgi (supplemental Fig. S4). Subsequent FRET measurements showed that all the main N-glycosyltransferases tested form enzyme homomers (FRET+, 12–58%) (Fig. 1B). Interestingly, the same enzymes were also found to form functionally relevant (i.e. sequentially acting) heteromeric complexes in the cells (Fig. 1C). The medial Golgi enzymes GlcNAcT-I and GlcNAcT-II (FRET+, 16.4 ± 1.5% (mean ± S.D.; n = 3)) (Fig. 1C) form one of these complexes and is likely involved in the processing of high mannose type glycans to complex type glycans. Previously, different approaches have been used to show that these two enzymes interact (9, 25). The complex may also involve Golgi mannosidase II (9) and other GlcNAcTs as well (26), although these were not studied here.
FIGURE 1.
Glycosyltransferase complexes in the N-glycosylation pathway. A, schematic representation of the N-glycosylation pathway and the enzymes involved (for enzyme names, see supplemental Table S1 and the KEGG pathway database. B, homomeric interactions. C, heteromeric interactions. All values are expressed as the mean of FRET+ (± S.D.) from three independent measurements using COS7 cells. FRET gates were set to exclude FRET negative cells (black). FRET+ cells are marked as red dots. D, inhibition of FRET+ signal by competing enzyme constructs. COS7 cells were transfected using the indicated plasmid constructs and quantified by flow cytometry as described under “Experimental Procedures.” The small decrease in FRET+ cells seen with GlcNAcT-II and AE2 co-expression is likely caused by the slightly reduced protein synthesis due to co-expression of a competing or non-competing construct. E, localization of the FRET+ signal as analyzed by FRET microscopy. Briefly, COS7 cells transfected with mCer- and mVen-tagged GalT-I (first row) or ST6Gal-I (second row) were fixed and examined by fluorescence microscopy. The FRET signal was determined from the images using the built-in FRET analysis program (Youvan). Note that the FRET signal between homomeric (first and second rows) and heteromeric (third row) enzyme pairs is detected in the Golgi region only. A non-interacting enzyme pair (GalT-I/GlcNAcT-II; fourth row) does not show any FRET signal. Scale bar, 10 μm.
The trans-Golgi enzymes GalT-I and ST6Gal-I (FRET+, 16.7 + 0.8%), or GalT-I and ST3Gal-III (FRET+, 18.4 + 0.4%) were also found to form heteromeric complexes with each other (Fig. 1C). These two complexes are likely responsible for the termination of N-glycan chains with galactose and sialic acid, differing only in the type of sialic acid linkage they make. In contrast, no interaction (FRET+, <5%) was detected between the medial Golgi enzyme GlcNAcT-II and the trans-Golgi enzyme GalT-I (Fig. 1C). The two sialyltransferases (ST6Gal-I and ST3Gal-III) also did not interact with each other. Cross-testing of all other possible enzyme combinations did not reveal any other heteromeric interactions between the N-glycosyltransferases tested (data not shown). The specificity of these interactions was verified by co-expressing both competitive and non-competitive enzyme/protein constructs together with the FRET enzyme constructs. Co-expression of non-tagged GalT-I or ST6Gal-I was able to significantly inhibit the interaction between the two trans-Golgi enzymes by 82 and 75%, respectively, whereas co-expression of either the GlcNAcT-II or AE2 (a non-relevant Golgi membrane protein (27, 28) was not (p > 0.05; Fig. 1D). FRET microscopy also confirmed that both the homomeric and heteromeric GalT-I·ST6Gal-I complexes localize almost exclusively in the Golgi, i.e. the compartment where they are known to function (Fig. 1E). No signal was detected in the Golgi between non-interacting GlcNAc T-II and GalT-I, as expected. Taken together, the results show that all the main N-glycosyltransferases form either homo- or heteromeric complexes in the Golgi of live cells.
Formation of O-Glycosyltransferase Complexes
Mucin type O-glycans are the main products of the O-glycosylation pathway, and alterations in them are important, e.g. for cancer progression (29–31). Their synthesis involves a number of both initiating and elongating O-glycosyltransferases, of which the various core structure-forming enzymes are best known (Fig. 2, see supplemental Table S1). Thus far, no complexes have been reported between any of the O-glycosyltransferases either in vivo or in vitro.
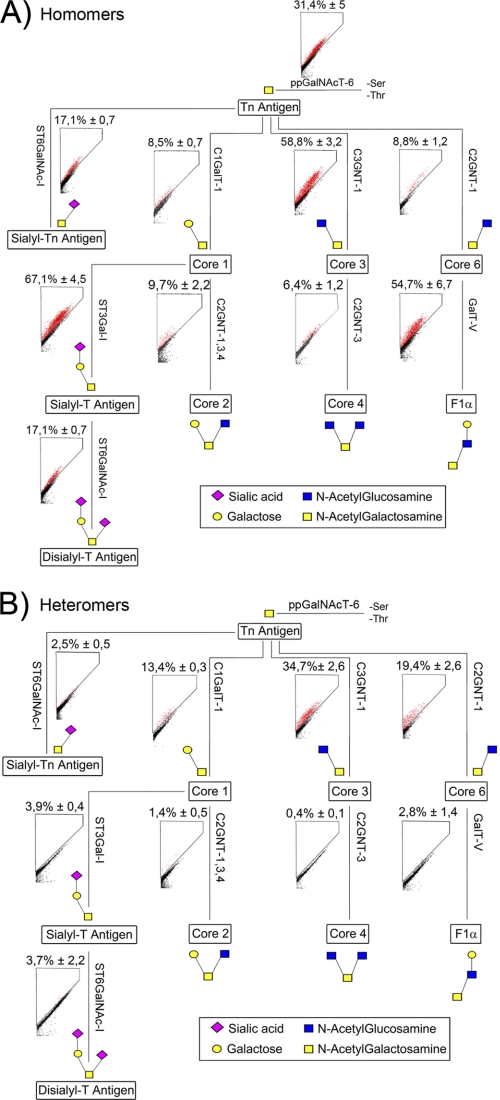
FIGURE 2.
Glycosyltransferase complexes in the O-glycosylation pathway. Schematic representation of the mucin-type core O-glycan structures and the enzymes involved (for enzyme names, see supplemental Table S1 and the KEGG pathway database). Enzyme names are given along their respective reaction paths. A, FRET measurements of the homomeric interactions were done as described in the legend to Fig. 1. All of the values are represented as percentages of FRET+ COS7 cells (mean ± S.D. from three independent measurements). B, heteromeric interactions. FRET measurements were done as described in the legend to Fig. 1. Localization data of all tested O-glycosyltransferases are shown in supplemental Fig. S5.
To investigate whether O-glycosyltransferases also form complexes in live cells, we first confirmed that the FRET enzyme constructs localized correctly in the Golgi of COS7 cells (supplemental Fig. S5). Subsequent FRET measurements revealed that all of the O-glycosyltransferases tested also form homodimers (Fig. 2A). Screening of potential heteromeric interactions between sequentially acting enzymes showed a clear interaction between the initiating ppGalNAcT-6 (the Tn-antigen-synthesizing enzyme) and each of the core 1, 3 and 6 forming enzymes (Fig. 2B). No interaction however, was detected between the initiating ppGalNAcT-6 and the enzymes that convert the initial core structures 1, 3 and 6 to core structures 2, 4 and F1α, respectively (data not shown). Moreover, we did not detect any interaction between the core 1 and 2, core 3 and 4, or core 6 and F1α forming enzymes (Fig. 2B), nor between the core forming enzymes and the sialyltransferases ST3Gal-I and ST6GalNAc-I (Fig. 2B) that synthesize the sialyl-Tn, sialyl-T and disialyl-T core structures. Moreover, cross-testing of all other potential interactions between O-glycosyltransferases did not reveal any additional interactions (data not shown). These findings show that in mucin-type core O-glycosylation, only the enzymes that are responsible for the synthesis of core 1, 3 and 6 structures form heteromeric complexes, whereas the enzymes involved in further elongation of O-glycans form only homomeric complexes. These data, however, do not exclude the possibility that the core 2, 4 and F1α forming enzymes may form heteromeric complexes with the enzymes that are required for the elongation of O-glycans.
N- and O-Glycosyltransferases Do Not Interact
Because both the N- and O-glycosyltransferases form complexes in live cells, it was important to test whether they form complexes with each other as well. Therefore, potential N- and O-glycosylation enzyme pairs were selected on the basis of their ability to use the same acceptor and donor substrate sugars. Cross-testing potential interactions between N- and O-GlcNAc-transferases (Fig. 3A), between N-galactosyl- and O-sialyltransferases (and vice versa, Fig. 3B), and between the initial O-GalNAc-transferase and N-Gal- or N-GlcNAc-transferases (Fig. 3C), however, revealed no significant FRET signal between any of the enzyme pairs tested. Testing of all other combinations between N- and O-glycosyltransferases (supplemental Table S3) gave similar results. Thus, N- and O-glycosyltransferases do not interact with each other, but rather, form heteromeric complexes only with the enzymes that operate in the same pathway. These data indicate that the two pathways are physically distinct from each other.
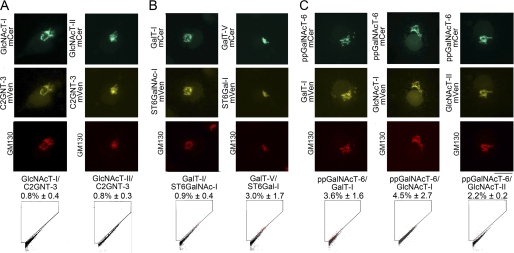
FIGURE 3.
Enzyme complexes between the N- and O-glycosylation enzymes. COS7 cells were transfected with plasmids encoding selected potentially interacting FRET enzyme pairs between N- and O-glycosyltransferases. The names of enzyme pairs tested are given below each micrograph. The localization of the enzyme pairs in the Golgi membranes (marked by GM130 antibody) was confirmed by fluorescence microscopy. A, O- and N- GlcNAc transferases. B, galactosyl- and sialyltransferases. C, GalNAc- and galactosyl- or GlcNAc transferases. The FRET signal (expressed as percentages of FRET+ cells, mean ± S.D.; n = 3) between the indicated enzymes was measured by flow cytometry as described in the legend to Fig. 1.
Functional Relevance of the Complexes
To assess the functional relevance of the glycosyltransferase complexes, we measured first the in vivo cell surface sialylation state in transfected COS7 cells by using the recently established biotin-streptavidin fluorescence-based protocol (20). In comparison to non-transfected cells (Fig. 4A), overexpression of GalT-I alone did not significantly increase cell surface sialylation (1.2-fold), whereas ST6Gal-I overexpression resulted in a 3.7-fold increase (p < 0.001). Co-expression of GalT-I and ST6Gal-I together increased the sialylation state 7.6-fold, compared with ST6Gal-I expression alone (p < 0.001). However, no increase was detected upon co-expression of ST6Gal-I with GlcNAcT-II or AE2 compared with ST6Gal-I alone (p values>0.05; Fig. 4A). Similarly, GalT-I and ST3Gal-III co-expression resulted in a marked increase (p < 0.001) in the cell surface sialic acid content in transfected cells relative to non-transfected cells or cells that were transfected with only one enzyme construct (Fig. 4A).
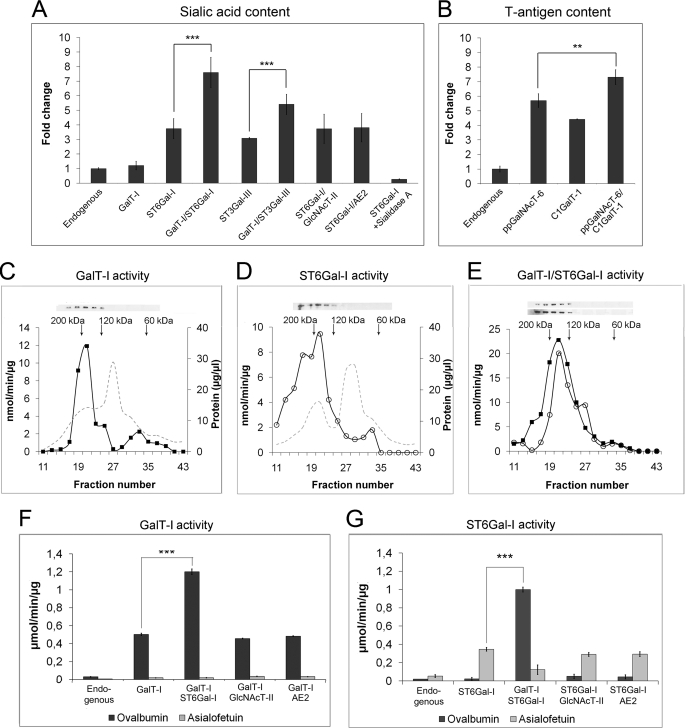
FIGURE 4.
Functional relevance of the enzyme complexes. COS7 cells were transfected with the indicated enzyme constructs and stained for sialic acid content (A) and T-antigen expression (B) as described under “Experimental Procedures.” Prior to T-antigen measurements, N-glycans were removed by peptide N-glycosidase F. Quantification was done by using flow cytometry. All measurements were made in triplicate and are expressed as fold changes relative to endogenous level (mean ± S.D.; n = 3). Shown are size-exclusion chromatography and in vitro activities of the fractions in cells expressing GalT-I alone (C), ST6Gal-I alone (D), or both GalT-I and ST6Gal-I (E). COS7 cells transfected with the appropriate plasmids were solubilized, fractionated, and subjected to enzyme activity measurements as described under “Experimental Procedures.” Enzyme activities are expressed as μmol/min/μg protein. Solid squares denote GalT-I activity values in fractions, and open circles denote ST6Gal-I activity. Arrows denote the standard proteins used to calibrate the column (Bio-gel A0.5m, Bio-Rad). Shown are GalT-I (F) and ST6Gal-I (G) activities of total cell lysates. Activities were measured as described above and are expressed as the mean (μmol/min/μg protein ± S.D.) from three independent measurements. **, p < 0.01; ***, p < 0.001.
The in vivo functional relevance of the O-glycosyltransferase complexes was assessed next by measuring the activity of the core 1 synthesizing complex using cell surface T-antigen expression level as a marker. Cells were treated with peptide N-glycosidase F to remove cell surface N-glycans prior to quantification. Overexpression of the initiating ppGalNAcT-6 or C1GalT-1 alone increased cell surface T-antigen expression by 5.7- and 4.4-fold, relative to non-transfected cells (p < 0.001; Fig. 4B), respectively. The increase observed with ppGalNAcT-6 is probably due to the increased availability of the Tn-antigen (Fig. 2B) for further galactosylation by endogenous C1GalT-1. Co-expression of ppGalNAcT-6 and C1GalT-1 resulted in a more pronounced increase in T-antigen expression (7.3-fold) relative to non-transfected cells. This increase was statistically significant (p < 0.01) when compared with cells expressing either ppGalNAcT-6 or C1GalT-1 alone.
The data presented above indicate that the enzymes are active in live cells. Whether the activity increase is due to complex formation, rather than the presence of monomeric enzymes, was assessed next. GalT-I and ST6Gal-I were selected as our target enzymes, mainly because this enzyme pair exhibited the highest activity upon co-expression and because their enzymatic activities can be readily measured using the established protocols. Fractionation of the cell lysates by size-exclusion chromatography showed that both the GalT-I and ST6Gal-I activities obtained from single (Fig. 4, C and D) or double-transfected (Fig. 4E) cells eluted in the fractions that correspond in size to homo- and heteromeric enzyme tetramers (∼180–190 kDa), and not to enzyme monomers (40–60 kDa). Western blotting (Fig. 4, C–E) of the fractions confirmed that the enzyme proteins also eluted in the same fractions.
To directly compare the enzymatic activities of the enzyme homo- and heteromers, we measured their in vitro activities in total cell lysates from GalT-I- and/or ST6Gal-I-expressing cells. Excess ovalbumin and asialofetuin were used as relevant acceptor proteins, respectively. As expected, overexpression of GalT-I alone increased galactosyltransferase activity against ovalbumin by 17.2-fold (p < 0.001; Fig. 4F), relative to the endogenous GalT-I activity. Interestingly, co-expression of ST6Gal-I, a later enzyme in the N-glycosylation pathway (Fig. 1A), increased GalT-I activity by 41-fold relative to the endogenous GalT-I activity. This represents a 2.4-fold increase over cells that express GalT-I alone (p < 0.001). Immunoblotting of GalT-I and ST6Gal-I proteins confirmed that this activity increase was not due to a higher enzyme protein expression level in the cells (supplemental Fig. S6). In contrast to ST6Gal-I, no increase in GalT-I activity was detected upon co-transfection with the non-interacting GlcNAcT-II enzyme or AE2, the two control proteins used in the measurements (p > 0.05; Fig. 4F). In all cases, GalT-I activity against the non-relevant acceptor, asialofetuin, was negligible. These results, together with the fact that a later enzyme (ST6Gal-I) can increase the enzymatic activity of the earlier enzyme (GalT-I) in the pathway, suggest that these terminal N-glycosyltransferases act cooperatively in N-glycan synthesis.
Sialyltransferase activity in the cell lysates was measured accordingly. Expression of ST6Gal-I alone (Fig. 4G) increased sialyltransferase activity against excess asialofetuin by 6.2-fold relative to non-transfected cells. ST6Gal-I activity was not increased upon co-expression with GalT-I, GlcNAcT-II, nor AE2 (p > 0.05; Fig. 4G), as expected because the N-glycans in asialofetuin contain terminal galactose residues and cannot be utilized by these enzymes. ST6Gal-I activity against ovalbumin was also negligible in all cases, consistent with the presence of terminal GlcNAcs in the N-glycans of ovalbumin (32–34). However, when ST6Gal-I was co-expressed with GalT-I, we observed a 15-fold increase in ST6Gal-I activity against ovalbumin relative to non-transfected cells. This represents a 2.4-fold higher sialyltransferase activity than the ST6Gal-I activity measured from single transfected cells against excess asialofetuin (p < 0.001; Fig. 4G). Although this GalT-I-mediated ST6Gal-I activity increase against ovalbumin could be explained just by increased galactosylation of the N-glycans of ovalbumin and their use as an acceptor for the sialyltransferase, it should be noted that both homomeric and heteromeric enzyme activities were measured in the presence of excess acceptor proteins. Therefore, we consider these data as additional evidence for the cooperative functioning of these two trans-Golgi enzymes. The findings are consistent with the in vivo and size-exclusion chromatography data (Fig. 4, A and E), showing that the enzymes have higher enzymatic activity when co-expressed.
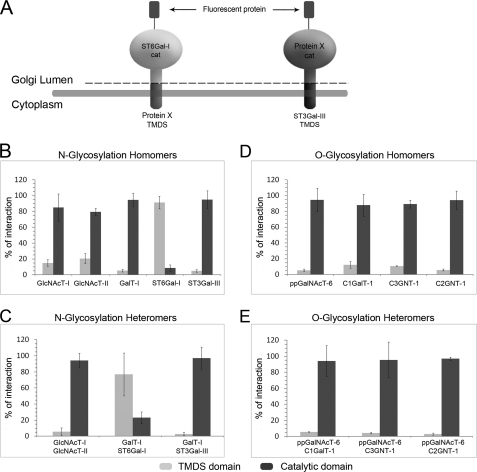
Characterization of Interacting Domains
Most, if not all, glycosyltransferases are type II membrane proteins consisting of an N-terminal transmembrane domain (with a short cytoplasmic tail), a stem region, and a luminal catalytic domain. To assess whether the catalytic or the TMDS is responsible for the interactions, we constructed (Fig. 5A) plasmid vectors that allowed shuffling of these domains with the corresponding domains from the other enzymes. FRET measurements showed (Fig. 5B) that homomeric interactions between N-glycosyltransferases are mediated mainly (>70%) by the catalytic domains of these enzymes. This holds true also for the medial Golgi enzymes GlcNAcT-I or GlcNAcT-II, although their interaction may partially involve the TMDS domains as well. The only exception was ST6Gal-I, in which the TMDS region appears to be mainly responsible for the homomer formation. For most enzymes, the same domains were also found to be involved in the formation of heteromeric complexes (Fig. 5C). The medial Golgi enzymes GlcNAcT-I and II interacted mainly via their catalytic domains, as did also GalT-I and ST3Gal-III. In contrast, the interaction between GalT-I and ST6Gal-I was mediated mainly by their TMDS domains. All of the O-glycosyltransferase homo- and heteromeric interactions (Fig. 5D and E) were found to be preferentially (>85%) mediated by the catalytic domains of the enzymes.
FIGURE 5.
Identification of interacting domains. A, schematic presentation of the chimeric constructs used. Preliminary screening tests are shown in supplemental Fig. S3. B and C, homo- and heteromeric interactions between N-glycosyltransferases in COS7 cells. D and E, homo- and heteromeric interactions between O-glycosyltransferases in COS7 cells. In each case, the FRET+ signal is expressed as percentages of the total interaction mediated by the TMDS and catalytic domains together (mean ± S.D.; n = 3). The FRET+ cells were quantified by flow cytometry as described in the legend to Fig. 1.
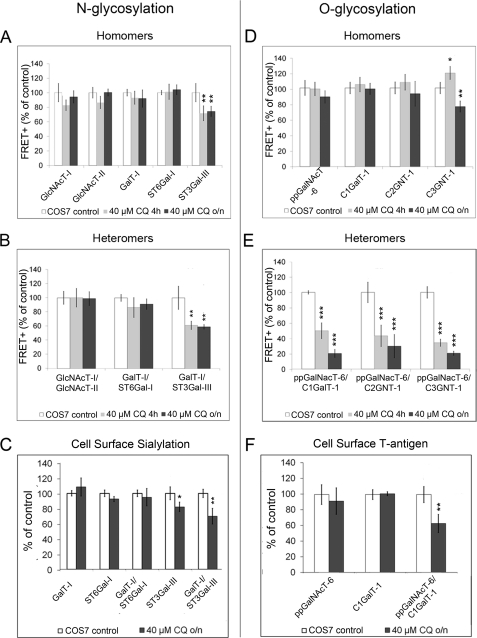
pH Sensitivity of Enzyme Complexes
Previously, we have reported that a slight drug-induced increase in Golgi pH results in mislocalization of ST3Gal-III (but not of ST6Gal-I) into the endosomal compartments, probably reflecting differences in complex formation (35). Therefore, we tested the pH sensitivities of the enzyme homo- and heteromers in live cells by using FRET. We treated cells with 40 μm chloroquine, which raises Golgi pH by ∼0.4 units (36), i.e. less than the increase found in cancer cells. FRET measurements showed that most of the N-glycosyltransferase homomers were insensitive to this pH increase. The only exception was ST3Gal-III homomers (Fig. 6A), which showed a slight but significant (p < 0.01) decrease in complex formation. N-glycosyltransferase heteromers behaved similarly, in that only the complex involving ST3Gal-III was significantly affected by the drug (p < 0.01; Fig. 6B). No marked differences were found between short (4 h) and overnight chloroquine treatment. Drug treatment also induced a concomitant and statistically significant decrease in the cell surface sialylation state in cells overexpressing ST3Gal-III and GalT-I/ST3Gal-III, relative to non-treated cells (p values<0.05; Fig. 6C), which indicates that there is a direct relationship between complex formation and the proper synthesis of sialylated N-glycans.
FIGURE 6.
pH sensitivity of the glycosyltransferase complexes. COS7 cells were treated with 40 μm chloroquine (CQ) either for 4 h or overnight (o/n). All of the results are presented as percentages of nontreated COS7 cells (mean ± S.D.; n = 3). A and B, homo- and heteromeric interactions between N-glycosyltransferases. The FRET+ signal between the indicated enzymes measured by flow cytometry as described in the legend to Fig. 1. C, cell surface sialic acid content in live cells. Quantification was performed as described in the legend to Fig. 4A. D and E, homo- and heteromeric interactions between O-glycosyltransferases. F, quantification of cell surface T-antigen expression in live cells using peanut agglutinin staining. The measurements were performed as described in the legend to Fig. 4B. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
O-Glycosyltransferase homomers were also mainly unaffected by the drug. Only the amount of C3GNT-1 homomers was slightly decreased (p < 0.05) after overnight drug treatment (Fig. 6D and supplemental Fig. S7). In contrast, all of the heteromers involved in core 1, 3, and 6 synthesis were highly pH-sensitive (Fig. 6E). Both 4-h and overnight treatment reduced the amount of enzyme heteromers by >50%. This decrease was not likely due to the reduced expression of the enzymes, as the 4-h treatment at the end of the transfection was nearly as effective as the overnight treatment. The decrease in the amount of ppGalNAcT-6 and C1GalT-1 heteromers at elevated Golgi pH also coincided with a reduction in T-antigen expression, the end product of these two enzymes (p < 0.01; Fig. 6F). Thus, the heteromeric O-glycosyltransferase complex appears to be required also for proper core glycosylation of cell surface O-glycans.
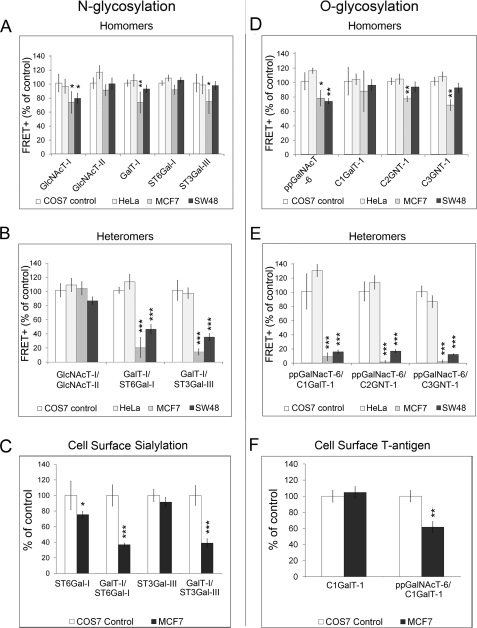
Complex Formation in Cancer Cells
The Golgi pH is also elevated in many cancer cells (36). Therefore, we anticipated that complex formation might be affected. To clarify this, we performed FRET measurements using acidification-defective cancer cells MCF7 and SW48 (36) as targets. COS7 and HeLa cells, which both have a normal Golgi pH, were used as control cells. We found that the amount of homomeric N-glycosyltransferase complexes and the heteromeric complex between the medial Golgi enzyme GlcNAcT-I and GlcNAcT-II was only slightly lower in acidification-defective cancer cells than in control cells (Fig. 7, A and B). However, the amount of heteromeric GalT-I·ST3Gal-III complex was reduced by more than 50% in MCF7 and SW48 cells (Fig. 7B), relative to control cells. The fact that the amount of the ST6Gal-I and GalT-I complex was also markedly reduced despite its pH insensitivity (Fig. 6) was unexpected but may indicate that micro-environmental factors other than the Golgi pH may also regulate complex formation. Nevertheless, the low amount of these two heteromeric sialyltransferase complexes in MCF7 cells that was not due to either mislocalization (supplemental Fig. S8) or lower expression level of the enzyme constructs (supplemental Fig. S9), was also found to correlate with reduced sialylation of cell surface glycans (Fig. 7C).
FIGURE 7.
Glycosyltransferase complexes in acidification-defective cancer cells. The FRET+ signal between the indicated enzymes was measured by flow cytometry as described in the legend to Fig. 1. All of the results are presented as percentages of the FRET+ values of COS7 cells (mean ± S.D.; n = 3). A and B, homo- and heteromeric interactions between N-glycosyltransferases. C, cell surface sialic acid content in live cells. Quantification was performed as described in the legend to Fig. 4A. D and E, homo- and heteromeric interactions between O-glycosyltransferases. F, quantification of cell surface O-linked T-antigen using peanut agglutinin staining in MCF7 cells compared with COS7 control cells. The measurements were performed as described in the legend to Fig. 4B. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Similar to the results with N-glycosyltransferases, the amount of homomeric O-glycosyltransferases complexes (Fig. 7D) was not markedly altered in acidification-defective cancer cells (FRET+, >70% of control). In contrast, all the heteromeric interactions were markedly reduced both in MCF7 and SW48 cells (FRET+, <20% of control) (Fig. 7E). The low amount of the core 1 (T-antigen) synthesizing complex also coincided with the reduced expression of the cell surface T-antigen (p < 0.01; Fig. 7F). Collectively, the above results indicate a direct relationship between heteromeric complex formation and proper synthesis of cell surface N- and O-glycans.
DISCUSSION
In this report, we have mapped for the first time all the main Golgi glycosyltransferase interactions within both the N- and O-glycosylation pathways in living cells. We show that the glycosyltransferases tested form Golgi-localized homomeric and heteromeric complexes within each pathway (Figs. 1 and 2). No complexes, however, were detected between the enzymes that operate in different pathways (Fig. 3), showing that the two pathways are physically distinct. Our size-exclusion chromatography data (Fig. 4, C–E) also confirmed that the glycosyltransferases tested exist mostly as complexes and not as enzyme monomers in live cells.
Several lines of evidence indicate that the complexes are functionally relevant. We showed that all enzyme complexes tested were found to be enzymatically active in vivo (Fig. 4. A and B). Their overexpression either as homomers or as heteromers increased terminal sialylation and the production of T-antigen in live cells. Heteromeric complexes were also found to form only between consecutively acting enzymes in either pathway. In addition, we showed that the GalT-I·ST6Gal-I complex had higher activity than either of the enzyme homomers (Fig. 4, C–G), which suggests that heteromeric complex formation is required for the activity increase and that these two enzymes function cooperatively. Moreover, we showed that disruption of heteromeric interactions by raising Golgi pH with chloroquine, correlated with reduced cell surface sialylation and T-antigen expression in the cells (Fig. 6). Finally, a similar correlation between impaired complex formation and glycosylation was also detected in acidification defective cancer cells (Fig. 7). Thus, these results suggest that the heteromeric complexes are functionally more relevant than enzyme homomers and likely responsible for the various glycosylation reactions in a living cell.
The functional relevance of the homomeric complexes is more difficult to interpret. One possibility is that certain homomeric complexes may also be involved in glycan synthesis. Alternatively, their formation may be required only for the endoplasmic reticulum-to-Golgi transport, consistent with the observed pH insensitivity of the complexes. Moreover, it is also possible that they are required for correct localization of the enzymes in the Golgi in the absence of an interacting partner. To clarify the functional relevance of the enzyme homomers, further studies are thus required. For example, multicolor FRET would resolve whether the homomeric or heteromeric interaction is preferred in vivo or if the homomeric complexes form at all in the presence of interacting partner(s). Other approaches could also involve knock-out or knockdown studies to confirm what role enzyme complexes have in glycan synthesis. Comparison of the enzymatic activities of isolated homomeric and heteromeric enzyme complexes would also resolve whether or not homo- and heteromeric complexes are equally active.
Collectively, our results help explain the overall functioning of the Golgi glycosylation pathways in higher eukaryotes. According to our results, the formation of enzyme complexes and their organization into specific assembly lines facilitates the efficient synthesis of different glycan structures with high precision and speed. Thus, the heteromeric complexes in the N-glycosylation pathway are likely involved in the processing of high mannose-type glycans to complex-type glycans (GlcNAcT-I·GlcNAcT-II) and termination of N-glycans with galactose and either α2,6-linked (GalT-I·ST6Gal-I) or α2,3-linked sialic acid (GalT-I·ST3Gal-III). The heteromeric O-glycosyltransferase complexes, in turn, are likely involved in the synthesis of the main mucin-type core structures. Although not assessed here, we anticipate that other ppGalNAc transferases likely form similar complexes with the core structure forming enzymes, allowing efficient site-specific core glycosylation of acceptor proteins. The fact that some O-glycosylation enzymes were found to form only homomeric complexes (Fig. 2) is not inconsistent with this view as they may form as yet unidentified heteromeric complexes with later enzymes in the pathway.
The proposed functional model involving heteromeric enzyme complexes allows more efficient and faster glycan synthesis by reducing the number of various acceptor recognition steps, and by increasing the affinity, specificity and processivity of the enzymes involved (Fig. 4, F and G). It also prevents potential intervention by competing enzymes. This is well illustrated by the mucin-type core O-glycosylation, where several core structure forming enzymes can potentially compete with each other for the same GalNAc acceptor (29). The presence of a preformed heteromeric complex (Fig. 2) prevents competing reactions (such as sialylation of the Tn-antigen) from occurring, thus favoring the synthesis of complex-specific core structures. Complex formation also allows the observed overlapping distribution of glycosyltransferases within the Golgi stack (37) without the loss of specificity.
The suggested model also provides an explanation for certain cancer-associated glycosylation abnormalities (29, 38–41). We showed that the elevated Golgi pH inhibits the formation of heteromeric termination complexes, which in turn results in altered glycosylation of cell surface constituents (Fig. 6). This is illustrated, e.g. by the pH-induced disassembly of the GalT-I·ST3Gal-III complex whereby the competitive GalT-I·ST6Gal-I complex becomes dominant and will terminate N-glycans with an α2,6-linked sialic acid, instead of α2,3-linked sialic acid. Such a change is observed also with the carcinoembryonic antigen during cancer progression (42). In addition, at elevated Golgi pH, the reduced amount of the core 1-synthesizing complex (Figs. 6F and 7F) can explain the increased expression of Tn- and sialyl-Tn-antigens (29) by the competing ST6GalNAc-I sialyltransferase. Collectively, these findings emphasize that proper Golgi acidity is crucial for the correct glycosylation of cellular glycoconjugates.
In conclusion, the results illustrate the important role of enzyme complexes in the synthesis of N- and O-glycans. Besides increasing reaction specificity and speed, complex formation also allows the modulation of glycan synthesis during cell growth, differentiation, and embryonic development, similar to the dynamic interactions that regulate other cellular processes. Several key issues such as the functional relevance of enzyme homomers and the identity of factors that regulate or affect the observed interplay between Golgi glycosyltransferases still remain to be solved. Unraveling these issues will lead to a better understanding of glycan synthesis in general, as well as to the development of new diagnostic and therapeutic tools for glycosylation-associated diseases, including cancer.
Supplementary Material
This work was supported by The Academy of Finland (to S. K.) and the Finnish Glycoscience Graduate School (to A. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S9.
- GalT-I
- β-1,4-galactosyltransferase I
- ST6Gal-I
- α-2,6-sialyltransferase I (for enzyme names and EC numbers, see supplemental Table S1)
- mCer
- monomeric Cerulean
- TMDS
- transmembrane/stem domain
- mVen
- monomeric Venus.
REFERENCES
- 1. Good M. C., Zalatan J. G., Lim W. A. (2011) Science 332, 680–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hart G. W. (1992) Curr. Opin. Cell Biol. 4, 1017–1023 [DOI] [PubMed] [Google Scholar]
- 3. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2008) Essentials of Glycobiology, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 4. Dunphy W. G., Brands R., Rothman J. E. (1985) Cell 40, 463–472 [DOI] [PubMed] [Google Scholar]
- 5. Kornfeld R., Kornfeld S. (1985) Ann. Rev. Biochem. 54, 631–664 [DOI] [PubMed] [Google Scholar]
- 6. de Graffenried C. L., Bertozzi C. R. (2004) Curr. Opin. Cell Biol. 16, 356–363 [DOI] [PubMed] [Google Scholar]
- 7. Young W. W., Jr (2004) J. Membr. Biol. 198, 1–13 [DOI] [PubMed] [Google Scholar]
- 8. Nilsson T., Au C. E., Bergeron J. J. (2009) FEBS Lett. 583, 3764–3769 [DOI] [PubMed] [Google Scholar]
- 9. Nilsson T., Slusarewicz P., Hoe M. H., Warren G. (1993) FEBS Lett. 330, 1–4 [DOI] [PubMed] [Google Scholar]
- 10. Jungmann J., Munro S. (1998) EMBO J. 17, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCormick C., Duncan G., Goutsos K. T., Tufaro F. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bieberich E., MacKinnon S., Silva J., Li D. D., Tencomnao T., Irwin L., Kapitonov D., Yu R. K. (2002) Biochemistry 41, 11479–11487 [DOI] [PubMed] [Google Scholar]
- 13. Hassinen A., Rivinoja A., Kauppila A., Kellokumpu S. (2010) J. Biol. Chem. 285, 17771–17777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banning C., Votteler J., Hoffmann D., Koppensteiner H., Warmer M., Reimer R., Kirchhoff F., Schubert U., Hauber J., Schindler M. (2010) PLoS ONE 5, e9344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thyrock A., Stehling M., Waschbüsch D., Barnekow A. (2010) Biochem. Biophys. Res. Commun. 396, 679–683 [DOI] [PubMed] [Google Scholar]
- 16. Giraudo C. G., Maccioni H. J. F. (2003) J. Biol. Chem. 278, 40262–40271 [DOI] [PubMed] [Google Scholar]
- 17. Rizzo M. A., Springer G. H., Granada B., Piston D. W. (2004) Nat. Biotechnol. 22, 445–449 [DOI] [PubMed] [Google Scholar]
- 18. Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002) Nat. Biotechnol. 20, 87–90 [DOI] [PubMed] [Google Scholar]
- 19. Kokkonen N., Rivinoja A., Kauppila A., Suokas M., Kellokumpu I., Kellokumpu S. (2004) J. Biol. Chem. 279, 39982–39988 [DOI] [PubMed] [Google Scholar]
- 20. Zeng Y., Ramya T. N., Dirksen A., Dawson P. E., Paulson J. C. (2009) Nature Methods 6, 207–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Campbell B. J., Rowe G. E., Leiper K., Rhodes J. M. (2001) Glycobiology 11, 385–393 [DOI] [PubMed] [Google Scholar]
- 22. Fahrenkrug J., Falktoft B., Georg B., Rask L. (2009) Biochemistry 48, 5142–5148 [DOI] [PubMed] [Google Scholar]
- 23. Hansske B., Thiel C., Lübke T., Hasilik M., Höning S., Peters V., Heidemann P. H., Hoffmann G. F., Berger E. G., von Figura K., Körner C. (2002) J. Clin. Invest. 109, 725–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Legaigneur P., Breton C., El Battari A., Guillemot J. C., Auge C., Malissard M., Berger E. G., Ronin C. (2001) J. Biol. Chem. 276, 21608–21617 [DOI] [PubMed] [Google Scholar]
- 25. Opat A. S., Houghton F., Gleeson P. A. (2000) J. Biol. Chem. 275, 11836–11845 [DOI] [PubMed] [Google Scholar]
- 26. Huang H. H., Stanley P. (2010) J. Cell Biol. 190, 893–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kellokumpu S., Neff L., Jämsä-Kellokumpu S., Kopito R., Baron R. (1988) Science 242, 1308–1311 [DOI] [PubMed] [Google Scholar]
- 28. Holappa K., Suokas M., Soininen P., Kellokumpu S. (2001) J. Histochem. Cytochem. 49, 259–269 [DOI] [PubMed] [Google Scholar]
- 29. Brockhausen I. (2006) EMBO Rep. 7, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tarp M. A., Clausen H. (2008) Biochim. Biophys. Acta 1780, 546–563 [DOI] [PubMed] [Google Scholar]
- 31. Tian E., Ten Hagen K. G. (2009) Glycoconj. J. 26, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berger E. G., Mandel T., Schilt U. (1981) J. Histochem. Cytochem. 29, 364–370 [DOI] [PubMed] [Google Scholar]
- 33. Gerber A. C., Kozdrowski I., Wyss S. R., Berger E. G. (1979) Eur. J. Biochem. 93, 453–460 [DOI] [PubMed] [Google Scholar]
- 34. Berger E. G., Kozdrowski I., Weiser M. M., van den Eijnden D. H., Schiphorst W. E. C. M. (1978) European Journal of Biochemistry 90, 213–222 [DOI] [PubMed] [Google Scholar]
- 35. Rivinoja A., Hassinen A., Kokkonen N., Kauppila A., Kellokumpu S. (2009) J. Cell. Physiol. 220, 144–154 [DOI] [PubMed] [Google Scholar]
- 36. Rivinoja A., Kokkonen N., Kellokumpu I., Kellokumpu S. (2006) J. Cell. Physiol. 208, 167–174 [DOI] [PubMed] [Google Scholar]
- 37. Rabouille C., Hui N., Hunte F., Kieckbusch R., Berger E. G., Warren G., Nilsson T. (1995) J. Cell Sci. 108, 1617–1627 [DOI] [PubMed] [Google Scholar]
- 38. Dube D. H., Bertozzi C. R. (2005) Nat. Rev. Drug Discov. 4, 477–488 [DOI] [PubMed] [Google Scholar]
- 39. Fuster M. M., Esko J. D. (2005) Nat. Rev. Cancer. 5, 526–542 [DOI] [PubMed] [Google Scholar]
- 40. Hakomori S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10231–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lau K. S., Dennis J. W. (2008) Glycobiology 18, 750–760 [DOI] [PubMed] [Google Scholar]
- 42. Yamashita K., Totani K., Kuroki M., Matsuoka Y., Ueda I., Kobata A. (1987) Cancer Res. 47, 3451–3459 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.