Abstract
Type IV P-type ATPases (P4-ATPases) are putative phospholipid flippases that translocate phospholipids from the exoplasmic (lumenal) to the cytoplasmic leaflet of lipid bilayers and are believed to function in complex with CDC50 proteins. In Saccharomyces cerevisiae, five P4-ATPases are localized to specific cellular compartments and are required for vesicle-mediated protein transport from these compartments, suggesting a role for phospholipid translocation in vesicular transport. The human genome encodes 14 P4-ATPases and three CDC50 proteins. However, the subcellular localization of human P4-ATPases and their interactions with CDC50 proteins are poorly understood. Here, we show that class 5 (ATP10A, ATP10B, and ATP10D) and class 6 (ATP11A, ATP11B, and ATP11C) P4-ATPases require CDC50 proteins, primarily CDC50A, for their exit from the endoplasmic reticulum (ER) and final subcellular localization. In contrast, class 2 P4-ATPases (ATP9A and ATP9B) are able to exit the ER in the absence of exogenous CDC50 expression: ATP9B, but not ATP11B, was able to exit the ER despite depletion of CDC50 proteins by RNAi. Although ATP9A and ATP9B show a high overall sequence similarity, ATP9A localizes to endosomes and the trans-Golgi network (TGN), whereas ATP9B localizes exclusively to the TGN. A chimeric ATP9 protein in which the N-terminal cytoplasmic region of ATP9A was replaced with the corresponding region of ATP9B was localized exclusively to the Golgi. These results indicate that ATP9B is able to exit the ER and localize to the TGN independently of CDC50 proteins and that this protein contains a Golgi localization signal in its N-terminal cytoplasmic region.
Keywords: ATPases, Endosomes, Golgi, Membrane, Protein Translocation, Protein-Protein Interactions, Subcellular Organelles
Introduction
In eukaryotic cells, the lipid bilayer of the plasma membrane as well as membranes of secretory and endocytic compartments exhibits asymmetric lipid distributions; aminophospholipids, phosphatidylserine (PS),3 and phosphatidylethanolamine are concentrated in the cytoplasmic leaflet (1, 2). For example, in resting human red blood cells, PS and phosphatidylethanolamine are restricted primarily to the inner leaflet of the plasma membrane, whereas phosphatidylcholine and sphingomyelin are exposed on the cell surface (3, 4). Regulated exposure of PS in the outer leaflet occurs in many biological processes, such as apoptotic cell death, platelet coagulation reactions, and the fusion of muscle (4–7); similarly, phosphatidylethanolamine is exposed on the surface of the cleavage furrow during cytokinesis (8). In addition, phospholipid asymmetry of the bile canalicular membrane is critical to membrane integrity and normal bile secretion by hepatocytes (9); loss of phospholipid asymmetry due to mutations in the human FIC1/ATP8B1 (a member of the P4-ATPase family) gene causes a liver disease, progressive familial intrahepatic cholestasis (10).
P4-ATPases are a subfamily of P-type ATPases and have been implicated in flipping aminophospholipids from the exoplasmic (lumenal) leaflet to the cytoplasmic leaflet (11–15). The yeast P4-ATPases (Drs2p, Neo1p, Dnf1p, Dnf2p, and Dnf3p) are all involved in protein transport in the secretory and endocytic pathways albeit at different stages (16). P4-ATPases also play essential roles in membrane trafficking in other organisms, including Caenorhabditis elegans and Arabidopsis thaliana (17, 18). P4-ATPases form heteromeric complexes with members of the CDC50 protein family (14, 19). Mutations in the CDC50 genes in yeast and Arabidopsis phenocopy P4-ATPase mutations and disrupt transport and asymmetry of aminophospholipids (14, 17, 20, 21). The yeast CDC50 proteins Cdc50p, Lem3p, and Crf1p associate with Drs2p and Dnf1p/Dnf2p, and Dnf3p, respectively (14, 19). In contrast, the Neo1p P4-ATPase does not associate with either Cdc50p or Lem3p (14); furthermore, unique among the five yeast P4-ATPases, deletion of the NEO1 gene alone is lethal (22). Recently, the specificity of interactions of human class 1 P4-ATPases (ATP8A1, ATP8A2, ATP8B1, ATP8B2, ATP8B3, and ATP8B4) with CDC50 proteins and the requirement of CDC50 proteins for the subcellular localization of the ATPases have been characterized (23–26). However, the functional association with CDC50 proteins and subcellular localization of other classes of human P4-ATPases remain largely unknown.
Here, we systematically characterized the functional interactions of human P4-ATPases with CDC50 proteins using HeLa cells. We found that ATP9B, which is a putative orthologue of yeast Neo1p, requires neither CDC50A nor CDC50B for its localization to the trans-Golgi network (TGN), although most human P4-ATPases require CDC50 proteins (primarily CDC50A). We also characterized the physical interactions of class 2 (ATP9A and ATP9B) and class 6 (ATP11A, ATP11B, and ATP11C) P4-ATPases with CDC50 proteins, determined their subcellular localizations, and identified the Golgi localization signal in ATP9B.
EXPERIMENTAL PROCEDURES
RT-PCR
Total RNA from HeLa and RPE1 cells was isolated using an RNeasy minikit (Qiagen) and subjected to RT-PCR analysis using a SuperScript III One-Step RT-PCR system (Invitrogen). Total RNA of mouse testis was a gift from Yuki Okada, Kyoto University. The primers used are shown in supplemental Table S1.
Cloning
A full-length cDNA of ATP11C was obtained by amplification of HeLa cell total RNA by RT-PCR. The full-length cDNAs of human ATP8A1 (IMAGE clone 40036536), ATP9B (IMAGE clone 40123958), ATP10A (IMAGE clone 6258931), and ATP10D (IMAGE clone 40082621) were purchased from Open Biosystems (Huntsville, AL), and cDNA of ATP10B (ORK01674) was obtained from Kazusa DNA Research Institute (Kisarazu, Chiba, Japan). The partial cDNA of human ATP8B1 (IMAGE clone 3609288) was purchased from Open Biosystems; those of ATP9A (ORK00109), ATP11A (ORK04210), and ATP11B (ORK04211) were obtained from Kazusa DNA Research Institute. Missing cDNA regions were obtained by amplification of HeLa total RNA and ligated to the partial cDNAs to construct full-length cDNAs. The full-length P4-ATPase cDNAs were cloned into the pENTR3C vector (Invitrogen). The region encompassing the attR1 site, ccdB gene, chloramphenicol resistance gene, and attR2 site (DEST) was amplified from pcDNA6.2/V5-DEST (Invitrogen) by PCR. BglII and XhoI artificial restriction sites were introduced at the 5′- and 3′-ends of the PCR product and cloned into the pCAGGS-neodelEcoRI expression vector (27) with a C-terminal HA tag. Transfer of the genes to expression vectors was performed using the Gateway system (Invitrogen). Human CDC50A was amplified by RT-PCR from HeLa total RNA and cloned into the pTAC1 vector by TA cloning (BioDynamics Laboratory). Human CDC50A and open reading frames of human CDC50B (Open Biosystems) were subcloned into pcDNA3, pcDNA3 with an N-terminal FLAG tag, and pIRES2-EGFP (Clontech).
Antibodies and Reagents
Sources of antibodies used in the present study were as follows: polyclonal rabbit anti-TGN46 (28), a kind gift from Minoru Fukuda, Burnham Institute; monoclonal mouse anti-transferrin receptor (TfnR) (H68.4), Zymed Laboratories Inc.; monoclonal mouse anti-GM130, anti-EEA1, and anti-Lamp-1, BD Biosciences; monoclonal rat anti-HA (3F10), Roche Applied Science; monoclonal mouse anti-FLAG (M2) and polyclonal rabbit anti-FLAG, Sigma-Aldrich; Alexa Fluor 488-conjugated monoclonal mouse anti-CD147 (HIM6), BioLegend; Alexa Fluor-conjugated secondary antibodies, Molecular Probes; and Cy3-conjugated and horseradish peroxidase-conjugated secondary antibodies, Jackson ImmunoResearch Laboratories.
Cell Culture, siRNA-mediated Knockdown, and Immunofluorescence Analysis
Culture of HeLa cells and transfection of expression plasmids were performed as described previously (29, 30). RPE1 cells were grown in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F-12 medium supplemented with 10% heat-inactivated fetal calf serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Preparation of pools of siRNA for CDC50A and CDC50B and knockdowns using these siRNA pools were performed as described previously (31, 32) (33). Briefly, a pool of siRNA directed against the mRNA region spanning nucleotide residues 597–1086 of human CDC50A or 599–1056 of human CDC50B (when the A residue of the initiation Met codon is assigned as residue 1) was prepared using a BLOCK-iT RNAi TOPO transcription kit and a BLOCK-iT Dicer RNAi kit (Invitrogen). HeLa cells were transfected with the siRNA pool using Lipofectamine 2000 (Invitrogen) and incubated for 24 h. The transfected cells were then transferred to a culture dish containing coverslips, incubated for a further 48 h, and processed for immunofluorescence and RT-PCR analyses. The immunofluorescence staining was described previously (30) and was visualized using an Axiovert 200MAT microscope (Carl Zeiss, Thornwood, NY) for epifluorescence images or an LSM510 META microscope (Carl Zeiss) or FV1000 microscope (Olympus, Melville, NY) for confocal images.
Immunoprecipitation
HeLa cells were transfected using FuGENE 6 or X-tremeGENE 9 (Roche Applied Science) with different combinations of expression vectors for P4-ATPase and CDC50 and grown for 2 days. Prior to cell lysis, protein expression was induced by treating the cells with 4 mm sodium butyrate for 12–16 h. The cells were then lysed in lysis buffer (20 mm Hepes, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40) containing a CompleteTM protease inhibitor mixture (Roche Applied Science) at 4 °C for 30 min. The lysates were centrifuged at maximum speed for 20 min at 4 °C in a microcentrifuge to remove cellular debris and insoluble materials. The supernatant was incubated at 4 °C for 15 min with an anti-HA antibody and further incubated at 4 °C for 16 h with Protein G-coupled Dynabeads (Invitrogen). After washing, beads were incubated at 37 °C for 2 h in SDS sample buffer, and the supernatant was subjected to immunoblot analysis using rat anti-HA and mouse anti-FLAG or mouse anti-TfnR antibodies.
RESULTS
Expression of P4-ATPases and CDC50 Proteins in HeLa Cells
To investigate the interactions of human P4-ATPases with CDC50 proteins and their subcellular localization in HeLa cells, we first determined whether these genes are endogenously expressed in HeLa cells. RT-PCR analyses revealed the presence of mRNA for all but one P4-ATPase; ATP8A2 was not expressed in HeLa cells but was highly expressed in human retinal pigment epithelial (RPE1) cells (Fig. 1). HeLa cells expressed CDC50A and CDC50B but not CDC50C, indicating that both CDC50A and CDC50B are potential interacting partners of P4-ATPases; because CDC50C expression is restricted to spermatocytes and spermatids (25, 26, 34–36), total RNA of mouse testis was subjected to RT-PCR as a positive control of CDC50C mRNA expression using a set of primers designed to amplify both human and mouse CDC50C mRNAs.
FIGURE 1.
Gene expression of P4-ATPase and CDC50 in HeLa cells. RT-PCR was performed using total RNA isolated from HeLa and RPE1 cells and mouse testis. In control reactions, RNA was omitted. Primers are shown in supplemental Table S1. Primers for CDC50C were designed to amplify both human and mouse mRNAs; the product size of human and mouse is 236 and 194 base pairs, respectively.
Translocation of P4-ATPases from ER upon Exogenous Expression of CDC50A or CDC50B
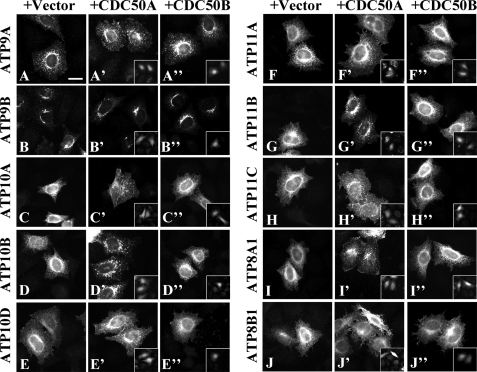
Recent studies have revealed that CDC50 proteins co-immunoprecipitate with class 1 P4-ATPases (ATP8A1, ATP8A2, ATP8B1, ATP8B2, ATP8B3, and ATP8B4) and are required for transport of these P4-ATPases from the ER (23–25, 37). We first sought to determine whether other P4-ATPases require CDC50A and/or CDC50B for their translocation from the ER to specific cellular compartments. We transiently expressed C-terminally HA-tagged P4-ATPases either alone (Fig. 2, A–J) or in combination with untagged CDC50A or CDC50B expressed on a bicistronic transcript along with GFP (CDC50A-IRES-EGFP or CDC50B-IRES-EGFP) (Fig. 2, A′–J′ and A″–J″, respectively) in HeLa cells; the insets in Fig. 2, A′–J′ and A″–J″, show EGFP signals, which indirectly indicate CDC50 expression. In the absence of exogenous CDC50 expression, class 5 (ATP10A, ATP10B, and ATP10D) and class 6 (ATP11A, ATP11B, and ATP11C) P4-ATPases were predominantly localized to the ER (Fig. 2, C–H, and supplemental Fig. S1), overlapping almost completely with the ER marker protein calnexin (see Fig. 4D, panel a, and supplemental Fig. S1). ATP8A1 and ATP8B1 were also localized to the ER when they were expressed alone (Fig. 2, I and J, and supplemental Fig. S1) as reported previously (24, 25). By contrast, the staining for class 2 P4-ATPases (ATP9A and ATP9B) was observed on intracellular organelles (Fig. 2, A and B) and showed only limited overlap with calnexin (see Fig. 4A, panel a, and supplemental Fig. S1) even in the absence of exogenous CDC50 expression. ATP9A was localized to peripheral punctate and perinuclear structures, whereas distribution of ATP9B was limited to perinuclear Golgi-like structures (Fig. 2, A and B, respectively). The subcellular distribution of ATP9A and ATP9B was not apparently altered by coexpression of either CDC50A or CDC50B (Fig. 2, A–A″ and B–B″, respectively). These results indicate that unlike other HA-tagged P4-ATPases ATP9A-HA and ATP9B-HA are able to exit the ER without exogenous CDC50 expression. Upon coexpression of CDC50A, significant translocation of other HA-tagged P4-ATPases was observed even though these P4-ATPases were overexpressed: ATP10A, ATP10D, ATP11C, and ATP8B1 were predominantly localized to the plasma membrane (Fig. 2, C′, D′, H′, and J′, and supplemental Fig. S2); ATP10B and ATP11B were primarily localized to punctate structures in the cytoplasm (Fig. 2, D′ and G′); ATP11A was predominantly localized to the plasma membrane and secondarily to some punctate structures (Fig. 2F′ and supplemental Fig. S2); and ATP8A1 was localized to punctate structures in the cytoplasm with some observed on the plasma membrane (Fig. 2I′ and supplemental Fig. S2). In contrast, only a few P4-ATPases were translocated upon coexpression of CDC50B: ATP8B1 was localized to the plasma membrane as seen with CDC50A coexpression (Fig. 2J″) (23–25) and ATP11C was localized to the plasma membrane but to a much lesser extent (Fig. 2, H′ and H″). Other P4-ATPases (ATP10A, ATP10B, ATP10D, ATP11A, ATP11B, and ATP8A1) were apparently retained in the ER even upon CDC50B coexpression (Fig. 2, C″, D″, E″, F″, G″, and I″). Thus, these C-terminally HA-tagged P4-ATPases primarily require CDC50A for their translocation from the ER toward specific cellular destinations. These results and those of previous reports (23–25) show that CDC50 proteins are indispensable for export of most P4-ATPases from the ER but may be negligible for translocation of ATP9A and ATP9B.
FIGURE 2.
Translocation of human P4-ATPases upon CDC50 coexpression. HeLa cells were transiently co-transfected with an expression vector for C-terminally HA-tagged P4-ATPase and a control IRES-EGFP vector (A–J) or an expression vector for CDC50A-IRES-EGFP (A′–J′) or CDC50B-IRES-EGFP (A″–J″). After 48 h of transfection, cells were processed for immunofluorescence microscopy. Insets indicate CDC50A- or CDC50B-expressing cells identified by EGFP signals. Bar, 20 μm.
FIGURE 4.
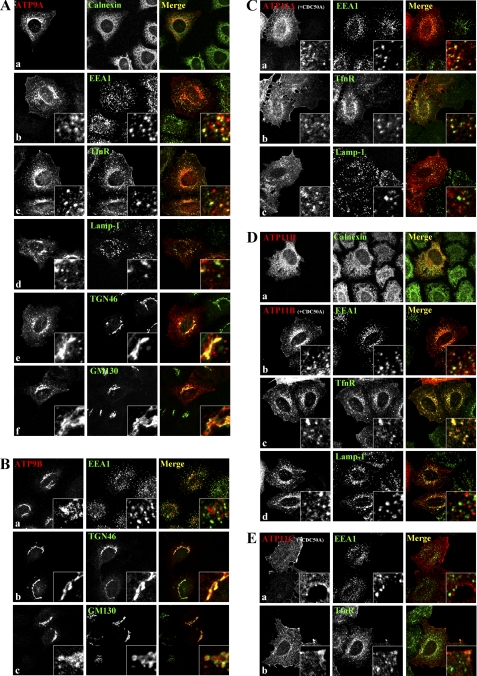
Subcellular localization of human P4-ATPases in HeLa cells. HeLa cells were transiently transfected with an expression vector encoding HA-tagged ATP9A (A, panels a–f), ATP9B (B, panels a–c), or ATP11B (D, panel a) alone or co-transfected with FLAG-CDC50A and an expression vector for HA-tagged ATP11A (C, panels a–c), ATP11B (D, panels b–d), or ATP11C (E, panels a and b). Samples were doubly stained for HA and an organelle marker as indicated.
CDC50 Proteins Are Not Co-immunoprecipitated with ATP9A and ATP9B
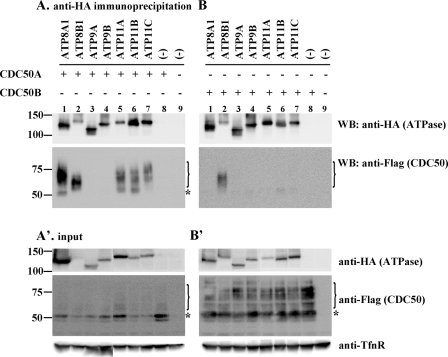
We next investigated whether CDC50A and CDC50B physically interact with P4-ATPases, including ATP9A and ATP9B. To this end, HeLa cells were transiently transfected with different combinations of a C-terminally HA-tagged P4-ATPase and an N-terminally FLAG-tagged CDC50 construct; total cell lysates were immunoprecipitated with anti-HA antibody and processed for immunoblotting with anti-FLAG antibody. Expression of the tagged proteins was confirmed by immunoblotting of total cell lysates (see Fig. 3, A′ and B′, input panels). ATP8A1-HA co-immunoprecipitated with FLAG-CDC50A but not FLAG-CDC50B (Fig. 3, A and B, lane 1), whereas ATP8B1-HA co-immunoprecipitated with both CDC50 proteins (Fig. 3, A and B, lane 2) consistent with a previous report (24). ATP11A-HA, ATP11B-HA, and ATP11C-HA co-immunoprecipitated with FLAG-CDC50A but not FLAG-CDC50B (Fig. 3, A and B, lanes 5–7). These results are in keeping with the data in Fig. 2, which demonstrates that exogenous expression of CDC50A, but not CDC50B, results in translocation of ATP8A1, ATP11A, ATP11B, and ATP11C from the ER, whereas both CDC50A and CDC50B are able to translocate coexpressed ATP8B1. In contrast to the class 1 and class 6 P4-ATPases, ATP9A-HA and ATP9B-HA failed to co-immunoprecipitate with FLAG-CDC50A and FLAG-CDC50B (Fig. 3, A and B, lanes 3 and 4), suggesting that neither ATP9A nor ATP9B forms a stable complex with CDC50 proteins in the cell. These results are in line with the data in Fig. 2 showing that ATP9A and ATP9B are able to exit the ER in the absence of coexpressed CDC50.
FIGURE 3.
Co-immunoprecipitation analysis of interactions between P4-ATPases and CDC50 proteins. HeLa cells were transfected with an expression vector encoding either FLAG-CDC50A (A, lane 8) or FLAG-CDC50B (B, lane 8) alone or in combination with that encoding HA-tagged P4-ATPase as indicated (lanes 1–7). As a control, HeLa cells were transfected with an empty vector (lane 9). After 48 h of transfection, cells were lysed and immunoprecipitated with anti-HA antibody. Bound materials (A and B) and inputs (A′ and B′) were subjected to SDS-PAGE and immunoblotting using anti-HA, anti-FLAG, or anti-TfnR antibody. A bracket and an asterisk indicate positions of highly glycosylated forms and an endoglycosidase H-sensitive, ER-localized form of CDC50 proteins, respectively (see supplemental Fig. S3). WB, Western blot.
It is noteworthy that in co-immunoprecipitations with HA-tagged class 1 and class 6 P4-ATPases the bands corresponding to CDC50 proteins appear as a smear (Fig. 3, A and B, indicated by a bracket) in marked contrast to relatively sharp bands around 50 kDa in the corresponding lanes of the input panels (Fig. 3, A′ and B′, indicated by an asterisk). CDC50A is known to be N-glycosylated (24, 26). The 50-kDa band was sensitive to endoglycosidase H (supplemental Fig. S3A), whereas the higher molecular weight bands were N-glycosidase F-sensitive but endoglycosidase H-resistant (supplemental Fig. S3, A and B). Likewise, CDC50B has an evolutionarily conserved N-glycosylation site (36). Therefore, the smeared bands at higher molecular weight probably represent highly glycosylated forms of CDC50 proteins, and the 50-kDa bands represent ER-localized forms. These results suggest that P4-ATPases preferentially form complexes with highly glycosylated forms of CDC50 proteins.
Endomembrane localization of class 2 and 6 P4-ATPases
We next determined the subcellular localizations of class 2 and class 6 P4-ATPases; presumably, these locations represent the sites where the proteins serve their ultimate function. To this end, HeLa cells were transfected with an expression plasmid for HA-tagged P4-ATPase alone (in the case of class 2) or in combination with FLAG-CDC50A (in the case of class 6) and doubly immunostained for HA and each of several organelle markers. As described above, the staining for ATP9A was found on punctate structures throughout the cytoplasm as well as perinuclear Golgi-like structures (see Fig. 2A). The punctate ATP9A staining overlapped extensively with an early endosomal marker, EEA1 (Fig. 4A, panel b), and partially with an early/recycling endosomal marker, TfnR (A, panel c), but not significantly with a late endosomal/lysosomal marker, Lamp-1 (A, panel d). On the other hand, the perinuclear ATP9A staining overlapped extensively with TGN46 (Fig. 4A, panel e) but exhibited limited overlap with a cis-Golgi marker, GM130 (A, panel f). Thus, ATP9A is localized to the early/recycling endosomes but not late endosomes and to the TGN rather than the cis-Golgi. Unlike ATP9A, ATP9B was localized exclusively to the perinuclear region (see Fig. 2B) but not on EEA1-positive early endosomes (Fig. 4B, panel a). The ATP9B staining overlapped significantly with TGN46 (Fig. 4B, panel b) but was adjacent to GM130 (B, panel c), suggesting that ATP9B mainly localizes to the TGN. The Golgi localization of transiently expressed ATP9A and ATP9B was not affected by treating cells with cycloheximide (data not shown), excluding the possibility that the Golgi signals represent a transient localization along the exocytic pathway.
When coexpressed with CDC50A, ATP11A localized mainly to the plasma membrane (supplemental Fig. S2) and secondarily to punctate structures in the cytoplasm (Fig. 4C, panel a). The punctate staining for ATP11A overlapped significantly with TfnR (C, panel b) and partially with EEA1 (Fig. 4C, panel a), suggesting that a subpopulation of ATP11A associates with early/recycling endosomal compartments. The peripheral punctate structures positive for ATP11B were superimposed on TfnR-positive endosomes and overlapped partially with EEA1-positive structures but not with Lamp-1-positive structures (Fig. 4D, panels b–d), indicating that ATP11B is mainly localized to recycling endosomal compartments. ATP11C signals were observed predominantly at the plasma membrane and did not overlap with any endomembrane markers examined, including EEA1, TfnR, and Lamp-1 (Fig. 4E, panels a and b, and data not shown). Because exogenous CDC50A tends to be overexpressed and is largely localized to the ER in HeLa cells, we could not determine the specific localization of CDC50A in HeLa cells by immunofluorescence analysis (supplemental Fig. S2).
CDC50 Knockdown Does Not Affect Localization of ATP9B
Although our data indicate that exogenously expressed CDC50 proteins are not critical for the translocation of ATP9A and ATP9B (Figs. 2B and 4B), it remained a possibility that endogenous CDC50 may contribute to transport of the class 2 P4-ATPases. To address this possibility, we asked whether knockdown of endogenous CDC50A and/or CDC50B by RNAi affects P4-ATPase localization. To circumvent issues arising from overexpression, we first tried to establish HeLa cell lines stably expressing these P4-ATPases and succeeded in establishing a cell line expressing ATP9B-HA. We also established a cell line stably expressing ATP11B-HA for use as a control.
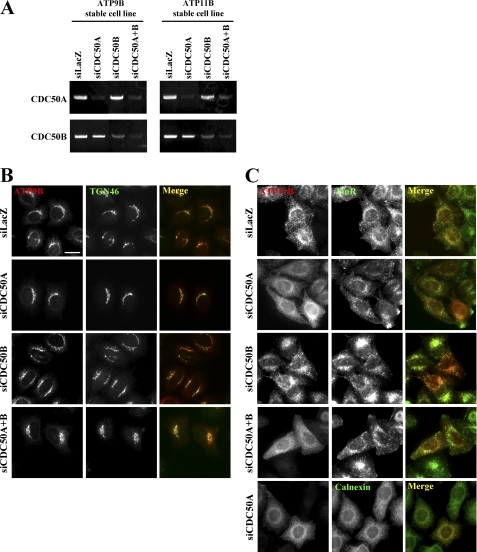
Because antibodies that can detect endogenous levels of CDC50A or CDC50B are not available, specific and efficient knockdown of CDC50A or CDC50B in the stable cell lines was confirmed by RT-PCR (Fig. 5A). As shown in Fig. 5B, neither knockdown of CDC50A or CDC50B alone nor knockdown of both genes simultaneously affected the ATP9B localization to the TGN. In contrast, knockdown of CDC50A alone or both CDC50A and CDC50B, but not CDC50B alone, caused redistribution of ATP11B from the recycling endosomal compartments (Fig. 5C). The ATP11B staining in the CDC50A-knocked down cells was colocalized with calnexin (Fig. 5C, bottom panels). These observations demonstrate that CDC50 is not essential for the ER exit or TGN localization of ATP9B, whereas CDC50A, but not CDC50B, is required for ER exit and/or recycling endosomal localization of ATP11B.
FIGURE 5.
Depletion of CDC50A does not affect Golgi localization of ATP9B but abolishes endosomal localization of ATP11B. HeLa cells stably expressing HA-tagged ATP9B (A, left panels, and B) or ATP11B (A, right panels, and C) were treated with a pool of siRNA for LacZ, CDC50A, CDC50B, or both CDC50A and CDC50B. After 72 h, cells were lysed to isolate total RNA, which was processed for RT-PCR using primers shown in supplemental Table S1 (A). Alternately, cells were fixed, permeabilized, and immunostained for HA and either TGN46 (B), TfnR (C, upper four rows), or calnexin (C, bottom). Bar, 20 μm.
N-terminal Region of ATP9B Harbors Golgi Localization Signal
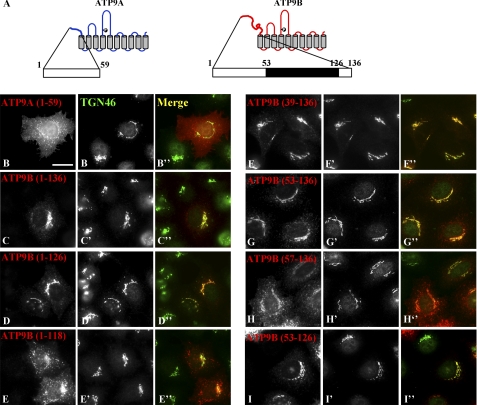
ATP9A and ATP9B exhibit a high sequence similarity with 75% identity at the amino acid level. However, their N-terminal cytoplasmic regions are divergent (supplemental Fig. S4, A and B). Because ATP9A is localized to the early/recycling endosomal compartments and the TGN, whereas ATP9B is localized exclusively to the TGN (Fig. 4, A and B), we speculated that the N-terminal variant regions give rise to the localization difference between ATP9A and ATP9B. To address this speculation, we constructed chimeric proteins of ATP9A and ATP9B, expressed them in HeLa cells (Fig. 6A), and counted the numbers of cells with different subcellular distributions of each protein (Fig. 6B). The ATP9BA construct in which the N-terminal cytoplasmic region of ATP9A was replaced with that of ATP9B (see supplemental Fig. S4B) localized exclusively to the Golgi apparatus (Fig. 6, A and B). On the other hand, the ATP9AB chimera in which the N-terminal region of ATP9B was replaced with that of ATP9A mislocalized to the ER. We do not know the exact reason for the mislocalization, but we speculate that ATP9AB may fail to fold properly at some point along its biosynthetic pathway. Taken together, these results suggest that the N-terminal cytoplasmic region of ATP9B contains a Golgi localization signal. Furthermore, we found that a construct encompassing the N-terminal cytoplasmic region of ATP9B alone (residues 1–136) can be localized to the Golgi apparatus (Fig. 7, A and C–C″), whereas that of ATP9A (residues 1–59) is distributed throughout the cytoplasm with some peripheral punctate structures (Fig. 7, A and B–B″). These observations further demonstrate that the N-terminal region of ATP9B contains a Golgi localization or Golgi retention signal.
FIGURE 6.
N-terminal cytoplasmic region of ATP9B is required for its Golgi localization. HeLa cells were transfected with an expression vector for HA-tagged ATP9A, ATP9B, or a chimeric construct of these proteins (ATP9AB or ATP9BA) schematically shown on the right side. Samples were doubly stained for HA and TGN46 and processed for immunofluorescence microscopy (A). Bar, 20 μm. Cells transfected and immunostained as in A were scored for distribution patterns of ER, ER + Golgi, Golgi, Golgi + puncta, and puncta (B). In each transfection, 200–350 cells were counted. The values are percentages of cells showing the specified distribution patterns of ATP9 constructs.
FIGURE 7.
Delineation of region critical for Golgi localization of ATP9B. HeLa cells were transfected with an expression vector for HA-tagged ATP9A(1–59) (B; schematically shown in A), ATP9B(1–136) (C; schematically shown in A), or a truncation mutant (D–I) as indicated. Samples were doubly stained for HA (B–I) and TGN46 (B′–I′) and processed for immunofluorescence microscopy. Merged images are also shown (B″–I″). Bar, 20 μm.
We next set out to delineate the minimal region of ATP9B required for its Golgi localization by truncating ATP9B(1–136). A construct with a 10-amino acid truncation at the C terminus, ATP9B(1–126), retained the ability to localize to the Golgi (Fig. 7, D–D″). However, a further eight-amino acid truncation induced a dramatic redistribution: the ATP9B(1–118) construct exhibited reduced localization to the Golgi and increased distribution to peripheral punctate structures (Fig. 7, E–E″) relative to ATP9B(1–136) (C–C″) and ATP9B(1–126) (D–D″). On the other hand, constructs with truncations of 39 or 53 amino acids from the N terminus (Fig. 7, F–F″ and G–G″, respectively) remained at the Golgi. However, a construct with a further four-amino acid truncation, ATP9B(57–136), was redistributed to punctate structures (Fig. 7, H–H″). Finally, we confirmed that the ATP9B(53–126) construct retained the ability to localize to the Golgi (Fig. 7, I–I″). Thus, the region encompassing amino acid residues 53–126 contains the minimal signal required for Golgi localization of ATP9B.
DISCUSSION
CDC50 proteins are indispensable for localization of class 1 P4-ATPases (ATP8A1, ATP8A2, ATP8B1, ATP8B2, and ATP8B4) to cellular compartments beyond the ER (24–26). In the present study, we found that class 5 (ATP10A, ATP10B, and ATP10D) and class 6 (ATP11A, ATP11B, and ATP11C) P4-ATPases also require CDC50 proteins for their exit from the ER and localization to specific cellular compartments where they perform their functions (Fig. 2). Although ATP8B1 was localized to the plasma membrane when coexpressed with either CDC50A or CDC50B, other P4-ATPases primarily require CDC50A (Fig. 2). In accordance with these observations, ATP8A1, ATP11A, ATP11B, and ATP11C were co-immunoprecipitated with CDC50A, whereas ATP8B1 could co-immunoprecipitate with either CDC50A or CDC50B (Fig. 3). Although a subpopulation of ATP10D was localized to the plasma membrane in the absence of exogenous CDC50A expression, its plasma membrane localization was more prominent in cells coexpressing CDC50A (Fig. 2, E–E″, and supplemental Fig. S2). When expressed at a low level in the absence of exogenous CDC50A expression, ATP8A1, ATP11A, ATP11B, and ATP11C all exhibited localization similar to that seen in cells where the P4-ATPase was expressed at a relatively high level in conjunction with exogenous CDC50A expression (see Fig. 5C, top panels, for ATP11B-HA stably expressed in HeLa cells; data not shown for others), suggesting that endogenous CDC50A might be sufficient to translocate some proportion of exogenous P4-ATPases from the ER.
Exogenously expressed ATP9A-HA and ATP9B-HA were able to localize to specific organelles beyond the ER independently of exogenous CDC50 expression (Fig. 2), whereas only a small proportion of each protein was associated with the ER. Moreover, knockdown of CDC50A, CDC50B, or both at once did not affect Golgi localization of stably expressed ATP9B-HA (Fig. 5B) even though CDC50A knockdown induced redistribution of stably expressed ATP11B-HA from endosomal compartments to the ER. Thus, it is likely that CDC50A is required for ER exit of ATP11B but not ATP9B. In addition, because neither ATP9A nor ATP9B co-immunoprecipitated with CDC50 proteins (Fig. 3), we conclude that unlike other P4-ATPases neither protein forms a stable complex with CDC50 proteins. In this context, it is interesting to note that in yeast P4-ATPases Drs2p, Dnf1p/Dnf2p, and Dnf3p associate with CDC50 family proteins Cdc50p, Lem3p, and Crf1p, respectively (14, 19); in contrast, Neo1p, which is phylogenetically related to human ATP9A and ATP9B (16, 22, 38), does not associate with either Cdc50p or Lem3p (14). Thus, the yeast Neo1p data are in agreement with our present data, showing that neither ATP9A nor ATP9B associates with CDC50A or CDC50B (Fig. 3).
A recent study in Arabidopsis has indicated that various plant P4-ATPases gain functionality when coexpressed with any of three different CDC50 proteins yet retain their distinct lipid substrate specificities irrespective of the CDC50 isoforms that bind to the P4-ATPases (39). In addition, murine ATP8A1 purified from baculovirus-infected insect cells retains PS-specific ATPase activity in the absence of CDC50 proteins (40). Moreover, yeast Drs2p purified with a substoichiometric amount of Cdc50p exhibits a PS-specific flippase activity in an in vitro reconstitution system (15). Overall, these studies indicate that the determinants of substrate specificity primarily reside in the P4-ATPases, not in the CDC50 proteins, and that flippase activity is retained in the absence of CDC50. Thus, the CDC50 proteins are likely to function as chaperone-like molecules that are required for exit of the P4-ATPase from the ER. However, Cdc50p is indirectly required for the ATPase activity of Drs2p (41). The intrinsic role of CDC50 proteins in the P4-ATPase complex remains to be determined.
We found that the distinct subcellular localizations of ATP9A and ATP9B can be attributed to the N-terminal cytoplasmic region of ATP9B. The ATP9BA chimera was localized exclusively to the Golgi apparatus as was wild-type ATP9B (Fig. 6). More intriguingly, the N-terminal region alone was able to localize to the Golgi; ultimately, we identified a region encompassing residues 53–126 that is minimally required for the Golgi localization of ATP9B. In this context, identification of interacting partners of the ATP9B N-terminal region will help to understand the cellular function of ATP9B.
Supplementary Material
Acknowledgments
We thank Minoru Fukuda and Yuki Okada for kindly providing anti-TGN46 antibody and total RNA of mouse testis, respectively.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan; the Special Coordination Fund for Promoting Science and Technology; the Targeted Proteins Research Program; the Takeda Science Foundation; and the Inamori Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- PS
- phosphatidylserine
- P4-ATPase
- type IV P-type ATPase
- TGN
- trans-Golgi network
- IRES
- internal ribosomal entry site
- EEA1
- early endosomal autoantigen 1
- TfnR
- transferrin receptor
- Lamp-1
- lysosomal-associated membrane protein-1
- ER
- endoplasmic reticulum
- EGFP
- enhanced GFP.
REFERENCES
- 1. Op den Kamp J. A. (1979) Annu. Rev. Biochem. 48, 47–71 [DOI] [PubMed] [Google Scholar]
- 2. van Meer G., Voelker D. R., Feigenson G. W. (2008) Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schrier S. L., Zachowski A., Devaux P. F. (1992) Blood 79, 782–786 [PubMed] [Google Scholar]
- 4. Zwaal R. F., Schroit A. J. (1997) Blood 89, 1121–1132 [PubMed] [Google Scholar]
- 5. Williamson P., Schlegel R. A. (2002) Biochim. Biophys. Acta 1585, 53–63 [DOI] [PubMed] [Google Scholar]
- 6. van den Eijnde S. M., van den Hoff M. J., Reutelingsperger C. P., van Heerde W. L., Henfling M. E., Vermeij-Keers C., Schutte B., Borgers M., Ramaekers F. C. (2001) J. Cell Sci. 114, 3631–3642 [DOI] [PubMed] [Google Scholar]
- 7. Leventis P. A., Grinstein S. (2010) Annu. Rev. Biophys. 39, 407–427 [DOI] [PubMed] [Google Scholar]
- 8. Emoto K., Kobayashi T., Yamaji A., Aizawa H., Yahara I., Inoue K., Umeda M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12867–12872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paulusma C. C., Groen A., Kunne C., Ho-Mok K. S., Spijkerboer A. L., Rudi de Waart D., Hoek F. J., Vreeling H., Hoeben K. A., van Marle J., Pawlikowska L., Bull L. N., Hofmann A. F., Knisely A. S., Oude Elferink R. P. (2006) Hepatology 44, 195–204 [DOI] [PubMed] [Google Scholar]
- 10. Folmer D. E., Elferink R. P., Paulusma C. C. (2009) Biochim. Biophys. Acta 1791, 628–635 [DOI] [PubMed] [Google Scholar]
- 11. Daleke D. L. (2003) J. Lipid Res. 44, 233–242 [DOI] [PubMed] [Google Scholar]
- 12. Tang X., Halleck M. S., Schlegel R. A., Williamson P. (1996) Science 272, 1495–1497 [DOI] [PubMed] [Google Scholar]
- 13. Pomorski T., Lombardi R., Riezman H., Devaux P. F., van Meer G., Holthuis J. C. (2003) Mol. Biol. Cell 14, 1240–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. (2004) Mol. Biol. Cell 15, 3418–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou X., Graham T. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16586–16591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Graham T. R. (2004) Trends Cell Biol. 14, 670–677 [DOI] [PubMed] [Google Scholar]
- 17. Poulsen L. R., López-Marqués R. L., McDowell S. C., Okkeri J., Licht D., Schulz A., Pomorski T., Harper J. F., Palmgren M. G. (2008) Plant Cell 20, 658–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruaud A. F., Nilsson L., Richard F., Larsen M. K., Bessereau J. L., Tuck S. (2009) Traffic 10, 88–100 [DOI] [PubMed] [Google Scholar]
- 19. Furuta N., Fujimura-Kamada K., Saito K., Yamamoto T., Tanaka K. (2007) Mol. Biol. Cell 18, 295–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen S., Wang J., Muthusamy B. P., Liu K., Zare S., Andersen R. J., Graham T. R. (2006) Traffic 7, 1503–1517 [DOI] [PubMed] [Google Scholar]
- 21. Kato U., Emoto K., Fredriksson C., Nakamura H., Ohta A., Kobayashi T., Murakami-Murofushi K., Kobayashi T., Umeda M. (2002) J. Biol. Chem. 277, 37855–37862 [DOI] [PubMed] [Google Scholar]
- 22. Prezant T. R., Chaltraw W. E., Jr., Fischel-Ghodsian N. (1996) Microbiology 142, 3407–3414 [DOI] [PubMed] [Google Scholar]
- 23. Paulusma C. C., Folmer D. E., Ho-Mok K. S., de Waart D. R., Hilarius P. M., Verhoeven A. J., Oude Elferink R. P. (2008) Hepatology 47, 268–278 [DOI] [PubMed] [Google Scholar]
- 24. van der Velden L. M., Wichers C. G., van Breevoort A. E., Coleman J. A., Molday R. S., Berger R., Klomp L. W., van de Graaf S. F. (2010) J. Biol. Chem. 285, 40088–40096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bryde S., Hennrich H., Verhulst P. M., Devaux P. F., Lenoir G., Holthuis J. C. (2010) J. Biol. Chem. 285, 40562–40572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coleman J. A., Molday R. S. (2011) J. Biol. Chem. 286, 17205–17216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Niwa H., Yamamura K., Miyazaki J. (1991) Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 28. Kain R., Angata K., Kerjaschki D., Fukuda M. (1998) J. Biol. Chem. 273, 981–988 [DOI] [PubMed] [Google Scholar]
- 29. Shin H. W., Kobayashi H., Kitamura M., Waguri S., Suganuma T., Uchiyama Y., Nakayama K. (2005) J. Cell Sci. 118, 4039–4048 [DOI] [PubMed] [Google Scholar]
- 30. Shin H. W., Morinaga N., Noda M., Nakayama K. (2004) Mol. Biol. Cell 15, 5283–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ishizaki R., Shin H. W., Mitsuhashi H., Nakayama K. (2008) Mol. Biol. Cell 19, 2650–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nishimoto-Morita K., Shin H. W., Mitsuhashi H., Kitamura M., Zhang Q., Johannes L., Nakayama K. (2009) J. Biol. Chem. 284, 10583–10592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamamoto H., Koga H., Katoh Y., Takahashi S., Nakayama K., Shin H. W. (2010) Mol. Biol. Cell 21, 2746–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Osada N., Hashimoto K., Hirai M., Kusuda J. (2007) Gene 392, 151–156 [DOI] [PubMed] [Google Scholar]
- 35. Xu P., Ding X. (2007) Acta Biochim. Biophys. Sin. 39, 739–744 [DOI] [PubMed] [Google Scholar]
- 36. Katoh Y., Katoh M. (2004) Oncol. Rep. 12, 939–943 [PubMed] [Google Scholar]
- 37. Coleman J. A., Kwok M. C., Molday R. S. (2009) J. Biol. Chem. 284, 32670–32679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Riekhof W. R., Voelker D. R. (2009) Biochim. Biophys. Acta 1791, 620–627 [DOI] [PubMed] [Google Scholar]
- 39. López-Marqués R. L., Poulsen L. R., Hanisch S., Meffert K., Buch-Pedersen M. J., Jakobsen M. K., Pomorski T. G., Palmgren M. G. (2010) Mol. Biol. Cell 21, 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paterson J. K., Renkema K., Burden L., Halleck M. S., Schlegel R. A., Williamson P., Daleke D. L. (2006) Biochemistry 45, 5367–5376 [DOI] [PubMed] [Google Scholar]
- 41. Lenoir G., Williamson P., Puts C. F., Holthuis J. C. (2009) J. Biol. Chem. 284, 17956–17967 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.