Abstract
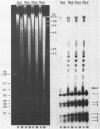
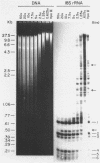
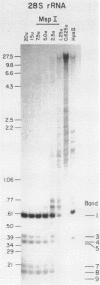
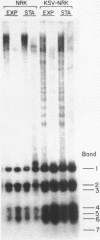
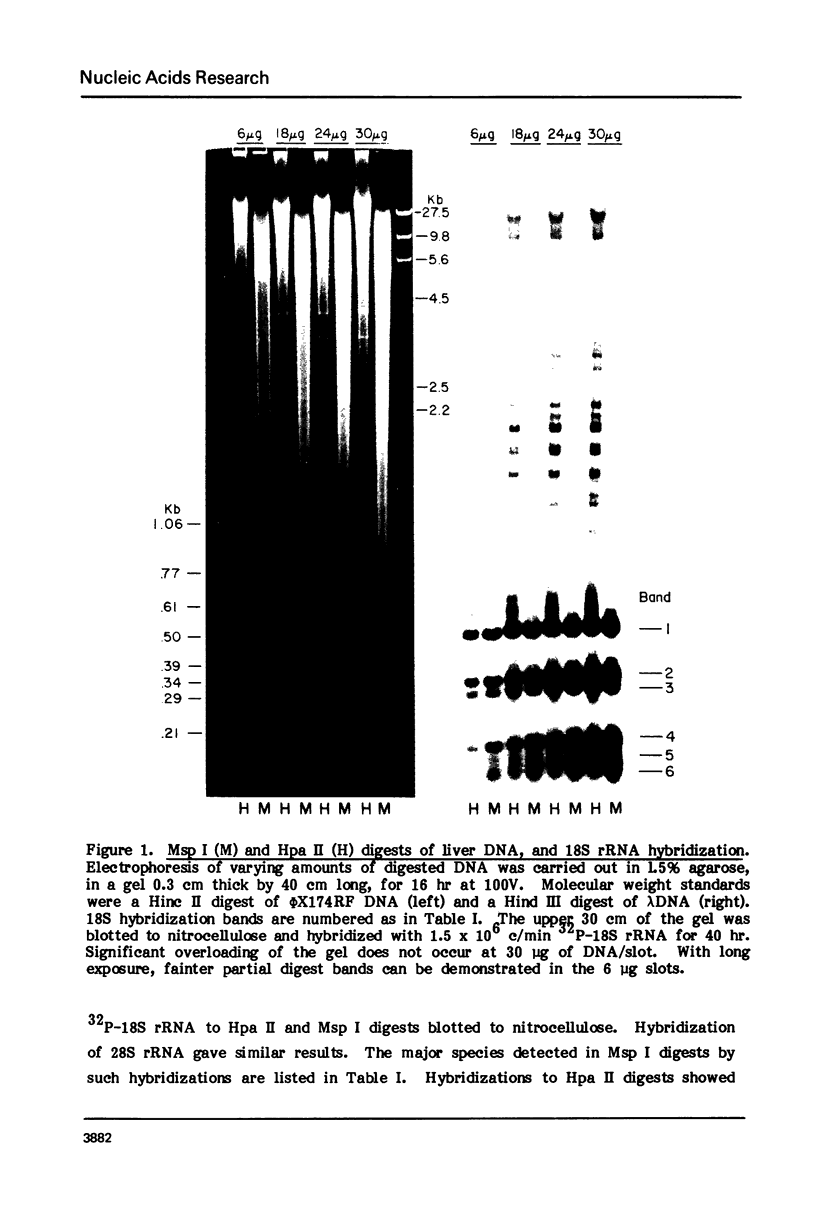
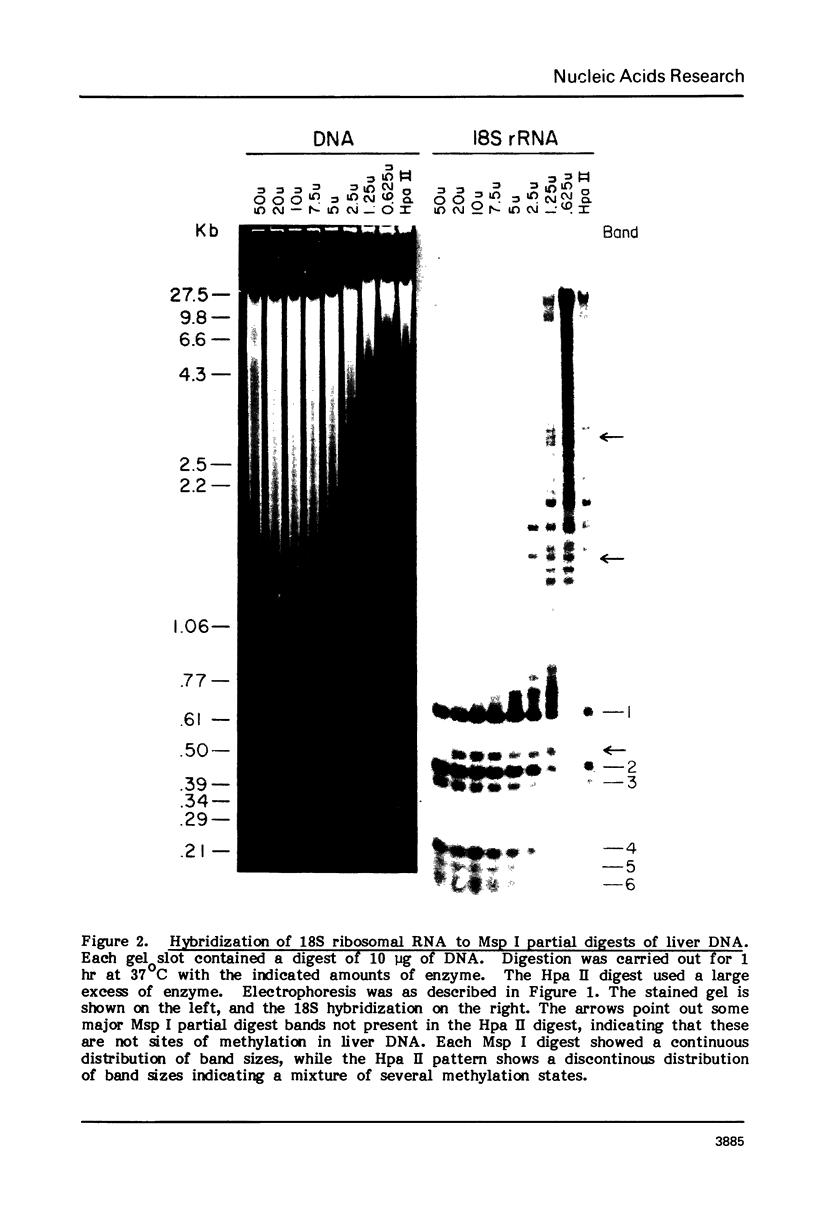
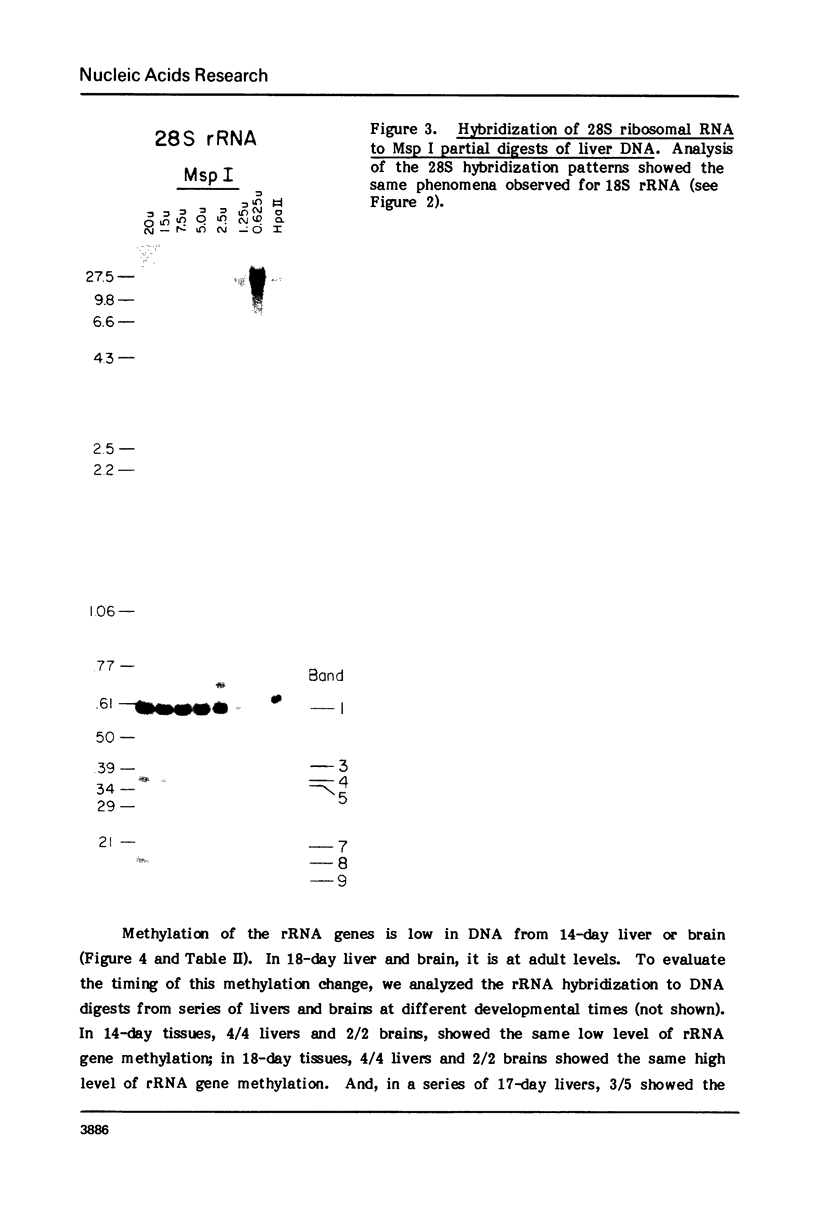
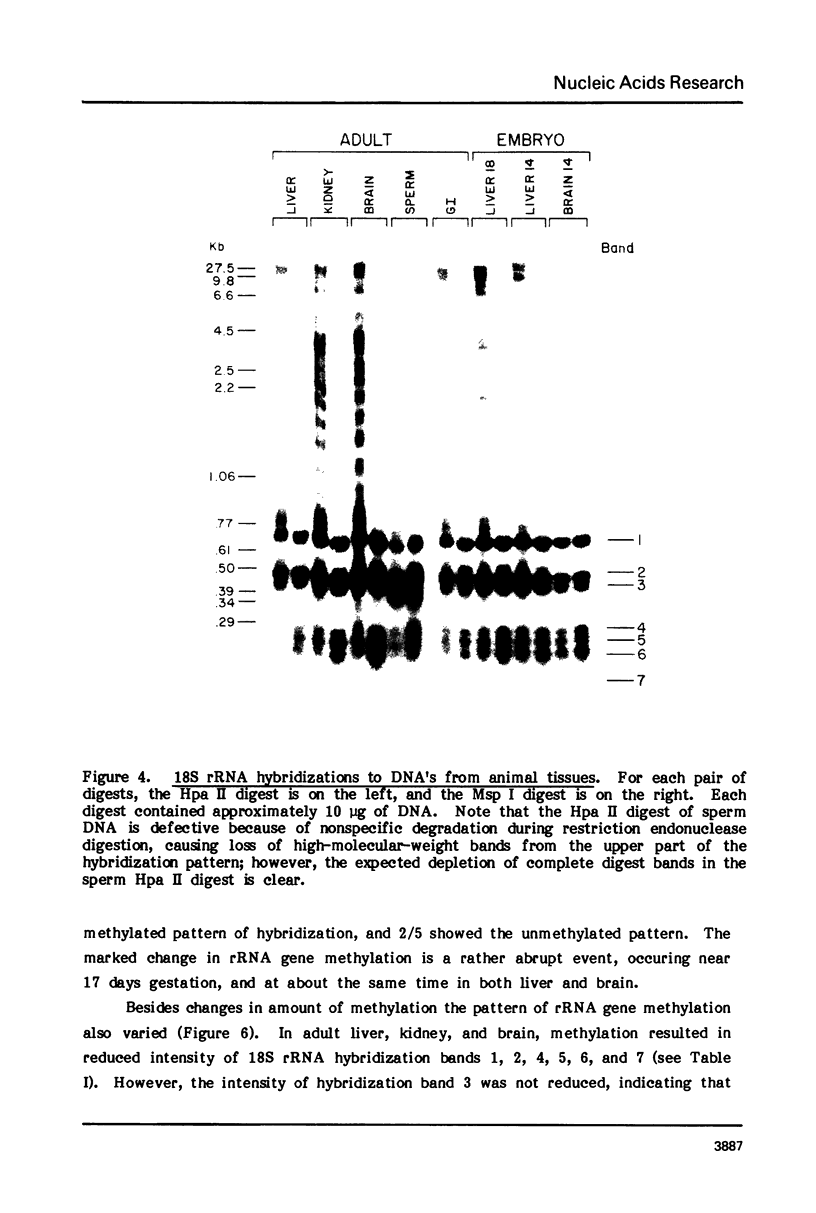
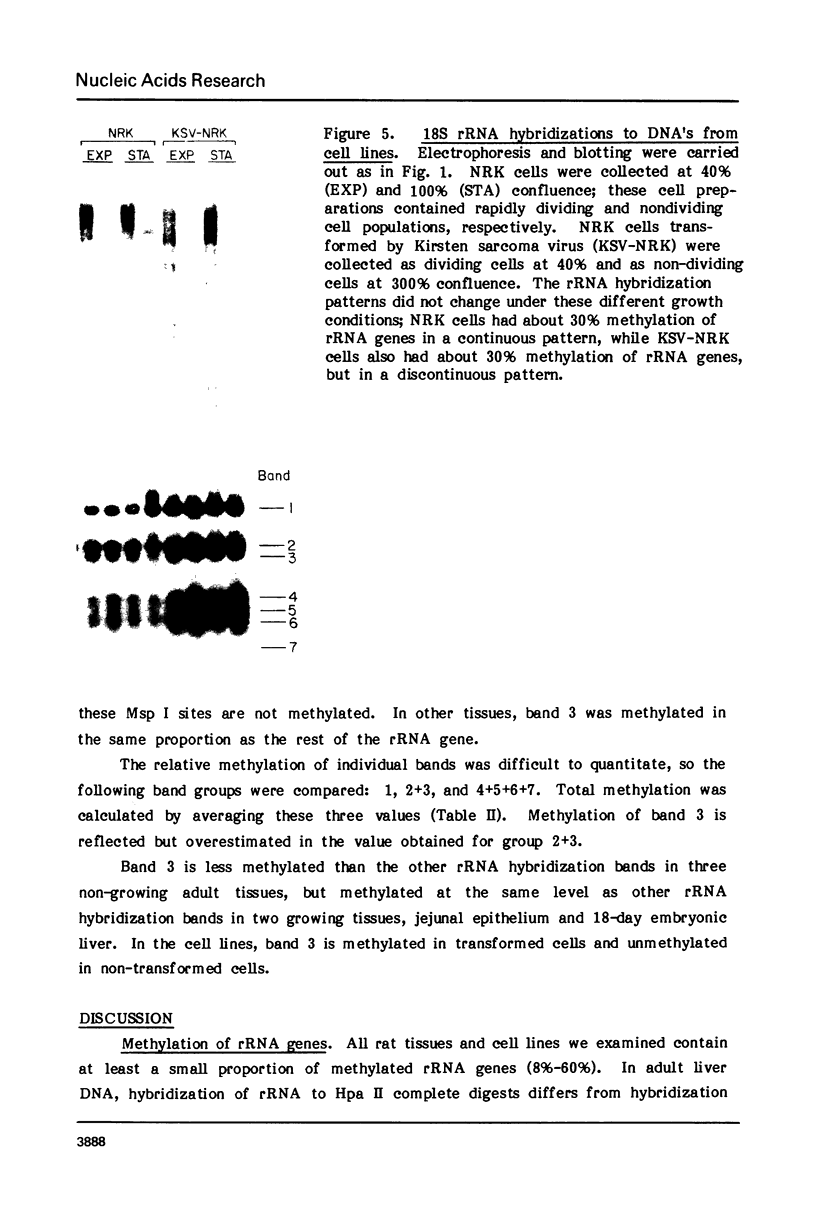
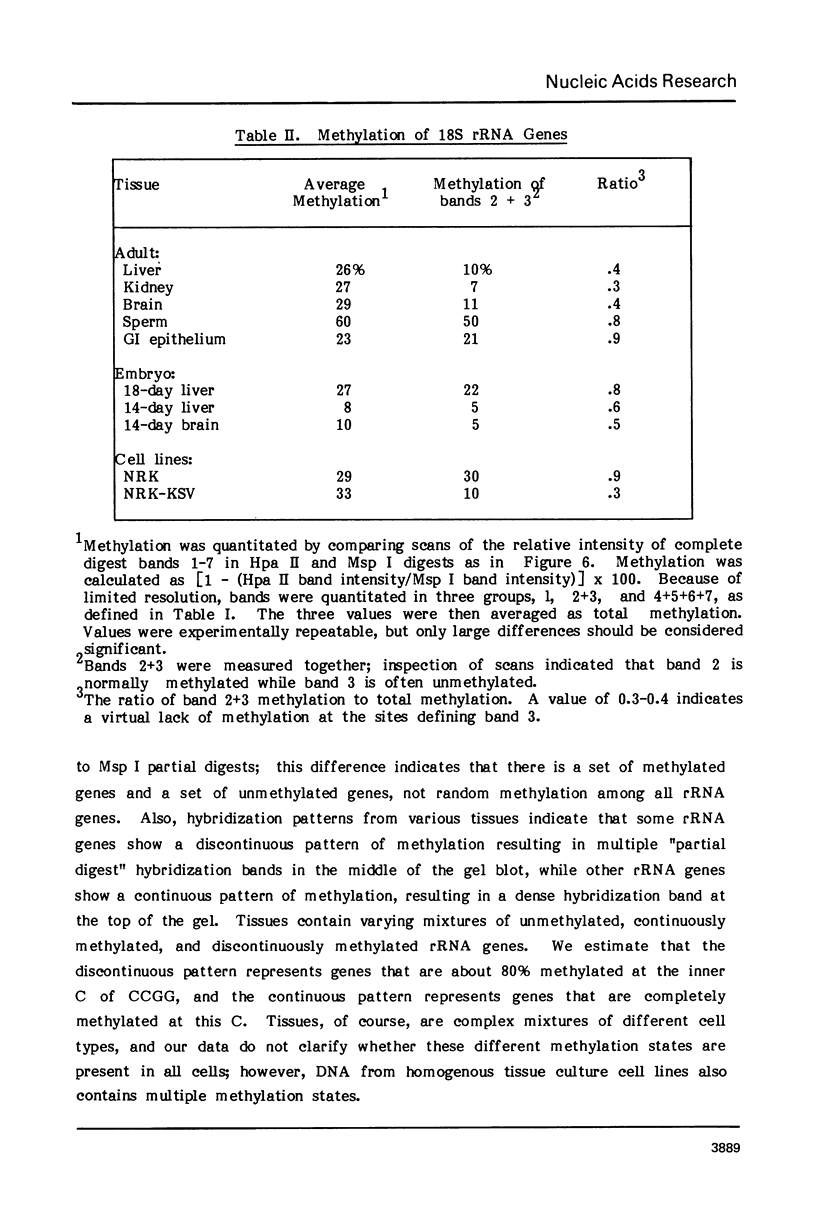
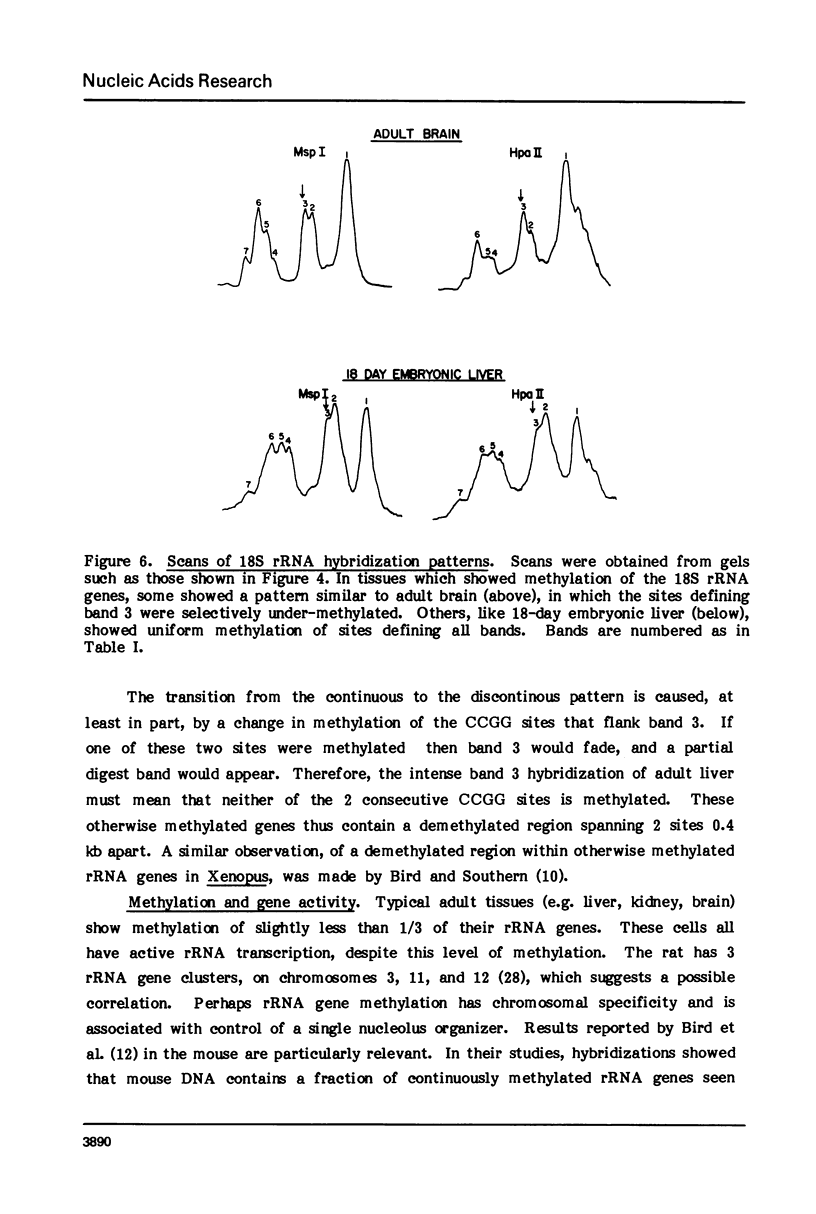
Both the pattern and level of rRNA gene methylation vary in the rat. This variation reflects stages in the maturation process and perhaps the level of gene expression in different tissues. We studied methylation at a common site, the inner cytosine of the sequence CCGG, by hybridizing 32P-rRNA to DNA digests obtained with endonuclease Msp I (which cleaves CCGG and CMCGG) and its isochizomer, HpaII (which cleaves only CCGG). In the liver, the changing pattern of rRNA gene methylation reflected the late stages of development: the rRNA genes were mostly unmethylated at 14 days gestation; by 18 days gestation, about 30% of them were methylated, and this level persisted into adulthood. In 18-day DNA, the methylation was uniform, but in adult DNA, the methylation pattern was discontinuous, because otherwise methylated genes contained a demethylated region. Similar developmental changes were observed in brain DNA. In a tissue culture cell line, the change from the continuous to the discontinuous pattern of the methylation could be induced by transformation with Kirsten sarcoma virus. And, in adult tissues, the lowest level of rRNA gene methylation was found in rapidly growing jejunal epithelium, and the highest level, in non-growing spermatozoa.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Parker B. A., Reiser J., Renart J., Stark G. R., Wahl G. M. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H., Gehring C. A. Methylated and unmethylated ribosomal RNA genes in the mouse. J Mol Biol. 1981 Oct 15;152(1):1–17. doi: 10.1016/0022-2836(81)90092-9. [DOI] [PubMed] [Google Scholar]
- Bird A. P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978 Jan 5;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Desai S. M., Hunt C., Locker J., Weiss S. B. Localization of the arginine tRNA gene to the D segment of T5 bacteriophage DNA. A new procedure for producing duplex DNA fragments. J Biol Chem. 1978 Sep 25;253(18):6544–6550. [PubMed] [Google Scholar]
- Desrosiers R. C., Mulder C., Fleckenstein B. Methylation of Herpesvirus saimiri DNA in lymphoid tumor cell lines. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3839–3843. doi: 10.1073/pnas.76.8.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippo P., Iaccarino M., Parisi E., Scarano E. Methylation of DNA in developing sea urchin embryos. J Mol Biol. 1968 Sep 14;36(2):195–208. doi: 10.1016/0022-2836(68)90375-6. [DOI] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Jelinek W., Molloy G., Fernandez-Munoz R., Salditt M., Darnell J. E. Secondary structure in heterogeneous nuclear RNA: involvement of regions from repeated DNA sites. J Mol Biol. 1974 Jan 25;82(3):361–370. doi: 10.1016/0022-2836(74)90597-x. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Günthert U., Trautner T. A. DNA methyltransferases affecting the sequence 5'CCGG. Nucleic Acids Res. 1981 Jun 25;9(12):2753–2759. doi: 10.1093/nar/9.12.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano Y., Maeda S., Sugiyama T. The location of ribosomal cistrons (rDNA) in chromosomes of the rat. Chromosoma. 1976 Mar 31;55(1):37–42. doi: 10.1007/BF00288325. [DOI] [PubMed] [Google Scholar]
- Locker J. Analytical and preparative electrophoresis of RNA in agarose-urea. Anal Biochem. 1979 Oct 1;98(2):358–367. doi: 10.1016/0003-2697(79)90154-4. [DOI] [PubMed] [Google Scholar]
- Panem S., Kirsten W. H. Release of mouse leukemia-sarcoma virus from synchronized cells. J Natl Cancer Inst. 1973 Feb;50(2):563–565. doi: 10.1093/jnci/50.2.563. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Weissbach A. DNA methylase from HeLa cell nuclei. Nucleic Acids Res. 1975 Oct;2(10):1669–1684. doi: 10.1093/nar/2.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Kitchin R. Selective silencing of eukaryotic DNA. Science. 1975 Aug 8;189(4201):426–433. [PubMed] [Google Scholar]
- Shiurba R., Nandi S. Isolation and characterization of germ line DNA from mouse sperm. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3947–3951. doi: 10.1073/pnas.76.8.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers K. D., Kirsten W. H. Long-term propagation of a murine erythroblastosis virus in vitro. J Natl Cancer Inst. 1968 May;40(5):1053–1065. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stumph W. E., Wu J. R., Bonner J. Determination of the size of rat ribosomal deoxyribonucleic acid repeating units by electron microscopy. Biochemistry. 1979 Jun 26;18(13):2864–2871. doi: 10.1021/bi00580a030. [DOI] [PubMed] [Google Scholar]
- Sugden B., De Troy B., Roberts R. J., Sambrook J. Agarose slab-gel electrophoresis equipment. Anal Biochem. 1975 Sep;68(1):36–46. doi: 10.1016/0003-2697(75)90676-4. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]