Background: The TRPA1 ion channel plays critical roles in pain and inflammatory processes.
Results: A TRPA1 structure is presented at 16-Å resolution with docked molecular models.
Conclusion: Channel activation could involve conformation changes resulting from ligands binding in a pocket proposed to bridge adjacent subunits.
Significance: Structural-functional understanding of TRPA1 is important for resolving fundamental pain and inflammatory mechanisms.
Keywords: Electron Microscopy (EM), Protein Purification, Protein Structure, Single Particle Analysis, TRP Channels, TRPA1 Ion Channel, Amphipols, Structure-Function Prediction
Abstract
Transient receptor potential ankyrin 1 (TRPA1) is a non-selective ion channel, which is expressed in nociceptor sensory neurons and transduces chemical, inflammatory, and neuropathic pain signals. Numerous non-reactive compounds and electrophilic compounds, such as endogenous inflammatory mediators and exogenous pungent chemicals, can activate TRPA1. Here we report a 16-Å resolution structure of purified, functional, amphipol-stabilized TRPA1 analyzed by single-particle EM. Molecular models of the N and C termini of the channel were generated using the I-TASSER protein structure prediction server and docked into the EM density to provide insight into the TRPA1 subunit organization. This structural analysis suggests a location for critical N-terminal cysteine residues involved in electrophilic activation at the interface between neighboring subunits. Our results indicate that covalent modifications within this pocket may alter interactions between subunits and promote conformational changes that lead to channel activation.
Introduction
The transient receptor potential (TRP)2 channel superfamily is one of the largest families of cation channels, having orthologs in most eukaryotes (1, 2). On the basis of amino acid sequence homology, a total of 28 members of the mammalian TRP family have been subdivided into six major branches: TRPV, TRPC, TRPM, TRPA, TRPP, and transient receptor potential mucolipin. TRP channels are predicted to be tetramers, with each subunit consisting of six transmembrane helices and cytoplasmic N and C termini of varying sizes (3–5). The pore, thought to form between transmembrane helix 5 (TM5) and TM6 of each monomer, becomes non-selectively permeable to cations in response to a wide variety of physical, chemical, and thermal stimuli (6, 7). The cytoplasmic termini of TRP channels contain different domains that are used to define the six subfamilies (8). To further understand TRP channel function and regulation, it is important to determine structures for multiple TRP family members and to unravel structural differences between them.
The founding member of the TRP superfamily was identified in Drosophila nearly 50 years ago (9). However an atomic resolution structure for a full-length TRP channel is not yet available. Isolated domains of TRP channels have been solved by x-ray crystallography or NMR, including the α-kinase domain of TRPM7 (10), the ankyrin repeat domains from the TRPV subfamily of proteins (11–15), the C-terminal cytoplasmic coiled-coil domains of TRPM7 and TRPP2 (16, 17), and the EF hand domain of TRPP2 (18, 19). These partial atomic structures are useful for understanding channel regulation/modulation and enhance the interpretation of moderate resolution full-length TRP channel structures (20).
Four full-length TRP channel structures have been studied by electron microscopy (20), including TRPV1 (21), TRPV4 (22), and TRPC3 (23, 24), and TRPM2 (25). Three-dimensional, cryo-EM structures of TRPV1 and TRPV4 were refined to 19-Å and 35-Å resolution, respectively, using the EMAN software package (26). These structures are remarkably similar in size, and both contain two distinct regions likely corresponding to the TM and cytoplasmic domains of the channel (21, 22). Furthermore, molecular modeling of the full-length TRPV1 channel showed similar subunit organization (27). Cryo-EM structures of TRPC and TRPM channels were dissimilar to the TRPV structures, which could be due to structural variance among TRP subfamilies or differences in protein preparation (8, 20). The molecular size and structural topology of TRPC subfamily members are predicted to be similar to the TRPV subfamily. However, the 15-Å cryo-EM structure of TRPC3, determined using the SPINNS algorithms (28–31), appears to diverge from the overall architecture found for the TRPV channels. The volume of the structure is more than two times larger than the expected size of the TRPC channel and features an open “lace-like” shape (23, 24). Using the same SPINNS algorithm, the structure of TRPM2 was solved at 28-Å resolution using negative-stain EM, and a “bell-like” architecture was found. The overall TRPM2 structure is more dense and smaller in size than TRPC3 structure although its molecular weight is almost two times larger than TRPC3 (25).
In short, previous structural analyses using electron microscopy have revealed disparate TRP channel topologies (see also reviews in Refs. 8, 20). This could be due to the overexpression systems used or the purification conditions, which involved various detergents (decyl maltoside, dodecyl maltoside) and detergent concentrations. It is conceivable that detergents could induce TRP channel structural variations. Therefore, we chose to use an amphipathic polymer (amphipol) A8-35 in place of detergent. A8-35 is thought to provide a more native-like environment for a membrane protein (32) and has been utilized in a variety of NMR and electron microscopy structural studies (33–38). One advantage of amphipols is their ability to bind tightly to the transmembrane portion of the protein, which allows the protein to remain soluble in buffers that contain no detergent or excess amphipol (39). Five EM structures have been determined using A8-35, including two structures for proton ATP synthase as well as structures of bacteriorhodopsin, mitochondrial complex I, and supercomplex B (I1III2IV1) (33–38). A8-35 was shown to preferentially interact with the TM portion of proteins (34), forming an approximately 10- to 20-Å belt along the TM regions that could be detected using EM (37, 40). These studies also demonstrate that detergent can be exchanged for A8-35 amphipol while preserving the fold and stability of membrane proteins (34).
In this study, we focused on the TRPA1 channel, the single mammalian member of the TRPA subfamily. TRPA1 contains between 14 and 17 N-terminal ankyrin repeats per subunit and a potential C-terminal coiled-coil domain (41). TRPA1 is of particular interest as a drug target (42) because of its expression in nociceptor sensory neurons (43) and its capacity to transduce a wide variety of noxious chemical stimuli into action potentials. TRPA1 is activated by a variety of electrophilic compounds, including endogenous inflammatory mediators, products of oxidative stress, and exogenous pungent chemicals. These electrophiles are thought to bind covalently to conserved cysteine and lysine residues in the N-terminal domain (44–49). Channel activation is also achieved by a variety of chemically unrelated non-reactive small molecules such as menthol, icilin, camphor, thymol, and tetrahyrocannabinoid (50–55). Because of its capacity to respond to such a wide variety of chemical compounds, TRPA1 is considered critical for noxious chemical sensation, inflammatory signaling, and physiological and pathophysiological pain sensation (56). Structural elucidation of TRPA1 would provide a valuable starting point for rational analgesic design.
Here we present the mouse TRPA1 structure determined from projections of negatively stained, solubilized TRPA1 complexes to 16-Å resolution. We overexpressed the mouse TRPA1 channel in Saccharomyces cerevisiae, purified the channel, and used Ca2+ flux assays to confirm the functionality of the reconstituted channel. Exchanging the detergent to A8-35 amphipol stabilized purified TRPA1 and allowed us to obtain highly monodisperse TRPA1 channel preparations suitable for single particle analysis. The overall molecular architecture of the channel resembles the topology of TRPV and other ion channels (8, 57), having a tetrameric assembly with a compact transmembrane domain and a basket-like cytoplasmic domain structure. To enhance the interpretation of the TRPA1 EM map, molecular models of TRPA1 N- and C-terminal domains were generated using the I-TASSER protein structure prediction server and docked into the EM density. This provided insight into the TRPA1 subunit organization and putative ligand binding pockets. Our structural analysis suggests a possible location for the critical cysteines involved in electrophilic activation.
EXPERIMENTAL PROCEDURES
Expression and Purification of TRPA1
Mouse TRPA1 cDNA was obtained from Dr. David Corey (Harvard University) and modified by PCR to include 5′ Spel and 3′ Mlul restriction enzyme sites. The C terminus was tagged with a 1D4 immunoaffinity epitope (TETSQVAPA) for purification purposes. This construct was subcloned into the yeast expression vector YEpHIS (58), which contains a constitutive PMA1 promoter, and then the protein was overexpressed in the protease-deficient S. cerevisiae yeast strain BJ5457 (59). Yeast cells were disrupted with a microfluidizer (M-110Y, Microfluidics, Newton, MA), and plasma membranes were isolated as described previously (59). Plasma membranes were solubilized by stirring for 1 h at 4 °C in 10 mm Fos-Choline 12 (FC12), 20 mm HEPES (pH 8.0), 150 mm NaCl, 1.0 mm DTT, 1 mm PMSF, and 10% glycerol. Insoluble material was removed by centrifugation at 100,000 × g for 45 min. The supernatant containing TRPA1 was mixed with CNBr-activated Sepharose 4B coupled to 1D4 antibody. After binding overnight at 4 °C, the column was packed and washed with 20 mm HEPES (pH 8.0), 150 mm NaCl, 1.0 mm DTT, 10% glycerol, and 3.0 mm FC12 (wash buffer). TRPA1 was eluted with 3 mg/ml 1D4 peptide (GenScript USA, Piscataway, NJ) added to the wash buffer. The protein was concentrated (Millipore Amicon Ultra 50K Ultracel) and subjected to size-exclusion chromatography using a Superose 6 column with wash buffer (BioLogic DuoFlow chromatography system, Bio-Rad). Purified protein was resolved by electrophoresis on a 4–20% Tris-glycine SDS-PAGE gel (Invitrogen) to analyze purity and apparent molecular weight.
Following purification of TRPA1, FC12 detergent was exchanged for A8-35 using methods described previously with some modifications (36). Briefly, TRPA1 in FC12 was incubated with A8-35 at a protein:amphipol ratio of 1:2.3 (w/w) for 4 h while gently rotating at 4 °C (see Fig. 1B). The sample was diluted below the critical micelle concentration of FC12 (1.5 mm) using detergent-free wash buffer (20 mm HEPES (pH 8.0), 150 mm NaCl, 10% glycerol, 1.0 mm DTT). Dialysis in a 20,000 molecular weight cutoff Floatalyzer G2 (SpectrumLabs) was performed overnight at 4 °C in a volume of detergent-free wash buffer that was 80–100 times the sample volume and contained a 250× excess of BioBeads relative to FC12 detergent in the sample (w/w). To remove any remaining detergent and verify the tetrameric assembly of TRPA1 in A8-35, gel filtration was performed on a Superose 6 column pre-equilibrated with detergent-free wash buffer. Approximately 30–50 μg of amphipol-stabilized TRPA1 was obtained per liter of yeast culture.
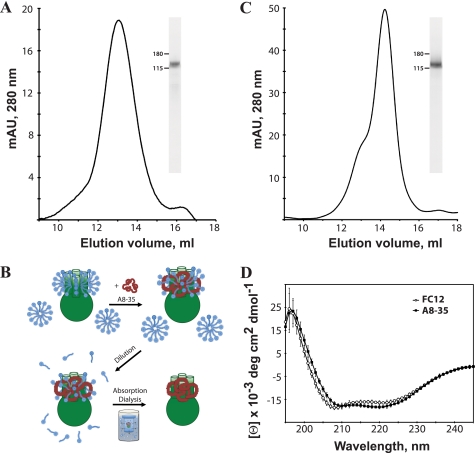
FIGURE 1.
Purification, amphipol exchange, and CD analysis of heterologously expressed mouse TRPA1. A, representative gel filtration chromatograph of mouse TRPA1 in FC12 detergent on Superose 6. The molecular mass of tetrameric TRPA1 with bound detergent is in close agreement with the observed peak at approximately 550 kDa. SDS-PAGE analysis of TRPA1 in FC12 (inset) after gel filtration indicates a purity > 95%. B, method and proposed mechanism of detergent-amphipol exchange for TRPA1 (see “Experimental Procedures” and Ref. 32). C, gel filtration chromatograph of mouse TRPA1 in A8-35 amphipols on Superose 6. A single peak elutes at approximately 500 kDa. SDS-PAGE analysis of TRPA1 in A8-35 (inset) after gel filtration indicates a purity > 95%. Fraction 14 was used for EM imaging. D, representative circular dichroism spectrum of purified TRPA1 in FC12 detergent (circles) and A8–35 amphipols (squares). Pronounced peaks near 210 and 222 nm indicate a folded protein of high α-helical content in both samples.
Reconstitution and Ca2+ Flux Assays
To test functionality of the purified channel extracted by detergent, Ca2+-flux measurements were performed on TRPA1 channels reconstituted into SUVs using a method described previously (21). In brief, 60 mg of soybean asolectin (L-α-phosphatidylcholine, Type II-S, Sigma-Aldrich) was solubilized with 3 ml of 400 mm n-octyl-β-d-glucopyranoside (Anatrace) in reconstitution buffer (25 mm Tris-HCl (pH 8.0), 150 mm NaCl, 5 mm CaCl2). Purified TRPA1 channel (0.5 mg) was mixed with the solubilized lipid and dialyzed against 5 liters of reconstitution buffer for 2–4 days at 4 °C using a 10,000 molecular weight cutoff Slidelyzer (Thermo Scientific). The Ca2+-containing vesicles were passed through Econo-Pac 10DG columns (Bio-Rad) and incubated with a chelating resin for 5 min (Chelex-100, Bio-Rad) to remove extravesicular Ca2+. A fluorescent Ca2+-indicator dye; Fura-2 (Invitrogen); HPLC-purified allicin (LKT Laboratories, St. Paul, MN); and all pipette tips, stir bars, and plastic ware used in the experiments were prepared in metal-free form by adding or soaking in Chelex-100 resin.
Fluorescence measurements were performed with a Quanta-Master 4 fluorimeter (Photon Technologies International). The vesicles were transferred into 2-ml quartz cuvettes, and 5 μm Fura-2 was added. With increasing Ca2+ concentration, the maximum fluorescence excitation of Fura-2 blue shifts so that the ratio of excitation intensity at 340 to 380 nm can be used to quantitate Ca2+ release from the vesicles. TRPA1-mediated vesicular calcium release was initiated by addition of 50 μm allicin, a TRPA1 activator especially prevalent in garlic (60). Allicin was added directly to the cuvette while stirring, and the excitation at 340 nm was continuously recorded at an emission wavelength of 510 nm. After the signal reached maximal intensity, an excitation spectrum from 300–400 nm was recorded and used to calculate Ca2+ concentration. The Ca2+-selective ionophore ionomycin (2 μm) was used to elicit maximum Ca2+ release (21).
Circular Dichroism Spectroscopy
Circular dichroism spectra were collected on an Aviv 215 spectrometer at 10 °C using a 1-mm path length quartz cuvette. For each sample, three sequential scans were taken at 1-nm increments from 200–250 nm. Protein concentrations ranged between 0.8–1.0 μm and were determined on a Beckman Coulter Du 530 spectrophotometer using an extinction coefficient of ϵ = 109,015 m−1cm−1 at 280 nm, which was calculated using ExPasy ProtParam. The assay buffer contained 20 mm sodium phosphate (pH 7.8), 150 mm NaCl, 1.0 mm DTT, 10% glycerol, and 3 mm FC12. Scans were corrected by subtracting the buffer-only trace. Secondary structural analysis was conducted using the CONTINLL and SELCON3 algorithms of the CDPro software suite (61, 62). A reference set suitable for membrane proteins was selected for the calculations (IBasis 10, SMP56) (62).
Electron Microscopy, Image Processing, and Modeling
A 3-μl sample of purified, A8-35 stabilized TRPA1 was adsorbed on carbon film-coated copper grids, washed with three droplets of pure water, and subsequently stained with 1% uranyl acetate. Data were acquired on an FEI Tecnai F20 TWIN microscope operated at 200 keV under low-dose conditions (20 electrons/Å2). Images were collected at a nominal magnification of ×55,000 at various defocus values between −0.5 to −1.5 μm and recorded with a Gatan US4000 UHS charge-coupled device camera (4K×4K). Particles (8,505 total) were boxed using the e2boxer interactive procedure built into the EMAN2 software package (63). Contrast transfer function correction was accomplished using phase-flipped particles with a high-pass filter using the new methodology developed in the EMAN2 software package. Initial models were produced with e2initialmodel using C1 and C4 symmetry on the basis of the generated reference-free class averages from the e2refined2d program. The final 3D map was refined using standard iterative projection-matching, class-averaging and Fourier reconstruction in EMAN2 (63). The mouse TRPA1 protein sequence was submitted to the I-TASSER online modeling program, which generates three-dimensional structure models by multiple-threading alignments and iterative structural assembly simulations (64–66). TRPA1 map segmentation and domain fitting was carried out using methods implemented in Segger, a segmenting tool available through the Chimera visualization software (67, 68).
RESULTS
Expression, Purification, and Functional Analysis of TRPA1
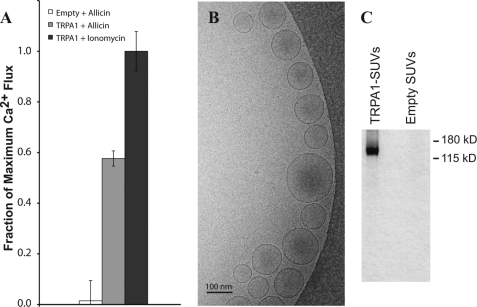
A heterologous yeast S. cerevisiae expression system, which has been successfully utilized for several membrane proteins, including TRPV1 (21, 59), was used to express and purify the TRPA1 channel. 1D4 monoclonal antibody affinity chromatography (21) was employed to purify the 1D4-tagged TRPA1 protein, and gel-filtration was performed to isolate the tetrameric form. The gel filtration profile showed a clear peak corresponding to the 514-kDa tetrameric channel with approximately two bound FC12 detergent micelles (Fig. 1A). After purification, TRPA1 was reconstituted into Ca2+-containing SUVs by detergent dialysis (21), and a fluorescence assay was used to measure Ca2+-flux through the TRPA1 channel following activation by allicin (a membrane-permeable TRPA1 ligand; see “Experimental Procedures”). Fig. 2A shows the fractional Ca2+ release from empty (1.5 ± 8.0%) and TRPA1-containing (57.7 ± 3.0%) vesicles upon addition of 50 μm allicin normalized to the maximum Ca2+ release upon addition of 2 μm ionomycin. Maximal reconstitution efficiency required an excess of lipid to protein and results in the formation of some liposomes that do not contain a channel (21, 69). On the basis of previous methods for estimating channel reconstitution efficacy (70), we calculated that there is approximately one TRPA1 channel in 50% of the vesicles. These results demonstrate that TRPA1 is a functional channel after extraction and purification in FC12 detergent. Electron microscopy verified vesicle formation (Fig. 2B), and SDS-PAGE analysis of the vesicles confirmed the presence of TRPA1 (Fig. 2C).
FIGURE 2.
Agonist-induced Ca2+ efflux from TRPA1 small unilamellar vesicles. TRPA1 was reconstituted in soybean asolectin vesicles containing intravesicular Ca2+ by detergent dialysis as described under “Experimental Procedures,” and Ca2+ efflux was measured in response to allicin addition. A, the Ca2+ efflux data indicate changes in the normalized Fura-2 (5 μm) fluorescence ratio (340/380 nm) following addition of 50 μm allicin to empty vesicles (1.5 ± 8.0%, white), vesicles with purified TRPA1 (57.7 ± 3.0%, gray), and after addition of 2 μm ionomycin (100 ± 7.8%, dark gray) to release the entire intravesicular calcium store. Data are normalized to the maximal fluorescence ratio observed after addition of ionomycin (n = 5). The fluorescence ratio prior to allicin addition has been subtracted, and was about 30% of the maximal fluorescence after ionomycin addition. B, cryo-EM micrograph of TRPA1 SUVs on a carbon-coated grid. C, SDS-PAGE of TRPA1-containing and empty SUVs showing TRPA1 incorporation.
TRPA1 Stabilization with A8-35 Amphipol
After purification, the FC12 detergent was exchanged for A8-35, using previously published methods (36) with some modifications (see Fig. 1B and “Experimental Procedures”). The gel filtration profile confirmed the formation of the TRPA1-A8–35 complex, eluting in a sharper peak than the FC12-solubilized TRPA1 at an apparent molecular mass of 500 kDa corresponding to a tetrameric complex (Fig. 1C). In addition, circular dichroism spectroscopy was used to verify global folding of the purified tetramer and to measure the secondary structure composition of the extracted, purified channel. The far ultraviolet CD spectrum of TRPA1 had negative peaks near 210 and 222 nm, consistent with a protein of high α-helical content (Fig. 1D) (71, 72). Using the CDPro software suite, the secondary structure composition of TRPA1 was estimated from the CD spectra. The CONTINLL and SELCON3 algorithms both estimate that TRPA1 is composed of 50–60% α-helices, 5–10% β-sheets, 15–20% turns, and 20–30% unordered structure (35–50% total coiled structure). The secondary structure did not significantly change after exchange of FC12 detergent with A8-35 amphipol. The secondary structure composition prediction from the I-TASSER program (64) was similar to the results obtained from experimental approaches (53% α-helices, 2% β-sheets, and 44% total coiled structure).
Electron Microscopy and Three-dimensional Reconstruction of TRPA1
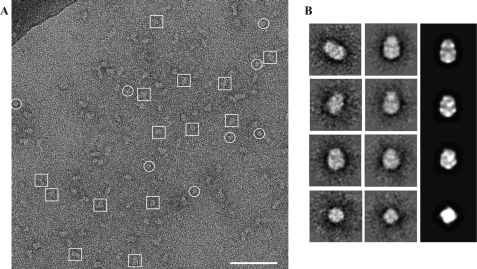
In the gel filtration chromatograph of TRPA1-A8-35, a small shoulder is visible in the peak, likely representing a small fraction of TRPA1 bound to FC12. To avoid heterogeneity, we used only fraction 14 for EM imaging. The A8-35-stabilized TRPA1 protein was highly monodisperse, as indicated by single-particle EM images, which show a distribution of TRPA1 molecules in both side views (Fig. 3A, boxed areas) and top or bottom views (Fig. 3A, circled areas). Particle images (8505 in total) were analyzed using the EMAN2 software package (63). After contrast transfer function correction, reference-free class averages and initial models were generated. From reference-free class averaging, we deduced that the protein consists of two domains and has four-fold symmetry (Fig. 3B and supplemental Fig. S1), supporting the biochemical evidence for the tetrameric assembly of TRPA1. Because initial models generated with and without four-fold symmetry were in agreement, we continued image processing with four-fold symmetry imposed on the data using standard iterative projection matching, class averaging, and Fourier reconstruction in EMAN2 (63). The consistency between reference-free class averages, reference-based class averages, and reprojections of the 3D reconstruction supports the accuracy of the map (Fig. 3B). The resolution was determined by splitting data into even and odd halves and computing the Fourier shell correlation in EMAN2, which gave a 0.5 value of approximately 16 Å (supplemental Fig. S1).
FIGURE 3.
Single-particle electron microscopy of TRPA1 in A8-35 amphipol. A, representative CCD image of purified TRPA1 rendered soluble by exchange into A8-35 and subsequently stained with 1% uranyl acetate on a carbon-coated copper grid. A distribution of “side views” (boxes) and “top views” (circles) are indicated. Scale bar = 70 nm. The darkened top left corner is a portion of the grid bar. B, representative selections of reference-free class averages (left panel), four-fold symmetry imposed class averages (center panel) and their back-projections (right panel).
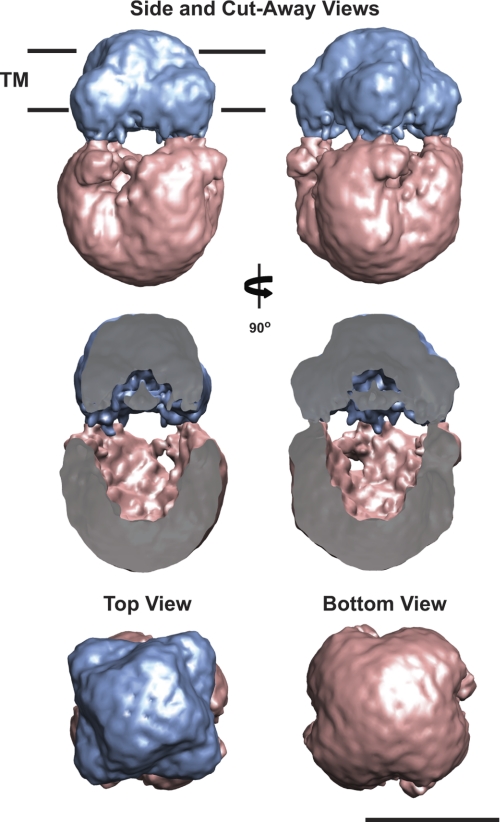
The TRPA1 structure represented in Fig. 4 is at a volume threshold corresponding to a protein molecular mass of 525 kDa, which is approximately the size of the TRPA1 tetramer with an 11-kDa A8-35 belt. The structure is 195 Å tall and has two distinct regions. One region, most likely the TM region, is 70 Å tall and 100 Å wide (Fig. 4, blue). The other, the expected cytoplasmic region, is 130 Å tall and 120 Å wide (Fig. 4, pink). The primary sequence analysis predicts that 22% of the amino acids are within the TM and extracellular loop regions, and the remainder of the sequence is contained in the cytoplasmic N terminus (64%) and C terminus (14%). The dual regions were assigned using segmentation and fitting methods implemented in Segger, available in the visualization software Chimera (67, 68). Furthermore, the TRPA1 TM region clearly has two distinct outlines (Fig. 4, top view): an upper TM portion of the TRPA1 channel (70 Å × 70 Å) and a wider volume in the lower portion of the TM (100 Å × 100 Å). The former would comprise the TRPA1 exterior TM region, whereas the latter possibly represents an A8-35 belt interacting with the TM region of TRPA1 (Fig. 4 and supplemental movie S1). The expected cytoplasmic region of TRPA1 presents a compact, basket-like architecture reported previously for other ion channels (57).
FIGURE 4.
Three-dimensional reconstruction of TRPA1. The final 3D reconstruction of TRPA1 is presented at a threshold corresponding to a molecular mass of 525 kDa. The proposed transmembrane domain (TM, blue) and cytoplasmic domain (CP, pink) are indicated. Side, cut-away, top, and bottom views of the reconstruction are shown. The cut-away view shows the internal cavity in the TRPA1 EM density. Segmentation was accomplished using Segger. Scale bar = 10 nm.
Molecular Modeling and Docking into the TRPA1 EM Map
To interpret the TRPA1 EM map, we used molecular modeling and docking methods. For the TM domain, Dr. Fernandez Ballester provided his published molecular models for the TRPV1 channel in the closed and desensitized states, which were constructed using homology modeling based on the Kv1.2 K+ channel (27). Using Segger, the TM domain from both the closed and the desensitized TRPV1 models could be placed into the proposed TM region of TRPA1 as displayed in Fig. 5 and supplemental movie S2 (67, 68). For the cytoplasmic domains, the I-TASSER server was used to generate 3D models from the N- and C-terminal TRPA1 sequences. The I-TASSER server generates 3D atomic models by conducting multiple folding simulations on the basis of templates that it identifies as structural homologs in the Protein Data Bank (PDB) (64). All models corresponding to the N terminus of TRPA1 (residues 1–721) predicted an ordered region of 12 ankyrin repeats (residues 31–437) and a more flexible downstream region (residues 438–721). The I-TASSER server used the crystal structure of the 12 ankyrin repeat stack from human ankyrinR (PDB 1N11) and the crystal structure of the ankyrin repeat domain of TRPV2 (PDB 2ETB) as template structures to assemble the N-terminal model of TRPA1. We utilized Segger for fitting and manual placement of these N-terminal models into the TRPA1 electron microscopy map. Although the TRPA1 reconstruction does not have sufficient resolution to allow unambiguous fitting (67, 68), the I-TASSER predicted model for the 12 ankyrin repeats (residues 1–667) could be placed within the cytoplasmic domain of the density map, leaving an open cavity in the center of this domain, as was observed in the EM density map (Figs. 4 and 5 and supplemental movies S1 and S2).
FIGURE 5.
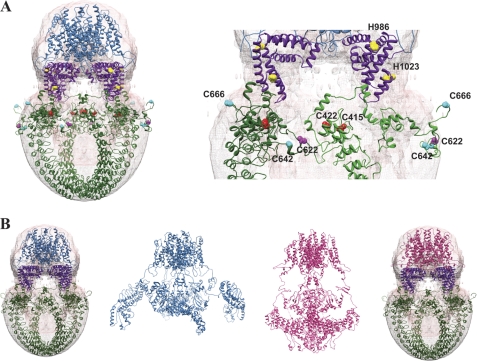
Model TRPA1 placement into EM 3D reconstruction. A, the modeled TRPV1 TM domain in the closed state (blue ribbon, Ref. 27) was fit into the proposed TM domain of the 3D structure (pink) using the Segger program in Chimera. The N-terminal (green ribbon) and C-terminal (purple ribbon) sequences were modeled using I-TASSER and placed within the proposed cytoplasmic domain. Conserved cysteines involved in electrophilic activation in the mouse (Cys-415, Cys-422, Cys-622), human (Cys-622, Cys-642, Cys-666), and Drosophila (Cys-622, Cys-642) homologs are highlighted in the zoomed-in view (ball-and-stick structures and labeled, right panel). Histidines involved in zinc activation are also indicated (yellow). The EM map has been deposited into the EMDB (accession number EMD-5334). B, comparison of the TRPA1 3D reconstruction map fitted with TM domain of the closed TRPV1 model (blue ribbon, full structure, see Ref. 27) and desensitized TRPV1 model (magenta ribbon, full structure, see Ref. 27) placed into the 3D electron microscopy reconstruction of TRPA1 along with the I-TASSER models of the N termini (green ribbon) and C termini (purple ribbon).
I-TASSER only returned one model with a suitable confidence score for the C terminus (residues 964–1125). This model was based on the crystal structure of the Mycobacterium tuberculosis SecA protein (PDB code 1NKT), and was manually placed into the 3D density map immediately below the predicted TM region within the region linking it to the cytoplasmic domains (Fig. 5).
Mutagenesis studies have identified five cysteines and two histidine residues important for TRPA1 activation by electrophilic compounds and Zn2+, respectively (44, 46, 73). The locations of these residues are highlighted in the modeled N- and C termini fitted into the TRPA1 density map (Fig. 5). This arrangement generates testable hypotheses for channel activation mechanisms in future studies. Taken together, this TRPA1 map reconstruction with protein modeling provides the first insight into the overall TRPA1 channel organization.
DISCUSSION
TRP channels are a family of cation channels that are involved in many sensory transduction pathways, including vision, hearing, touch, pain, and thermo- and mechano-sensation. In this study, we have determined the structure of one of the members of the TRP family, mouse TRPA1, using single-particle electron microscopy on purified protein heterologously expressed in S. cerevisiae. We confirmed that the expressed and purified TRPA1 protein forms functional channels within reconstituted vesicles. The TRPA1 structure provides clear evidence for a tetrameric assembly of TRPA1 with features resembling ankyrin repeats in the cytoplasmic portion of the protein. Furthermore, molecular modeling of the TRPA1 channel allows dissection of the subunit organization of the channel and reveals a possible ligand-binding pocket for pain-evoking electrophilic compounds.
Previous EM structural studies on a subset of TRP channels revealed the overall architecture for the TRPV, TRPC, and TRPM subfamilies (21–25). However, these intermediate-resolution structures have generated debate because of unexplained, significant differences between them. Two possibilities for the structural disparities are profound evolutionary divergence or differences in protein preparation. All these structures were determined in the presence of detergent, which could potentially induce artifacts. For example, detergents have a tendency to adhere to both the TM and soluble domains of membrane proteins (74–76). This could introduce additional mass or structural disturbances, which could be the cause for the slight differences between the TRPV1 and TRPV4 3D reconstructions. Therefore, we chose to stabilize the TRPA1 channel with A8-35 amphipol. The amphipol A8-35 has proven to be an effective agent for maintaining membrane proteins in a soluble, stabilized state. Amphipols have a high affinity for the TM region of membrane proteins and preferentially localize to these sites, as shown by EM and NMR (39).
Overall, our amphipol-stabilized TRPA1 reconstruction is similar to the “hanging basket” reconstructions determined for the TRPV subfamily and other ion channels resolved by EM (21, 22, 57, 77–79). The size of our TRPA1 density is in agreement with its molecular mass, as was observed with the TRPV channel reconstructions. We were able to fit the molecular model of the TM region of the TRPV1 channel into the TM region of our TRPA1 reconstruction. The additional volumetric mass observed in the TRPA1 EM density (see Figs. 4 and 5) could represent the A8-35 amphipol molecules, and/or the size of the TRPA1 pore could be larger than the TRPV1 pore (80–82). The dimensions of the TRPA1 cytoplasmic domain are comparable with the TRPV1 and TRPV4 channel structures, given the larger molecular weight of TRPA1 (21, 22). In addition, the molecular models of the N and C termini of TRPA1 fit well into the 3D EM density of the channel, further validating our EM reconstruction.
TRPA1 plays a key role in the transduction of chemical, inflammatory, and neuropathic pain signals from the periphery to the brain. It has been found that diverse exogenous electrophilic compounds (allicin, cinnamaldehyde, allylisothiocyanate) and endogenous thiol-reactive inflammatory mediators (NO, H2O2, 4-hydroxynonenal, 15-deoxy-Δ12,14-prostaglandin J2) can activate TRPA1. Mutagenesis studies revealed that TRPA1 activation by electrophiles occurs though covalent modification of specific N-terminal cysteine residues (44–46). From the three species tested, five different cysteine residues have been proposed to be involved in channel activation (mouse TRPA1: Cys-415, Cys-422, and Cys-622 (46); human TRPA1: Cys-622, Cys-642, and Cys-666 (44); Drosophila TRPA1: Cys-622 and Cys-642 (45); mouse numbering throughout). Mutations of these critical cysteines abolished activation of TRPA1 by electrophilic compounds, but structurally unrelated non-electrophilic agonists were still able to activate the Cys-mutant TRPA1 channels (44–46). Subsequent studies on human TRPA1 showed that Cys-422 was also important for response to electrophilic inflammatory mediators in the human channel and that Cys-415 was important for response to both reactive and non-reactive activators (48), raising the possibility that all five cysteines play a conserved role in TRPA1 structure-function. Taking these studies together, it has been proposed that the N terminus of the channel serves as a biosensor for the detection of a large class of noxious environmental agents that triggers channel opening.
In our results, the molecular architecture of TRPA1 revealed by single-particle EM and molecular modeling suggests a possible arrangement of the N termini that would bring the more organized region of the ankyrin repeats domain (residues 31–437) into close proximity with the more flexible region (residues 438–667) of the adjacent monomer (Fig. 5). We observed that the important mouse TRPA1 cysteine residues (Cys-415, Cys-422, Cys-622) form a ligand-binding pocket within this predicted structure. More specifically, Cys-415 and Cys-422 from one subunit could interact with the Cys-622 located on a flexible loop on the adjacent subunit. Cys-642, another cysteine implicated in human and fly TRPA1 electrophilic activation, is also notably located on the same flexible loop as Cys-622, and Cys-666 is on a flexible loop that could also interact with this cysteine pocket. This suggests that the mouse, human, and fly homologues of the TRPA1 channel need not be markedly structurally divergent to account for the functional differences between them. It is currently not known how covalent cysteine modifications may translate into protein activation. Because the cysteine binding pocket in our model bridges adjacent subunits, it is possible that covalent modifications within this pocket could facilitate or disrupt interactions within or between subunits and promote conformational changes in the cytoplasmic domains of TRPA1 that lead to channel gating (83). This hypothesis should be taken with caution, given that the N-terminal domain was modeled separately from the TM domain. However, with this in mind, our proposed arrangement of the N termini in the TRPA1 structure is in agreement with the mutagenesis studies that identified these important residues (44–46). To further our understanding of TRPA1-mediated pain mechanisms, both structural and functional experiments will be required to dissect the details of how these ligands activate the channel leading to signal transduction.
In conclusion, we present a ligand-free structure of TRPA1 combined with molecular modeling that delineates subunit organization and presents proposed locations for ligand binding. Together, these complimentary techniques have allowed us to extend our understanding of the TRPA1 ion channel molecular architecture.
Supplementary Material
Acknowledgments
We thank Dr. Andreas Engel for support and help during this project and for access to the state-of-the-art electron microscopy facilities at the Cleveland Center for Structural and Membrane Biology. We also thank Heather Holdaway and Dr. Jason Mears for technical assistance during this project, Dr. Fernandez Ballester for providing the TRPV1 molecular model, and Dr. Phoebe Stewart and Dr. Michael Maguire for critical reading of this manuscript.
This work was supported by American Lung Association Biomedical Research Grant RG-166985-N (to V. Y. M.-B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and movies 1 and 2.
The density map (accession number EMD-5334) has been deposited in the Electron Microscopy Data Bank.
- TRP
- transient receptor potential
- TRPV
- transient receptor potential vanilloid
- TRPC
- transient receptor potential canonical
- TRPM
- transient receptor potential melastatin
- TRPA1
- transient receptor potential ankyrin 1
- TM
- transmembrane
- FC12
- Fos-Choline 12
- SUV
- small unilamellar vesicle
- 3D
- three-dimensional
- A8-35
- amphipathic polymer A8-35
- TRPP
- transient receptor potential polycystin
- I-TASSER
- iterative threading assembly refinement.
REFERENCES
- 1. Levine J. D., Alessandri-Haber N. (2007) Biochim. Biophys. Acta 1772, 989–1003 [DOI] [PubMed] [Google Scholar]
- 2. Venkatachalam K., Montell C. (2007) Annu. Rev. Biochem. 76, 387–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Montell C. (2005) Sci. STKE 2005, re3. [DOI] [PubMed] [Google Scholar]
- 4. Nilius B., Voets T., Peters J. (2005) Sci. STKE 2005, re8. [DOI] [PubMed] [Google Scholar]
- 5. Latorre R., Zaelzer C., Brauchi S. (2009) Q. Rev. Biophys. 42, 201–246 [DOI] [PubMed] [Google Scholar]
- 6. Wang Y. Y., Chang R. B., Waters H. N., McKemy D. D., Liman E. R. (2008) J. Biol. Chem. 283, 32691–32703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gees M., Colsoul B., Nilius B. (2010) Cold Spring Harb. Perspect. Biol. 2, a003962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li M., Yu Y., Yang J. (2011) Adv. Exp. Med. Biol. 704, 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cosens D. J., Manning A. (1969) Nature 224, 285–287 [DOI] [PubMed] [Google Scholar]
- 10. Yamaguchi H., Matsushita M., Nairn A. C., Kuriyan J. (2001) Mol. Cell 7, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 11. Jin X., Touhey J., Gaudet R. (2006) J. Biol. Chem. 281, 25006–25010 [DOI] [PubMed] [Google Scholar]
- 12. McCleverty C. J., Koesema E., Patapoutian A., Lesley S. A., Kreusch A. (2006) Protein Sci. 15, 2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lishko P. V., Procko E., Jin X., Phelps C. B., Gaudet R. (2007) Neuron 54, 905–918 [DOI] [PubMed] [Google Scholar]
- 14. Phelps C. B., Huang R. J., Lishko P. V., Wang R. R., Gaudet R. (2008) Biochemistry 47, 2476–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Phelps C. B., Wang R. R., Choo S. S., Gaudet R. (2010) J. Biol. Chem. 285, 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujiwara Y., Minor D. L., Jr. (2008) J. Mol. Biol. 383, 854–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu Y., Ulbrich M. H., Li M. H., Buraei Z., Chen X. Z., Ong A. C., Tong L., Isacoff E. Y., Yang J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11558–11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petri E. T., Celic A., Kennedy S. D., Ehrlich B. E., Boggon T. J., Hodsdon M. E. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9176–9181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schumann F., Hoffmeister H., Bader R., Schmidt M., Witzgall R., Kalbitzer H. R. (2009) J. Biol. Chem. 284, 24372–24383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moiseenkova-Bell V. Y., Wensel T. G. (2009) J. Gen. Physiol. 133, 239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moiseenkova-Bell V. Y., Stanciu L. A., Serysheva II, Tobe B. J., Wensel T. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7451–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shigematsu H., Sokabe T., Danev R., Tominaga M., Nagayama K. (2010) J. Biol. Chem. 285, 11210–11218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mio K., Ogura T., Kiyonaka S., Hiroaki Y., Tanimura Y., Fujiyoshi Y., Mori Y., Sato C. (2007) J. Mol. Biol. 367, 373–383 [DOI] [PubMed] [Google Scholar]
- 24. Mio K., Ogura T., Hara Y., Mori Y., Sato C. (2005) Biochem. Biophys. Res. Commun. 333, 768–777 [DOI] [PubMed] [Google Scholar]
- 25. Maruyama Y., Ogura T., Mio K., Kiyonaka S., Kato K., Mori Y., Sato C. (2007) J. Biol. Chem. 282, 36961–36970 [DOI] [PubMed] [Google Scholar]
- 26. Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 27. Fernández-Ballester G., Ferrer-Montiel A. (2008) J. Membr. Biol. 223, 161–172 [DOI] [PubMed] [Google Scholar]
- 28. Ogura T., Sato C. (2001) J. Struct. Biol. 136, 227–238 [DOI] [PubMed] [Google Scholar]
- 29. Ogura T., Iwasaki K., Sato C. (2003) J. Struct. Biol. 143, 185–200 [DOI] [PubMed] [Google Scholar]
- 30. Ogura T., Sato C. (2004) J. Struct. Biol. 145, 63–75 [DOI] [PubMed] [Google Scholar]
- 31. Ogura T., Sato C. (2004) J. Struct. Biol. 146, 344–358 [DOI] [PubMed] [Google Scholar]
- 32. Tribet C., Diab C., Dahmane T., Zoonens M., Popot J. L., Winnik F. M. (2009) Langmuir 25, 12623–12634 [DOI] [PubMed] [Google Scholar]
- 33. Tribet C., Audebert R., Popot J. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15047–15050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Popot J. L., Althoff T., Bagnard D., Banères J. L., Bazzacco P., Billon-Denis E., Catoire L. J., Champeil P., Charvolin D., Cocco M. J., Crémel G., Dahmane T., de la Maza L. M., Ebel C., Gabel F., Giusti F., Gohon Y., Goormaghtigh E., Guittet E., Kleinschmidt J. H., Kühlbrandt W., Le Bon C., Martinez K. L., Picard M., Pucci B., Sachs J. N., Tribet C., van Heijenoort C., Wien F., Zito F., Zoonens M. (2011) Annu. Rev. Biophys. 40, 379–408 [DOI] [PubMed] [Google Scholar]
- 35. Catoire L. J., Zoonens M., van Heijenoort C., Giusti F., Guittet E., Popot J. L. (2010) Eur. Biophys. J. 39, 623–630 [DOI] [PubMed] [Google Scholar]
- 36. Flötenmeyer M., Weiss H., Tribet C., Popot J. L., Leonard K. (2007) J. Microsc. 227, 229–235 [DOI] [PubMed] [Google Scholar]
- 37. Gohon Y., Dahmane T., Ruigrok R. W., Schuck P., Charvolin D., Rappaport F., Timmins P., Engelman D. M., Tribet C., Popot J. L., Ebel C. (2008) Biophys. J. 94, 3523–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilkens S. (2000) J. Bioenerg. Biomembr. 32, 333–339 [DOI] [PubMed] [Google Scholar]
- 39. Zoonens M., Catoire L. J., Giusti F., Popot J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8893–8898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Popot J. L., Berry E. A., Charvolin D., Creuzenet C., Ebel C., Engelman D. M., Flötenmeyer M., Giusti F., Gohon Y., Hong Q., Lakey J. H., Leonard K., Shuman H. A., Timmins P., Warschawski D. E., Zito F., Zoonens M., Pucci B., Tribet C. (2003) Cell Mol. Life Sci. 60, 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bessac B. F., Jordt S. E. (2008) Physiology 23, 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baraldi P. G., Preti D., Materazzi S., Geppetti P. (2010) J. Med. Chem. 53, 5085–5107 [DOI] [PubMed] [Google Scholar]
- 43. Anand U., Otto W. R., Facer P., Zebda N., Selmer I., Gunthorpe M. J., Chessell I. P., Sinisi M., Birch R., Anand P. (2008) Neurosci. Lett. 438, 221–227 [DOI] [PubMed] [Google Scholar]
- 44. Hinman A., Chuang H. H., Bautista D. M., Julius D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19564–19568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kang K., Pulver S. R., Panzano V. C., Chang E. C., Griffith L. C., Theobald D. L., Garrity P. A. (2010) Nature 464, 597–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Macpherson L. J., Dubin A. E., Evans M. J., Marr F., Schultz P. G., Cravatt B. F., Patapoutian A. (2007) Nature 445, 541–545 [DOI] [PubMed] [Google Scholar]
- 47. McNamara C. R., Mandel-Brehm J., Bautista D. M., Siemens J., Deranian K. L., Zhao M., Hayward N. J., Chong J. A., Julius D., Moran M. M., Fanger C. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13525–13530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takahashi N., Mizuno Y., Kozai D., Yamamoto S., Kiyonaka S., Shibata T., Uchida K., Mori Y. (2008) Channels 2, 287–298 [DOI] [PubMed] [Google Scholar]
- 49. Andersson D. A., Gentry C., Moss S., Bevan S. (2008) J. Neurosci. 28, 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cavanaugh E. J., Simkin D., Kim D. (2008) Neuroscience 154, 1467–1476 [DOI] [PubMed] [Google Scholar]
- 51. De Petrocellis L., Vellani V., Schiano-Moriello A., Marini P., Magherini P. C., Orlando P., Di Marzo V. (2008) J. Pharmacol. Exp. Ther. 325, 1007–1015 [DOI] [PubMed] [Google Scholar]
- 52. Karashima Y., Damann N., Prenen J., Talavera K., Segal A., Voets T., Nilius B. (2007) J. Neurosci. 27, 9874–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Doerner J. F., Gisselmann G., Hatt H., Wetzel C. H. (2007) J. Biol. Chem. 282, 13180–13189 [DOI] [PubMed] [Google Scholar]
- 54. Lee S. P., Buber M. T., Yang Q., Cerne R., Cortés R. Y., Sprous D. G., Bryant R. W. (2008) Br. J. Pharmacol. 153, 1739–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Story G. M., Peier A. M., Reeve A. J., Eid S. R., Mosbacher J., Hricik T. R., Earley T. J., Hergarden A. C., Andersson D. A., Hwang S. W., McIntyre P., Jegla T., Bevan S., Patapoutian A. (2003) Cell 112, 819–829 [DOI] [PubMed] [Google Scholar]
- 56. Macpherson L. J., Xiao B., Kwan K. Y., Petrus M. J., Dubin A. E., Hwang S., Cravatt B., Corey D. P., Patapoutian A. (2007) J. Neurosci. 27, 11412–11415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sokolova O. (2004) FEBS Lett. 564, 251–256 [DOI] [PubMed] [Google Scholar]
- 58. Figler R. A., Omote H., Nakamoto R. K., Al-Shawi M. K. (2000) Arch. Biochem. Biophys. 376, 34–46 [DOI] [PubMed] [Google Scholar]
- 59. Moiseenkova V. Y., Hellmich H. L., Christensen B. N. (2003) Biochem. Biophys. Res. Commun. 310, 196–201 [DOI] [PubMed] [Google Scholar]
- 60. Bautista D. M., Movahed P., Hinman A., Axelsson H. E., Sterner O., Högestätt E. D., Julius D., Jordt S. E., Zygmunt P. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12248–12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sreerama N., Woody R. W. (2000) Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 62. Sreerama N., Woody R. W. (2004) Protein Sci. 13, 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 64. Roy A., Kucukural A., Zhang Y. (2010) Nat. Protoc 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang Y. (2008) BMC Bioinformatics 9, 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Y. (2009) Proteins 77, 100–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 68. Pintilie G. D., Zhang J., Goddard T. D., Chiu W., Gossard D. C. (2010) J. Struct. Biol. 170, 427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cvetkov T. L., Prochaska L. J. (2007) Protein Expr. Purif. 56, 189–196 [DOI] [PubMed] [Google Scholar]
- 70. Heginbotham L., Kolmakova-Partensky L., Miller C. (1998) J. Gen. Physiol. 111, 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Greenfield N. J. (2006) Nat. Protoc. 1, 2876–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kelly S. M., Jess T. J., Price N. C. (2005) Biochim. Biophys. Acta 1751, 119–139 [DOI] [PubMed] [Google Scholar]
- 73. Hu H., Bandell M., Petrus M. J., Zhu M. X., Patapoutian A. (2009) Nat. Chem. Biol. 5, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Garavito R. M., Ferguson-Miller S. (2001) J. Biol. Chem. 276, 32403–32406 [DOI] [PubMed] [Google Scholar]
- 75. le Maire M., Champeil P., Moller J. V. (2000) Biochim. Biophys. Acta 1508, 86–111 [DOI] [PubMed] [Google Scholar]
- 76. Gohon Y., Popot J. L. (2003) Curr. Opin. Coll. Int. Sci. 8, 15–22 [Google Scholar]
- 77. Sokolova O., Kolmakova-Partensky L., Grigorieff N. (2001) Structure 9, 215–220 [DOI] [PubMed] [Google Scholar]
- 78. Orlova E. V., Papakosta M., Booy F. P., van Heel M., Dolly J. O. (2003) J. Mol. Biol. 326, 1005–1012 [DOI] [PubMed] [Google Scholar]
- 79. Chiu P. L., Pagel M. D., Evans J., Chou H. T., Zeng X., Gipson B., Stahlberg H., Nimigean C. M. (2007) Structure 15, 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Banke T. G. (2011) Brain Res. 1381, 21–30 [DOI] [PubMed] [Google Scholar]
- 81. Bobkov Y. V., Corey E. A., Ache B. W. (2011) Biochim. Biophys. Acta 1808, 1120–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Karashima Y., Prenen J., Talavera K., Janssens A., Voets T., Nilius B. (2010) Biophys. J. 98, 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Caterina M. J. (2007) Nature 445, 491–492 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.