Background: β-l-Arabinofuranosyl linkages are found in many plant biopolymers, but the degradation enzyme has never been found.
Results: A novel β-l-arabinofuranosidase was found in Bifidobacterium longum.
Conclusion: β-l-Arabinofuranosidase plays a key role in Bifidobacterium longum for β-l-arabinooligosaccharides usage.
Significance: The members of DUF1680 family might be used for the degradation of plant biopolymers.
Keywords: Carbohydrate, Carbohydrate Metabolism, Enzyme Catalysis, Enzyme Kinetics, Enzyme Purification, Gene Expression, Hydrolases, Site-directed Mutagenesis
Abstract
Pfam DUF1680 (PF07944) is an uncharacterized protein family conserved in many species of bacteria, actinomycetes, fungi, and plants. In a previous article, we cloned and characterized the hypBA2 gene as a β-l-arabinobiosidase in Bifidobacterium longum JCM 1217. In this study, we cloned a DUF1680 family member, the hypBA1 gene, which constitutes a gene cluster with hypBA2. HypBA1 is a novel β-l-arabinofuranosidase that liberates l-arabinose from the l-arabinofuranose (Araf)-β1,2-Araf disaccharide. HypBA1 also transglycosylates 1-alkanols with retention of the anomeric configuration. Mutagenesis and azide rescue experiments indicated that Glu-366 is a critical residue for catalytic activity. This report provides the first characterization of a DUF1680 family member, which defines a new family of glycoside hydrolases, the GH family 127.
Introduction
β-l-Arabinofuranosyl linkages with 1–4 arabinofuranosides are found in the sugar chains of extensin and solanaceous lectins (1, 2). Extensins and solanaceous lectins are members of the hydroxyproline (Hyp)2-rich glycoproteins (HRGPs) that are widely observed in plant cell wall fractions. Furthermore, terminal β-l-arabinofuranosyl residues have been found in arabinogalactan protein from the pollen of timothy grass (3), rhamnogalacturonan-II (4–6), olive arabinan (7), arabinoglucan from Angelica sinensis (8), and tomato arabinoxyloglucan (9). However, despite the broad distribution of β-l-arabinofuranosyl residues in plant cells, the degradative enzyme β-l-arabinofuranosidase has never been found.
Recently, we cloned a hypBA2 gene that encodes a novel β-l-arabinobiosidase from Bifidobacterium longum JCM 1217 on the basis of the sequence of BL0421 from B. longum NCC2705, which belongs to the glycoside hydrolase (GH) family 121 (10). The enzyme releases Araf-β1,2-Araf disaccharide (β-Ara2) from Araf-β1,2-Araf-β1,2-Arafβ-Hyp (Ara3-Hyp). Because released β-Ara2 should be hydrolyzed by its own enzyme for assimilation, we predicted that B. longum has a gene encoding β-l-arabinofuranosidase. BL0422 is part of a gene cluster with BL0421 and BL0420 and contains a domain of unknown function (DUF) 1680 family in the Pfam database (PF07944), which is a large family annotated as putative glycosyl hydrolases of unknown function.
In this study, we cloned the gene of a BL0422 ortholog from B. longum JCM 1217 and characterized the recombinant protein as a novel β-l-arabinofuranosidase. This is the first report of the characterization of a DUF1680 family member.
EXPERIMENTAL PROCEDURES
Materials
Extensin, potato lectin, Hyp-linked β-l-arabinooligosaccharides, β-Ara2, and Araf-β1,2-Arafβ-OMe (Ara2-Me) were prepared as described previously (10). Dansylated Hyp-linked β-l-arabinooligosaccharides were prepared as described by Gray (11). p-Nitrophenyl (pNP) substrates were obtained from Sigma. l-Arabinose was obtained from Wako Chemicals. The chemical structures of substrates are shown in supplemental Fig. S1. HypBA2-CΔ486 was expressed and purified as described previously (10).
Expression and Purification of Recombinant HypBA1
The genomic DNA of B. longum JCM 1217 was extracted using a FastPure DNA kit (Takara) and then used for PCR amplification of the gene for the BL0422 ortholog, hypBA1. The forward (5′-AAGGAGATATACATATGAACGTTACAATCACTTCCC-3′) and reverse (5′-TGCTCGAGTGCGGCCGCTCGACGCTGGAAGACA-3′) primers were designed from nucleotides 4–22 and 1959–1974, respectively, of BL0422 from B. longum NCC2705 to generate a C-terminal His6-tagged recombinant protein. The PCR amplification product of hypBA1 was cloned into the pET-23b vector (Novagen) with the In-Fusion Advantage PCR Cloning Kit (Clontech). The full-length hypBA1 gene was sequenced on an ABI 3100 DNA sequencer with a Big-Dye Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems). The resulting pET23b-hypBA1 plasmid was transformed into Escherichia coli BL21 (λDE3) cells, which were then grown at 20 °C by using the Overnight Express Autoinduction System (Novagen). Subsequently, the cell cultures were centrifuged, and the resultant pellet was resuspended in BugBuster protein extraction reagent (Novagen). The His-tagged proteins were purified on TALON metal affinity resin (Clontech), desalted by dialysis with a cellulose membrane (Wako), and concentrated using a 10-kDa ultrafiltration membrane (Millipore).
Enzyme Assays
The hydrolytic activity of the HypBA1 enzyme was assayed using dansylated cis-Araf-β1,2-Arafβ-Hyp (cis-Ara2-Hyp-DNS) as a substrate. The 40-μl reaction mixture contained 50 mm sodium acetate buffer (pH 4.5), 25 μm substrate, 5 mm Tris(2-carboxyethyl)phosphine (TCEP), and 0.17 milliunits·ml−1 of the HypBA1 enzyme. One unit of enzyme activity was defined as the amount of enzyme required to produce 1 μmol of cis-Ara-Hyp-DNS per minute. After incubating the reaction mixture at 37°C, the reaction was stopped by adding 10 μl of 5% trichloroacetic acid, and then analyzed by HPLC. The sample was applied to a Cosmosil 5C18-AR-II (2.5 × 250 mm, Nacalai) column at 30 °C with a mobile phase of methanol and 20 mm sodium phosphate (pH 2.5) (60:40, v/v) and a constant flow rate (1.0 ml·min−1). The elution was monitored by a fluorescence detector (FP-202, JASCO) with excitation and emission wavelengths of 365 and 530 nm, respectively. For TLC analysis of dansylated substrates, the spots on the plates were developed with a 3:1:1 mixture (v/v/v) of 1-butanol/acetic acid/water and then visualized with UV light.
Substrate Specificity of HypBA1
Arafβ-Hyp (Ara-Hyp), Ara2-Hyp, Ara3-Hyp, Araf-α1,3-Araf-β1,2-Araf-β1,2-Arafβ-Hyp (Ara4-Hyp), Ara2-Me, β-Ara2, and pNP-substrates were incubated at 37 °C for 16 h with 0.17 milliunits·ml−1 of HypBA1 enzyme in 100 μl of 50 mm sodium acetate buffer (pH 4.5). The reaction was stopped by boiling for 3 min. For TLC analysis, oligosaccharides were spotted on a silica gel 60 aluminum plate (Merck) using a 2:1:1 solvent mixture (v/v/v) of ethyl acetate/acetic acid/water. The sugars were visualized by spraying an orcinol-sulfate reagent onto the plate (12). For high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) analysis, oligosaccharides were analyzed with a CarboPac PA-1 column. The column was eluted at a flow rate of 1.0 ml·min−1 by using the following gradient: 0–5 min, 100% eluent A (0.1 m NaOH); 5–30 min, 0–100% eluent B (0.5 m sodium acetate and 0.1 m NaOH); and 30–35 min, 100% eluent B.
The pH dependence of enzyme activity was determined between pH 3.5 and 8.0 by using the following buffers: 50 mm sodium acetate (pH 3.5–6.0), 50 mm MES (pH 5.5–7.0), and 50 mm sodium phosphate (pH 6.5–8.0). The effect of temperature on enzyme activity was examined using 50 mm sodium acetate buffer (pH 4.5) at 15–50 °C.
Kinetic Analysis
The kinetic parameters of HypBA1 were determined using 0–750 μm β-Ara2, cis-Ara2-Hyp-DNS, and cis-Ara-Hyp-DNS as the substrates. In the case of β-Ara2, the 40 μl reaction mixture was incubated at 37°C for 10 min, and then stopped by adding 10 μl of 500 mm NaOH. The amount of liberated l-arabinose was quantified by HPAEC-PAD as described above, using an l-arabinose standard. In the case of cis-Ara-Hyp-DNS, liberated cis-Hyp-DNS were analyzed according to the same procedure used for cis-Ara2-Hyp-DNS.
Transglycosylation of Ara2-Hyp
The transglycosylation reactions were performed using Ara2-Hyp as a donor and 1-alkanols as acceptors. Thirty nanomoles of Ara2-Hyp were incubated at 37 °C for 3 h with 340 milliunits·ml−1 of HypBA1 in 100 μl of 50 mm sodium acetate buffer (pH 4.5) with 5 mm TCEP and 20% methanol, ethanol, or 1-propanol as an acceptor. Subsequently, the reaction products were analyzed by TLC with a 2:1:1 solvent mixture of ethyl acetate/acetic acid/water (v/v/v). The sugars were visualized by spraying an orcinol-sulfate reagent onto the plate (12). For structural analysis, the transglycosylation product from the reaction in 20% methanol was purified by HPLC on a Cosmosil Sugar-D (4.6 × 250 mm, Nacalai) column at 30 °C with a mobile phase of acetonitrile and water (75/25, v/v) and a constant flow rate (1.0 ml·min−1). The elution was monitored by a refractive index (RI) detector (RI-8022, TOSOH), and the fraction that contained the transglycosylation product was collected. 1H and 13C NMR spectra were measured with a JMM-ECA600KS spectrometer (JEOL).
Site-directed Mutagenesis and Chemical Rescue
The QuikChange Site-directed Mutagenesis kit (Stratagene) was used to introduce amino acid substitutions into HypBA1 by using the primers shown in supplemental Table S1. After confirmation of the desired mutations by DNA sequencing, these mutant enzymes were expressed and purified according to the same procedure used for the wild-type enzyme. The effect of external nucleophile of the E366A mutant was investigated by adding 0–400 mm of sodium azide in 40 μl of 50 mm sodium acetate buffer (pH 4.5), 7.5 μg of E366A mutant, and 25 μm cis-Ara2-Hyp-DNS as a substrate. After incubating at 37 °C for 1 h, the reaction was stopped by adding 10 μl of 5% trichloroacetic acid, and then analyzed by HPLC as described above.
Bacterial Strains and Culture Conditions
The Bifidobacterium strains grown in Gifu anaerobic medium (GAM) broth (Nissui) were B. longum JCM 1217 and JCM 7054, B. longum subsp. infantis JCM 1222, B. pseudolongum JCM 1205, B. adolescentis JCM 1275, B. breve JCM 1192, and B. bifidum JCM 1254. The in vitro fermentation ability of β-Ara2 was tested using B. longum JCM 1217 and B. adolescentis JCM 1275 in peptone-yeast extract-Fildes (PYF) medium (13) containing 0.25% β-Ara2, glucose, or l-arabinose. The bacteria were cultured for 3 days at 37 °C under anaerobic conditions. The bacterial growth was judged from the decreased pH of the culture solution (14).
Assays of Bacterial Enzyme Activities
The cell cultures were centrifuged at 17,000 × g for 20 min, and the resultant pellets were washed with 50 mm Tris-HCl buffer (pH 6.8). Afterward, they were resuspended in 50 mm Tris-HCl buffer (pH 6.8) and sonicated with a Sonifier 250 (Branson). The cell lysates were incubated with 25 μm cis-Ara2-Hyp-DNS for 16 h at 37 °C and then analyzed by HPLC.
RESULTS
Expression and Purification of HypBA1
HypBA1 consisted of 658 amino acid residues exhibiting 98.9% identity with that of BL0422, and coincided with that of BLLJ_0211 from B. longum JCM 1217, for which the complete genome sequence is available (15). The recombinant HypBA1 protein was expressed at 20 °C as a soluble protein. SDS-PAGE showed that the purified recombinant HypBA1 protein migrated as a single band with an apparent molecular mass of 74 kDa (supplemental Fig. S2), which was in agreement with its calculated molecular mass of 74,329 Da. The final yield of the purified enzyme was 140 mg/liter of culture.
Substrate Specificity and General Properties of HypBA1
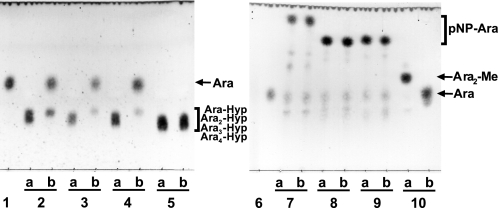
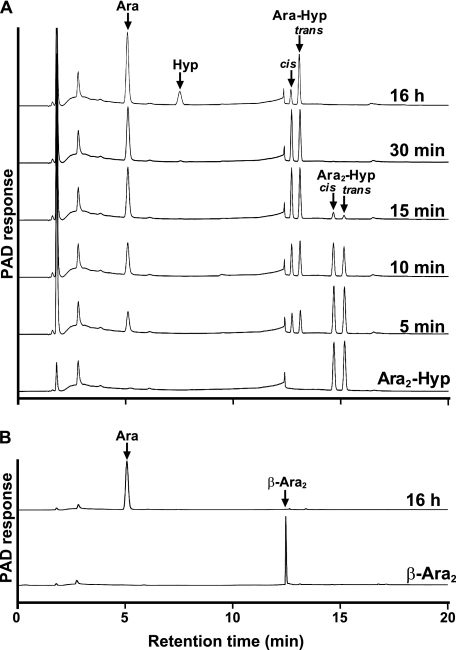
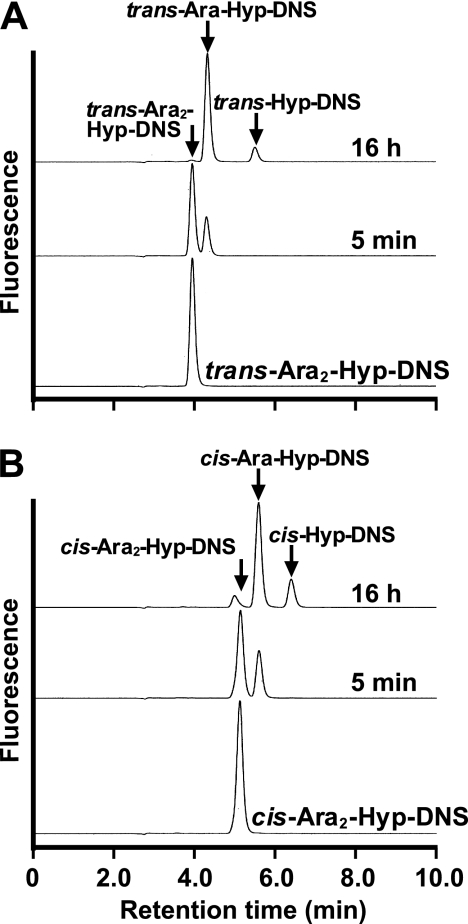
The enzymatic activity for dansylated cis-Ara3-Hyp-DNS was detected in the presence of β-mercaptoethanol, dithiothreitol, or TCEP, but not in the absence of reducing agents (supplemental Fig. S3). Several β-l-arabinooligosaccharides and synthetic pNP substrates were used to identify the substrate specificities for HypBA1 in the presence of TCEP. The enzyme released l-arabinose from Ara-Hyp, Ara2-Hyp, Ara3-Hyp, and Ara2-Me, but did not act on pNP-α-l-arabinopyranoside, pNP-α-l-arabinofuranoside, pNP-β-l-arabinopyranoside, or Ara4-Hyp (Fig. 1). HypBA1 also released l-arabinose from β-Ara2 (Fig. 2B). The suitable temperature and pH for cis-Ara2-Hyp-DNS were determined at 35 °C-40 °C and 4.5, respectively (supplemental Fig. S4). The specific activity of the purified enzyme was 2.1 units·mg−1 protein. The kinetic parameters for β-Ara2, cis-Ara2-Hyp-DNS and cis-Ara-Hyp-DNS are summarized in Table 1. The Km and kcat values for β-Ara2 and cis-Ara2-Hyp-DNS were within the same range, but the kcat value for cis-Ara-Hyp-DNS was 480-fold lower than that of cis-Ara2-Hyp-DNS. Consequently, the kcat/Km ratio of cis-Ara-Hyp-DNS was 670-fold lower than that of cis-Ara2-Hyp-DNS. HPAEC-PAD analysis showed that l-arabinose was released from Ara2-Hyp, and then the liberated Ara-Hyp gradually hydrolyzed to l-arabinose and Hyp (Fig. 2A). Likewise, both cis- and trans-Ara2-Hyp-DNS also hydrolyzed to Ara-Hyp-DNS, which then hydrolyzed to Hyp-DNS (Fig. 3). Under the conditions in which Ara3-Hyp could be degraded by HypBA2 and HypBA1 (supplemental Fig. S5A, lane 4), the reactivities for the glycoproteins were tested. Liberated sugars were detected from carrot extensin and potato lectin by HypBA2 but not by HypBA1 (supplemental Fig. S5). Furthermore, HypBA1 did not act on pNP-galacto-, gluco-, and xylo-pyranosides. The substrate specificity is summarized in supplemental Table S2. These results suggested that HypBA1 reacts with the liberated β-l-arabinooligosaccharides. Consequently, we classified the enzyme as an exo-acting β-l-arabinofuranosidase. The cleavage sites for HypBA1 are shown in supplemental Fig. S1.
FIGURE 1.
TLC analysis of HypBA1 reaction products. Substrates were incubated either without (lane a) or with (lane b) HypBA1 at 37 °C for 16 h. Ara-Hyp (lane 2), Ara2-Hyp (lane 3), Ara3-Hyp (lane 4), Ara4-Hyp (lane 5), pNP-α-l-arabinopyranoside (lane 7), pNP-α-l-arabinofuranoside (lane 8), pNP-β-l-arabinopyranoside (lane 9), and Ara2-Me (lane 10) were used as substrates. Lanes 1 and 6, l-arabinose standard.
FIGURE 2.
HPAEC-PAD analysis of HypBA1 reaction products. Ara2-Hyp (A) and β-Ara2 (B) were incubated with HypBA1 at 37 °C for up to 16 h. The double peaks correspond to cis and trans isomers.
TABLE 1.
Kinetic parameters of HypBA1 activity on different substrates
| Substrates | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | s−1mm−1 | |
| β-Ara2 | 0.85 ± 0.13 | 2.0 ± 0.20 | 2.3 |
| Ara2-Hypa | 0.31 ± 0.0013 | 6.3 ± 0.084 | 20 |
| Ara-Hypa | 0.43 ± 0.11 | 0.013 ± 0.0027 | 0.030 |
a cis isomers of dansylated substrates were used.
FIGURE 3.
HPLC chromatograms of HypBA1 reactions with dansylated substrates. The trans (A) and cis (B) isomers of Ara2-Hyp-DNS were incubated with HypBA1 at 37 °C for 5 min or 16 h.
Transglycosylation Activity of HypBA1
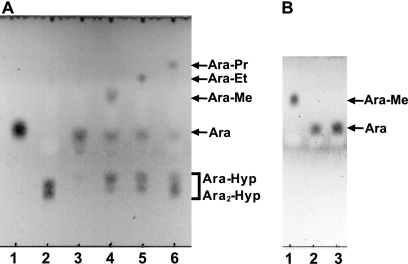
When 1-alkanols were used as the acceptors, the transglycosylation products were detected on TLC (Fig. 4A). The purified transglycosylation product (methyl l-arabinofuranoside) was hydrolyzed to l-arabinose by the HypBA1 treatment (Fig. 4B), which indicates that the methanol was linked by the β-anomeric form. The structure of this product was determined by 1H and 13C NMR (supplemental Fig. S6 and Table S3). The 1H NMR spectrum showed the anomeric proton as a doublet at 4.74 ppm with coupling constant J1,2 = 4.8 Hz. Furthermore, the 13C NMR spectra revealed that the transglycosylation product was found to be consistent with a methyl β-l-arabinofuranoside (Ara-Me) (16). These data indicated that HypBA1 is a retaining enzyme.
FIGURE 4.
Transglycosylation activity of HypBA1 in the presence of 1-alkanols. A, HypBA1 was incubated with Ara2-Hyp either in the absence (lane 3) or in the presence of 20% methanol (lane 4), ethanol (lane 5), or 1-propanol (lane 6) at 37 °C for 3 h. Lane 1, l-arabinose; lane 2, Ara2-Hyp. B, purified methyl l-arabinofuranoside was incubated without (lane 1) or with (lane 2) HypBA1 at 37 °C for 16 h. Lane 3, l-arabinose standard. Me, methyl; Et, ethyl; Pr, propyl.
Sequence Analysis of HypBA1
HypBA1 consisted of 658 amino acids that included DUF1680 without other sequence motifs (supplemental Fig. S7). HypBA1 was 38–98% identical to other DUF1680 members from bifidobacteria (supplemental Figs. S7 and S8). Duplicated DUF1680 members were found in the sequences of almost all Bifidobacterium species. HypBA1 (BLLJ_0211) constitutes a gene cluster with HypBA2 (BLLJ_0212) and a GH43 family member (BLLJ_0213) (supplemental Fig. S7). The gene cluster was conserved in B. longum NCC2705, B. longum subsp. infantis 157F, and B. pseudocatenulatum DSM 20438. In addition, the gene cluster without the GH43 family member was conserved in B. catenulatum DSM 16992 and B. dentium ATCC 27678.
Critical Amino Acid Residues of HypBA1
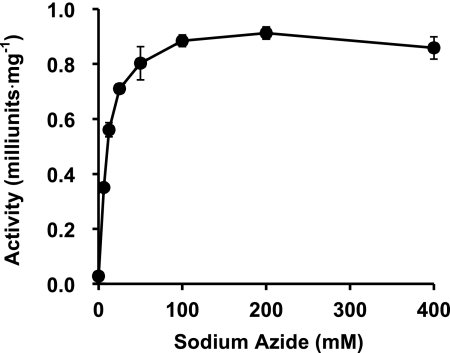
The candidate acidic amino acid residues were selected for site-directed mutagenesis studies based on multiple alignments and the HMM logo of the DUF1680 family in the Pfam data base (17). Alanine substitutions were introduced at the positions of Glu-322, Glu-338, and Glu-366, which are highly conserved among the HypBA1 homologues (indicated as asterisks in supplemental Fig. S8). The mutant enzymes were purified for the determination of specific activities. The E322A and E366A mutant enzymes were recovered in the soluble fractions with BugBuster. The E366A mutant enzyme exhibited a significant decrease in activity (0.0013%), and the E322A mutant showed 1.5% of the activity relative to the wild-type enzyme (Table 2). The E338A mutant enzyme was insoluble, and only a small amount of protein was recovered. Nonetheless, it exhibited 16% relative activity compared with the wild-type enzyme. The effect of external nucleophile on the activity of the E366A mutant was further investigated by using different concentrations of sodium azide. The activity of the mutant was rescued by the addition of azide (Fig. 5). In the presence of 200 mm sodium azide, the enzymatic activity was 33-fold greater than in the absence of external nucleophile. We also confirmed azide rescue by β-Ara2 as a substrate, whereas the glycosyl azide product was not observed on HPAEC-PAD and TLC (data not shown).
TABLE 2.
The specific activities of HypBA1 mutants
| Mutant enzymes | Specific activitya | Percentage of specific activity |
|---|---|---|
| milliunits mg−1 | % | |
| Wild type | 2100 | 100 |
| E322A | 32 | 1.5 |
| E338A | 340 | 16 |
| E366A | 0.028 | 0.0013 |
a Enzymatic activities were determined using the cis isomer of Ara2-Hyp-DNS.
FIGURE 5.
Effect of azide concentration on the activity of HypBA1 E366A mutant. Error bars show the standard deviation of triplicate measurements.
In Vitro Fermentability of β-Ara2 by B. longum
First, lysates of bifidobacterial cells grown in GAM medium were used as the enzyme source. β-l-Arabinofuranosidase activity was found in the cell lysate of B. longum JCM 1217 and B. longum JCM 7054 but not in that of B. adolescentis JCM 1275, B. breve JCM 1192, B. bifidum JCM 1254, B. pseudolongum JCM 1205, or B. longum subsp. infantis JCM 1222 (Fig. 6A). Moreover, enzymatic activity was not observed in the culture medium or in the bacterial cell suspensions for all Bifidobacterium strains described above (data not shown). The PYF medium containing 0.25% β-Ara2 was utilized as a carbohydrate source by B. longum JCM 1217 but not by B. adolescentis JCM 1275 (supplemental Table S4). In addition, β-Ara2 in the PYF medium was utilized by the fermentation of B. longum JCM 1217 (Fig. 6B). Furthermore, β-l-arabinofuranosidase activity was found in the cell lysate of B. longum JCM 1217 grown on β-Ara2 but not in the lysate of cultures grown in media containing glucose and l-arabinose (Fig. 6C). These data suggested that β-Ara2 is metabolized by β-l-arabinofuranosidase in B. longum.
FIGURE 6.
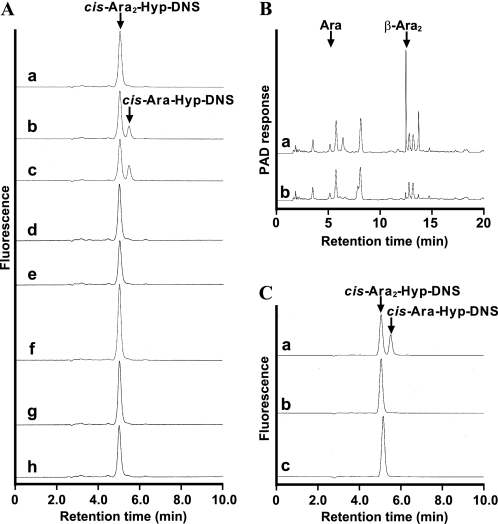
Detection of β-l-arabinofuranosidase activity in Bifidobacterium strains. A, HPLC analysis of reactions with cis-Ara2-Hyp-DNS and the cell lysates of Bifidobacterium strains grown on GAM medium. The Bifidobacterium strains that were incubated with cis-Ara2-Hyp-DNS are as follows: B. longum JCM 1217 (b), B. longum JCM 7054 (c), B. longum subsp. infantis JCM 1222 (d), B. pseudolongum JCM 1205 (e), B. adolescentis JCM 1275 (f), B. breve JCM 1192 (g), and B. bifidum JCM 1254 (h). Cis-Ara2-Hyp-DNS standard (a). B, HPAEC-PAD analysis of the culture PFY medium containing β-Ara2 before (a) and after (b) fermentation with B. longum JCM 1217. C, enzymatic activities of B. longum JCM 1217 grown on PFY medium containing β-Ara2 (a), l-arabinose (b), and glucose (c) as the carbohydrate sources.
DISCUSSION
The DUF1680 family has 597 members distributed among 315 species of enteric bacteria (i.e. Bifidobacterium, Bacteroides, Salmonella, Clostridium, and Esherichia), plant-pathogenic Xanthomonas, actinomycetes, fungi, and plants, as shown in the Pfam data base. The members of this family are hypothetical proteins of unknown function and have no sequence similarity with other glycoside hydrolase families. In this study, we cloned the gene encoding a member of the DUF1680 family and characterized its product as a novel β-l-arabinofuranosidase. Therefore, we propose that the enzyme be assigned to a new family of glycoside hydrolases, the GH family 127.
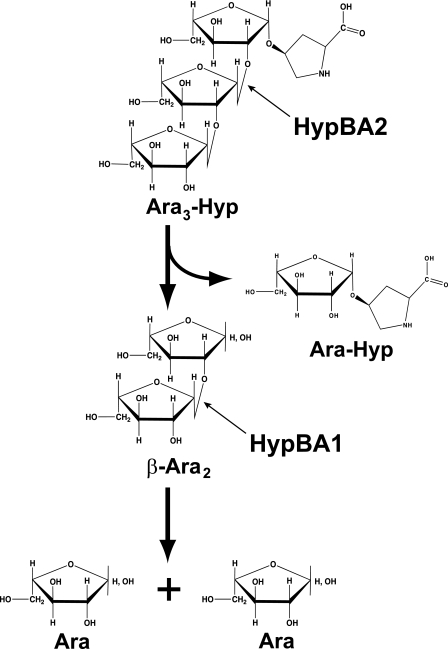
β-Ara2 was a suitable substrate for HypBA1 as well as Ara3-Hyp and Ara2-Hyp, which contain the Araf-β1,2-Araf structure at the nonreducing terminal. In extensins, β-l-arabinooligosaccharides are in close existence on repetitive Ser-Hyp4 motifs and contribute to protease resistance. It is thought that Hyp-linked β-l-arabinooligosaccharides do not occur naturally in the normal environment. Furthermore, HypBA1 did not directly release l-arabinose from extensin or potato lectin (supplemental Fig. S5). In addition, we showed that β-Ara2 was used as a carbohydrate source for B. longum, with enzymatic activity detected in the cell lysate (supplemental Table S4 and Fig. 6). Interestingly, the enzymatic activity was not detected in cells grown in the presence of l-arabinose or glucose. The amino acid sequence of HypBA1 lacks both a secretory signal and a transmembrane domain. Collectively, these results indicate that HypBA1 is an intracellular enzyme that degrades HypBA2-released β-Ara2, as schematically summarized in Fig. 7 and supplemental Fig. S9.
FIGURE 7.
Schematic drawing of the hydrolysis of Ara3-Hyp by HypBA1 and HypBA2.
Previously, we characterized an endo-α-N-acetylgalactosaminidase (BLLJ_0168) from B. longum JCM 1217, which releases Gal-β1,3-GalNAc (GNB) disaccharide from core-1 mucin-type O-glycans (18). Kitaoka et al. (19, 20) proposed a metabolic pathway for GNB from core-1 mucin-type O-glycans and Gal-β1,3-GlcNAc (LNB) from human milk oligosaccharides based on the characterization of the genes encoded in the GNB/LNB operon (BLLJ_1620-BLLJ_1626) of B. longum. Fushinobu et al. (21, 22) characterized the GNB/LNB-binding protein (BLLJ_1626) of an ATP-binding cassette (ABC)-type sugar transport system in the GNB/LNB pathway. As shown in supplemental Fig. S9, the contiguous genes of the hypBA1 gene (BLLJ_0211) have also been annotated as encoding subunits of a putative ABC-type sugar transport system (23): a solute-binding protein (BLLJ_0208) and 2 transmembrane subunits (BLLJ_0209 and BLLJ_0210). BLLJ_0208 exhibits 28% identity with the GNB/LNB-binding protein, and BLLJ_0209 and BLLJ_0210 also have >27% identity with the GNB/LNB transmembrane subunits (BLLJ_1624 and BLLJ_1625). Furthermore, the neighboring gene (BLLJ_0207) is predicted to be a LacI-type transcriptional regulator. Thus, we expect that BLLJ_0207 will regulate the gene cluster containing the β-Ara2 transport system (BLLJ_0208-BLLJ_0210) and the β-l-arabinooligosaccharides degradation enzymes (BLLJ_0211-BLLJ_0213) by internalizing β-Ara2.
The β-l-arabinooligosaccharides metabolic pathway in B. longum is predicted as shown in supplemental Fig. S9. First, a GH43 family member (BLLJ_0213) releases l-arabinose from extensin (Ara4-Hyp to Ara3-Hyp), and then HypBA2 (BLLJ_0212) releases β-Ara2 (Ara3-Hyp to Ara-Hyp) on the bifidobacterial cell surface. Next, the released l-arabinose and β-Ara2 are internalized into the bifidobacterial cell by uncharacterized transport system and predicted β-Ara2 transport system (BLLJ_0208-BLLJ_0210), respectively. Then, HypBA1 (BLLJ_0211) degrades β-Ara2 to l-arabinose. Furthermore, the l-arabinose metabolic enzymes for the conversion to d-xylulose-5-phosphate, which have been characterized in Corynebacterium glutamicum ATCC 31831 (24), exhibit 50–59% identity with those of B. longum JCM 1217: l-arabinose isomerase (BLLJ_0342), l-ribulokinase (BLLJ_0340), and l-ribulose 5-phosphate 4-epimerase (BLLJ_0341). As a result, HypBA1 plays a key role in B. longum for β-l-arabinooligosaccharides usage as a carbohydrate and energy source.
Recently, Fukuda et al. (15) reported that B. longum has an advanced ability for fructose uptake and acetate production, with the released acetate improving the intestinal defense mediated by epithelial cells. In addition to fructose, l-arabinose is a naturally found common carbohydrate and is found as a component of biopolymers such as hemicellulose and pectin. B. longum JCM 1217 encodes a number of candidates for the α-l-arabinofuranosidase gene, 11 members of the GH43 gene family, and 4 members of the GH51 gene family. Several reports indicate that B. longum has the ability to grow on l-arabinose and α-l-arabinooligosaccharides (14, 23, 25–27). We showed that B. longum also uses β-Ara2 as a carbohydrate source (supplemental Table S4). Several α- and β-l-arabinooligosaccharides degradation enzymes in B. longum might be involved in l-arabinose acquisition from plant polymers in the large intestine.
HypBA1 was identified as a retaining glycoside hydrolase, as described above. Hydrolysis by retaining glycoside hydrolases proceeds through a double-displacement mechanism with 2 catalytic residues. The catalytic residues typically utilized are either aspartate or glutamate residues. In the chemical rescue study, E366A mutant was rescued by the addition of azide, which suggests that Glu-366 is a catalytic residue for HypBA1. However, no glycosyl azide product was formed in the reaction mixture. A water molecule activated by azide ion might be reactivated E366A mutant without glycosyl azide production, as shown in GH43 β-xylosidase and GH14 β-amylase (28, 29).
B. longum JCM 1217 encodes 4 members of the DUF1680 family (BLLJ_0211, BLLJ_1826, BLLJ_1848, and BLLJ_0089), whereas B. longum NCC2705 encodes 2 members (BL0422 and BL0174) (supplemental Fig. S7). BL0422 constitutes a conserved gene cluster with the GH121 β-l-arabinobiosidase gene and the GH43-encoding gene as well as BLLJ_0211. BL0174 (98.8% identity with BLLJ_1826) is flanked by a gene cluster with 5 GH43 members and 1 α-galactosidase (BL0176-BL0190), whereas BLLJ_1826 is flanked by a small gene cluster without GH43 members (BLLJ_1824-BLLJ_1820). Interestingly, BLLJ_1848 constitutes a gene cluster with 5 duplicated GH43 members (BLLJ_1850-BLLJ_1854), in which the cluster is replaced by insertion sequences in B. longum NCC2705.
HRGPs that contain β-l-arabinooligosaccharides are widely distributed in land plants, mosses, ferns, and green algae (30). Furthermore, terminal β-l-arabinofuranosides are found in many plant biopolymers (3–8) and in yessotoxin from the dinoflagellate algae Protoceratium reticulatum (31, 32). Because DUF1680 family members are conserved in many species of bacteria, actinomycetes, fungi, and plants, they are thought to play a role in the effective degradation of plant biopolymers as well as HRGPs.
Supplementary Material
Acknowledgments
We thank Dr. M. Wakao and Dr. Y. Suda (Kagoshima University, Kagoshima, Japan) for help in measuring the NMR spectra. We thank Dr. S. Fushinobu (The University of Tokyo, Tokyo, Japan) for helpful discussions.
This work was supported in part by a Grant-in-aid for Young Scientists (B) 22780094 from the Japan Society for the Promotion of Science.
The nucleotide sequence of hypBA1 has been deposited in the DDBJ Database (Accession No. AB619598).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9 and Tables S1–S4.
- Hyp
- hydroxyproline
- HRGP
- hydroxyproline-rich glycoprotein
- Araf
- l-arabinofuranose
- DNS
- dansyl, 5-dimethylaminonaphthalene-1-sulfony
- DUF
- domain of unknown function
- GH
- glycoside hydrolase
- HPAEC-PAD
- high-performance anion-exchange chromatography with pulsed amperometric detection
- RI
- refractive index
- GAM
- Gifu anaerobic medium
- pNP
- p-nitrophenyl
- PYF
- peptone-yeast extract-Fildes.
REFERENCES
- 1. Kieliszewski M. J., Lamport D. T. A. (1994) Plant J. 5, 157–172 [DOI] [PubMed] [Google Scholar]
- 2. Kieliszewski M. J., Showalter A. M., Leykam J. F. (1994) Plant J. 5, 849–861 [DOI] [PubMed] [Google Scholar]
- 3. Brecker L., Wicklein D., Moll H., Fuchs E. C., Becker W. M., Petersen A. (2005) Carbohydr. Res. 340, 657–663 [DOI] [PubMed] [Google Scholar]
- 4. Mohnen D. (2008) Curr. Opin. Plant Biol. 11, 266–277 [DOI] [PubMed] [Google Scholar]
- 5. Pérez S., Rodríguez-Carvajal M. A., Doco T. (2003) Biochimie 85, 109–121 [DOI] [PubMed] [Google Scholar]
- 6. Whitcombe A. J., O'Neill M. A., Steffan W., Albersheim P., Darvill A. G. (1995) Carbohydr. Res. 271, 15–29 [DOI] [PubMed] [Google Scholar]
- 7. Cardoso S. M., Silva A. M., Coimbra M. A. (2002) Carbohydr. Res. 337, 917–924 [DOI] [PubMed] [Google Scholar]
- 8. Cao W., Li X. Q., Liu L., Yang T. H., Li C., Fan H. T., Jia M., Lu Z. G., Mei Q. B. (2006) Carbohydr. Polym. 66, 149–159 [Google Scholar]
- 9. York W. S., Kumar Kolli V. S., Orlando R., Albersheim P., Darvill A. G. (1996) Carbohydr. Res. 285, 99–128 [DOI] [PubMed] [Google Scholar]
- 10. Fujita K., Sakamoto S., Ono Y., Wakao M., Suda Y., Kitahara K., Suganuma T. (2011) J. Biol. Chem. 286, 5143–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gray W. R. (1967) Methods Enzymol. 11, 139–151 [Google Scholar]
- 12. Holmes E. W., O'Brien J. S. (1979) Anal. Biochem. 93, 167–170 [PubMed] [Google Scholar]
- 13. Yoshimoto M., Yamakawa O., Tanoue H. (2005) JARQ 39, 37–43 [Google Scholar]
- 14. Suzuki Y., Tanaka K., Amano T., Asakura T., Muramatsu N. (2004) J. Jpn. Soc. Hort. Sci. 73, 574–579 [Google Scholar]
- 15. Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K., Tobe T., Clarke J. M., Topping D. L., Suzuki T., Taylor T. D., Itoh K., Kikuchi J., Morita H., Hattori M., Ohno H. (2011) Nature 469, 543–547 [DOI] [PubMed] [Google Scholar]
- 16. Joseleau J.-P., Chambat G., Vignon M., Barnoud F. (1977) Carbohydr. Res. 58, 165–175 [Google Scholar]
- 17. Schuster-Böckler B., Schultz J., Rahmann S. (2004) BMC Bioinf. 5, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujita K., Oura F., Nagamine N., Katayama T., Hiratake J., Sakata K., Kumagai H., Yamamoto K. (2005) J. Biol. Chem. 280, 37415–37422 [DOI] [PubMed] [Google Scholar]
- 19. Kitaoka M., Tian J. S., Nishimoto M. (2005) Appl. Environ. Microbiol. 71, 3158–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishimoto M., Kitaoka M. (2007) Appl. Environ. Microbiol. 73, 6444–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fushinobu S. (2010) Biosci. Biotechnol. Biochem. 74, 2374–2384 [DOI] [PubMed] [Google Scholar]
- 22. Suzuki R., Wada J., Katayama T., Fushinobu S., Wakagi T., Shoun H., Sugimoto H., Tanaka A., Kumagai H., Ashida H., Kitaoka M., Yamamoto K. (2008) J. Biol. Chem. 283, 13165–13173 [DOI] [PubMed] [Google Scholar]
- 23. Parche S., Amon J., Jankovic I., Rezzonico E., Beleut M., Barutçu H., Schendel I., Eddy M. P., Burkovski A., Arigoni F., Titgemeyer F. (2007) J. Mol. Microbiol. Biotechnol. 12, 9–19 [DOI] [PubMed] [Google Scholar]
- 24. Kawaguchi H., Sasaki M., Vertès A. A., Inui M., Yukawa H. (2009) Appl. Environ. Microbiol. 75, 3419–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Biavati B., Scardovi V., Moore W. E. (1982) Int. J. Syst. Bacteriol. 32, 358–373 [Google Scholar]
- 26. Pastell H., Westermann P., Meyer A. S., Tuomainen P., Tenkanen M. (2009) J. Agric. Food Chem. 57, 8598–8606 [DOI] [PubMed] [Google Scholar]
- 27. Roy D., Ward P. (1990) J. Appl. Bacteriol. 69, 739–749 [DOI] [PubMed] [Google Scholar]
- 28. Miyake H., Otsuka C., Nishimura S., Nitta Y. (2002) J. Biochem. 131, 587–591 [DOI] [PubMed] [Google Scholar]
- 29. Shallom D., Leon M., Bravman T., Ben-David A., Zaide G., Belakhov V., Shoham G., Schomburg D., Baasov T., Shoham Y. (2005) Biochemistry 44, 387–397 [DOI] [PubMed] [Google Scholar]
- 30. Lamport D. T., Miller D. H. (1971) Plant Physiol. 48, 454–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Konishi M., Yang X., Li B., Fairchild C. R., Shimizu Y. (2004) J. Nat. Prod. 67, 1309–1313 [DOI] [PubMed] [Google Scholar]
- 32. Souto M. L., Fernández J. J., Franco J. M., Paz B., Gil L. V., Norte M. (2005) J. Nat. Prod. 68, 420–422 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.