Abstract
Using brain transcriptomic profiles from 853 individual honey bees exhibiting 48 distinct behavioral phenotypes in naturalistic contexts, we report that behavior-specific neurogenomic states can be inferred from the coordinated action of transcription factors (TFs) and their predicted target genes. Unsupervised hierarchical clustering of these transcriptomic profiles showed three clusters that correspond to three ecologically important behavioral categories: aggression, maturation, and foraging. To explore the genetic influences potentially regulating these behavior-specific neurogenomic states, we reconstructed a brain transcriptional regulatory network (TRN) model. This brain TRN quantitatively predicts with high accuracy gene expression changes of more than 2,000 genes involved in behavior, even for behavioral phenotypes on which it was not trained, suggesting that there is a core set of TFs that regulates behavior-specific gene expression in the bee brain, and other TFs more specific to particular categories. TFs playing key roles in the TRN include well-known regulators of neural and behavioral plasticity, e.g., Creb, as well as TFs better known in other biological contexts, e.g., NF-κB (immunity). Our results reveal three insights concerning the relationship between genes and behavior. First, distinct behaviors are subserved by distinct neurogenomic states in the brain. Second, the neurogenomic states underlying different behaviors rely upon both shared and distinct transcriptional modules. Third, despite the complexity of the brain, simple linear relationships between TFs and their putative target genes are a surprisingly prominent feature of the networks underlying behavior.
Keywords: Apis mellifera, gene regulation, social behavior, systems biology
Behavior is influenced by both heritable and environmental factors, sometimes via massive changes in brain transcriptomes (1). An emerging insight is that these changes induce shifts in “neurogenomic states” rather than activation of particular genes only in local neural circuits (2). This has led to the idea that distinct neurogenomic states underlie distinct behaviors (1), but it is not known how these states are defined or maintained. Further, the regulatory architecture of behaviorally relevant neurogenomic states has not been studied, and it is not known whether behavior is subserved by the kinds of transcriptional regulatory networks (TRNs) known for other phenotypes (3–6).
We applied tools and perspectives from molecular systems biology—used to study transcriptional regulation in the brain and elsewhere (3–6)—to transcript profiles from the BeeSpace Project, which used microarray analysis to study hereditary and environmental influences on brain gene expression and social behavior (Methods). This provided a unique aggregate dataset from a single laboratory (G.E.R.), using the same analytical platform, protocols, and analysis procedures (7). Because the natural behavioral repertoire of the honey bee (Apis mellifera) is perhaps the best studied of any nonhuman animal (8), we were able to analyze a rich set of naturalistic behavioral states.
We chose 27 pairwise comparisons among the 48 distinct behavior states that directly surveyed transcriptome responses to a heritable or environmental factor related to one or more of three ecologically important behavioral categories: aggression, hive defense; maturation, from working in the hive to foraging for nectar and pollen; and different foraging predispositions or types of experience, e.g., scouting for new floral resources or forming specific spatiotemporal memories for known resources, respectively (Methods and SI Appendix, Table S1). Nearly all genes were differentially expressed in at least one comparison; this broad survey thus captured natural variation across most of the brain transcriptome, without experimental genetic perturbation. SI Appendix, Tables S1 and S2 summarize the number of bees, experiments, and microarrays in this dataset and indicate which experiments have been published or will be described in forthcoming publications. Additional details of these experiments are in the SI Appendix, SI Methods.
We used a systems approach to create, test, and interpret a network model that predicts the expression of a large number of genes in the bee brain given the expression values of the TFs. Our strategy consisted of the following steps: (i) Integrate previous measurements of brain gene expression over a wide range of behavior states, which consist of behavioral responses to various genetic and environmental perturbations. (ii) Integrate expression from various conditions and develop a predictive behavioral TRN model from training data that links expression of putative target genes to the expression values of the TFs. (iii) Evaluate the TRN model in new test conditions and also on the basis of comparison with genomic data. (iv) Analyze the properties of the TRN with respect to other networks inferred for other phenotypes. (v) Identify key regulators/hubs in the TRN that predict behavior and may drive behavioral changes. (vi) Compare various subnetworks associated with aggression, maturation, and foraging.
Results and Discussion
Neurogenomic States Related to Behavior.
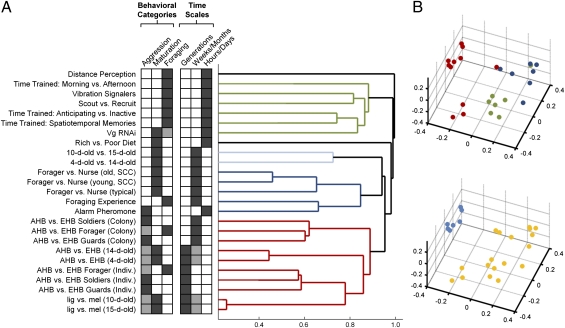
We first used hierarchical clustering to determine whether brain transcriptomic profiles tracked behavior. Each comparison was designed a priori to measure transcriptome responses to a heritable or environmental factor related to aggression, maturation, or foraging. Hierarchical clustering recapitulated this pattern; we identified three distinct clusters of transcriptomic profiles, corresponding to aggression, maturation, and foraging. This result demonstrates a strong relationship between behavior and brain gene expression, as it was obtained via unsupervised clustering (Fig. 1A and SI Appendix, Fig. S1).
Fig. 1.
Global analysis of results of transcriptome profile experiments of hereditary and environmental influences on brain gene expression and social behavior in adult honey bees. (A) Unsupervised hierarchical clustering groups brain transcriptional profiles generated by the comparisons listed in the Left column roughly into three major behavioral categories (P < 0.001, based on bootstrap) (SI Appendix, SI Text). Edges of the dendrogram are colored on the basis of results of clustering: red, aggression; blue, maturation; and green, foraging. This analysis also revealed differences due to the timescale over which a particular effect occurred; environmental factors exert effects within the lifespan, whereas heritable factors act across generations over evolutionary time. Shaded boxes show a priori classification of comparisons by behavioral categories and timescales on the basis of generally accepted knowledge of bee behavior (SI Appendix, Table S1). Some comparisons influence multiple categories or timescales; the primary effect of each comparison is denoted with a dark gray box, and the secondary focus is denoted with a light gray box. In subsequent analyses in this paper, we use only the primary effect for each comparison on the basis of prior knowledge in the literature (SI Appendix, Table S1). (B) MDS plot of transcriptional profiles clearly separates the different behavioral categories (same color coding as in A) and effects of environmental (yellow) and heritable (blue) factors. Comparisons involving queen mandibular pheromone and high- vs. low-pollen hoarding (Fig. 3) were not included in this analysis due to large numbers of missing values (full description of comparisons in SI Appendix, Table S1; additional statistics in SI Appendix, Fig. S1 and SI Methods). AHB, African honey bee; EHB, European honey bee; lig, A. mellifera ligustica; mel, A.m. mellifera; typical, typical colony; SCC, single-cohort colony (induces accelerated maturation); Vg, vitellogenin.
Clustering analysis also revealed differences between transcriptome responses to environmental or heritable factors (Fig. 1A). We found, for example, distinct subclusters of aggression-related comparisons corresponding to environmental (colony environment), or hereditary (individual genotype) factors. SI Appendix, Table S2 describes each of the comparisons and the degree of gene expression differences between the states.
The two results obtained with hierarchical clustering also were obtained independently with multidimensional scaling (MDS) (Fig. 1B). These findings indicate that distinct neurogenomic states underlie distinct behaviors and hint at differential effects of “nature” and “nurture.”
Brain Transcriptional Regulatory Network.
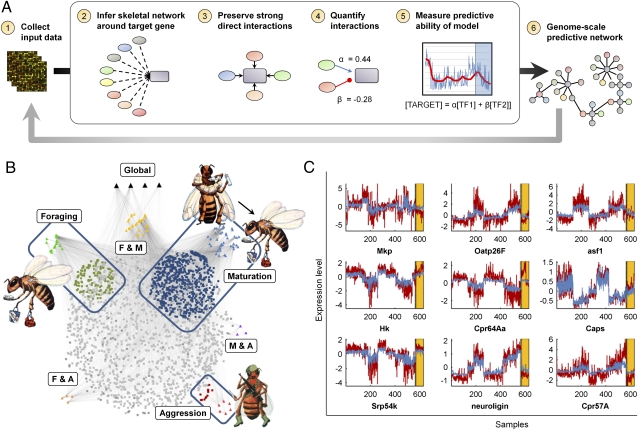
To explore the mechanisms potentially regulating behavior-specific neurogenomic states, we reconstructed a brain TRN model using an approach we call Analyzing Subsets of Transcriptional Regulators Influencing eXpression (ASTRIX). This approach is based on a combination of two well-known algorithms used for network inference (Fig. 2A and Methods). We generated a network of high-confidence putative TF (“target”)–gene interactions using Accurate Reconstruction of Cellular Networks (ARACNE) (9) and leveraged these interactions to predict expression in new conditions with Least Angle Regression (LARS) (10). In the training set, a subset of TF–gene relationships are identified that can accurately predict the target gene’s expression quantitatively, and then these interactions are moved forward to the test set for validation. Interactions were inferred from expression profiles and a list of 236 predicted bee TFs based on orthology with Drosophila melanogaster.
Fig. 2.
Reconstruction of a genome-scale model of a behaviorally related transcriptional regulatory network (TRN) from honey bee brain transcriptome profiles. (A) Flowchart outlining the ASTRIX approach: (1) input transcriptomic dataset; (2) “Skeletal” network inferred with ARACNE; high mutual information threshold (P < 10–6) retains only strong (putatively direct) interactions; (3) Data processing inequality (DPI) eliminates indirect interactions; (4) LARS fits a regression model for the regulation of each gene by transcription factors (TFs); (5) model accuracy for each gene assessed in training and test sets; and (6) accurately modeled genes retained in a parsimonious, quantitative model that predicts gene expression from TF expression. (B) A total of 190 TFs (triangles outside the sphere) are predicted to regulate 2,176 genes via 6,757 interactions (mean indegree = 3; mean outdegree = 35, Table 1, SI Appendix, Tables S2 and S11). Black triangles at the top represent the predicted global regulators: br, lilli, dl, and GB13780. The genes unique to each behavioral category (subnetwork; same color coding as in Fig. 1A) are highlighted along with their corresponding subnetwork specific TFs (within each box). TFs predicted to be involved in the regulation of two subnetworks are labeled between the boxes. (C) Plots show the accurate fit of the model (red) to actual expression data (blue) in training and test samples (Left and Right of the blue dotted line, respectively), for nine randomly selected genes with training correlation >0.8.
The bee brain TRN quantitatively predicts gene expression changes with remarkably high accuracy, even for behavioral states on which it was not trained (Fig. 2B and Table 1 and SI Appendix, Tables S2 and S11). It was able to model ∼25% (2,176) of the genes profiled (chosen on the basis of strong fit in the training set, Pearson correlation r > 0.8) and quantitatively predicted their expression with an average correlation of 0.87 in test sets (root mean square deviation, RMSD = 0.35, both in training and test sets). By contrast, a control experiment with TF expression permuted across states resulted in a 0.22 correlation, and zero genes predicted with >0.8 correlation, even in the training set (SI Appendix, Fig. S4). Stringent leave-one-state-out cross validation, with model prediction tested on behavior states for which it had not been trained, yielded similarly accurate results (RMSDtrain = 0.33, RMSDtest = 0.32); likewise for the 10-fold cross-validation test (Fig. 2C and SI Appendix, Figs. S2 and S3). The model’s success in predicting expression in new behavioral states suggests that there is a core set of TFs that regulate behavior-specific gene expression in the bee brain.
Table 1.
General features of the genome-scale model of a behaviorally related transcriptional regulatory network from honey bee brain transcriptome profiles
| Features | Values |
| Total genes in the network | 2,176 |
| TFs in the data set | 236 |
| TFs in the network | 190 |
| Interactions in the network | 6,757 |
| Mean indegree | 3 ± 1 |
| Mean outdegree | 35 ± 50 |
| Total modules | 190 |
| Modules enriched for GO/KEGG categories | 166 |
| Modules enriched for an upstream motif | 109 |
| Modules with enrichment for differentially expressed genes (DEG) | 108 |
| Global regulators | 4 |
| Correlation threshold | 0.8 |
| Correlation 10-fold CV (train and test) | 0.87 |
| Correlation background | 0 |
This TRN includes genes with known neurobiological and behavioral significance. Many neural-related Gene Ontology (GO) categories like synaptic transmission and neurotransmitter uptake were overrepresented among the TRN genes (SI Appendix, Table S4), indicating that features reflecting the general organization of the bee brain transcriptome are consistent with those of known brain processes. Genes in modules often shared annotated biological features: 87% (166 out of 190) of the modules were enriched for specific biological processes [based on GO or Kyoto Encyclopedia of Genes and Genomes annotations; SI Appendix, Table S5; false discovery rate (FDR) < 0.1]. Genes in modules also often shared evolutionarily conserved cis-regulatory motifs in upstream sequences (FDR < 0.1; SI Appendix, Table S5). Some modules also showed high similarity with previously described Drosophila networks (SI Appendix, Table S5), consistent with the existence of conserved modules of coregulated genes related to specific biological functions (11). As expected, genes in the TRN were enriched (P < 1e-10) for those that were strongly differentially expressed between behavioral states.
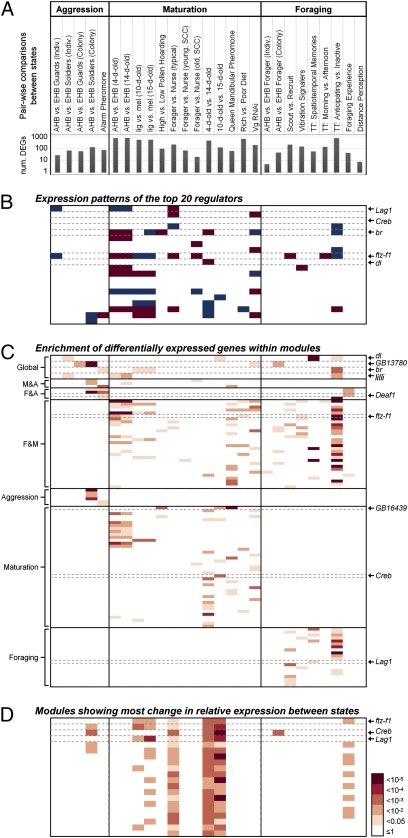
Having demonstrated predictive ability and biological relevance, we analyzed the TRN to explore the hypothesis that behaviorally related neurogenomic states arise, in part, from the coordinated action of TFs and their predicted target genes; the predicted targets were defined as those genes that share very high mutual information (P < 10−6) with a TF and have high predictive ability (r > 0.8). The top 20 most connected TFs (“hubs”) together were predicted to regulate 75% of the genes, and the top 3, 33% (SI Appendix, Figs. S5 and S6). Most of the top 20 hubs and many of their predicted targets were differentially expressed (FDR < 0.05) across many of the 27 comparisons (Fig. 3B and SI Appendix, Table S6). Only four TFs and their modules (sets of putative target genes predicted to be regulated by a specific TF) were active across all three behavioral categories (“global regulators”), i.e., their predicted target genes were significantly overrepresented among differentially expressed genes across all three behavioral categories. Other TFs and their modules were active only for a particular behavioral state within a category (Fig. 3C and SI Appendix, Table S7). State-specific TFs were significantly more common than predicted by chance (P < 1e-10), indicating that they also are a robust feature of this TRN. These results provide support for our hypothesis.
Fig. 3.
Global and behavior-specific regulators in the bee brain transcriptional regulatory network. (A) Extent of transcriptomic change (number of differentially expressed genes, DEGs) in each of the 27 pairwise comparisons between states that relate to aggression, maturation, or foraging. (B) Patterns of differential expression (FDR < 0.05) for the top 20 hubs in the TRN. TFs (rows) sorted by size of predicted gene modules. Blue, up-regulation; red, down-regulation. (C) TFs whose predicted target genes were “active” (significantly overrepresented among differentially expressed genes) across all three behavioral categories (“global regulators”) and TFs active only for aggression, maturation, or foraging (full description in SI Appendix, Table S7). Shaded boxes indicate the significance level for each enriched module (hypergeometric distribution). (D) TF-gene modules with the most significant between-state changes in the relative expression of module members (P values in legend at Right).
TFs playing key roles in the bee brain TRN include well-known regulators of neural and behavioral plasticity. Creb, an iconic regulator of neural plasticity in both invertebrates and vertebrates (12), is a top hub. Two other hubs are ftz-f1 and broad (br), which mediate responses to juvenile hormone (JH) (13), a hormone that regulates bee maturation, brain chemistry, and brain structure (14). The TRN also prominently features TFs better known in other contexts. For example, dorsal (dl), one of only four global regulators, is the insect ortholog of NF-κB (15), a well-known mediator of immune responses.
We were able to robustly and accurately model a large percentage (∼25%) of the brain transcriptome from whole-brain data. This might seem surprising, because neuron-, circuit-, and brain region-specific patterns of gene expression are well known (16). However, some hormones and neuromodulators can have widespread transcriptional effects on the brain (17). We used the results of two BeeSpace experiments that profiled gene expression in specific brain regions to explore whether performance of the TRN might be enhanced for a more homogenous population of cells. By contrast, the TRN was a better predictor of gene expression for whole-brain transcriptome profiles than for brain-region–specific profiles (Methods, SI Appendix, Fig. S10 and Table S10). These results suggest that additional factors that functionally link multiple cell types and brain regions may drive gene expression across the brain, including hormones, immune/inflammatory responses, and synaptic connections between neurons. Such factors are represented in the TRN by prominent hub genes (ftz-f1 and br; dl and Deaf1; and Creb, respectively. Other modules may reflect more general cellular functions, e.g., those involving general “housekeeping” (16); the largest module in the bee brain TRN is regulated by Lag1, a TF linked to metabolism (18). These and likely other reasons made it possible for the TRN to capture coherent patterns of transcriptional regulation, even for averaged values of gene expression across the brain.
Two intriguing dynamic properties of this TRN were detected, but their biological significance is unclear. First, whereas modules generally showed consistent gene expression across individuals within behavioral states (SI Appendix, Fig. S7), some modules exhibited significant changes in relative gene expression ordering between states (Fig. 3D). This “shuffling” was greatest for modules associated with hub genes (SI Appendix, Fig. S8) such as ftz-f1, Creb, and Lag1. Gene shuffling is associated with transcriptional “dysregulation” in diseased states (19), but such differences have not yet been detected in network analysis for normal states. Second, the shuffling varied depending on behavioral timescale. Increased shuffling was more often associated with longer-lasting behavioral states, e.g., those related to behavioral maturation, rather than more dynamic states, e.g., spatiotemporal floral memories (P = 0.008; Fig. 3D and SI Appendix, Table S8 and Fig. S9). Longer-lasting states also showed a significant increase in the number of TFs predicted to regulate each gene (P = 0.002; SI Appendix, Fig. S8). These results are reminiscent of the above-mentioned cluster analysis timescale differences and again hint at differences in transcriptional regulation over different timescales.
Validation of Transcriptional Regulatory Network.
Comprehensive validation of the predictions of this TRN would require manipulation of brain expression of multiple TFs with RNAi across multiple behavioral states under natural conditions. This is not yet feasible, although RNAi treatment has been used to test the effects of single genes in individual behavioral contexts in the field (20). Instead, partial validation for some of our results was obtained by four comparative bioinformatics analyses. (i) Genes in the modules of 14 TFs in the bee brain TRN had significant overlap with the direct, physical targets of the orthologous TF in Drosophila (11), (SI Appendix, Table S5, P < 10−15), compared with randomized bee TRN data, which had approximately two overlapping modules (SI Appendix, Fig. S12). (ii) The modules for 6 TFs in the bee brain TRN were enriched for the cis-regulatory motif recognized by their Drosophila ortholog (SI Appendix, Table S5). (iii) The prediction of state-specific TF activity was supported by results from an independent chromatin immunoprecipitation (ChIP-on-chip) study, which identified 16 of the TFs in the bee brain TRN linked to foraging and maturation as direct targets of Ultraspiracle. Ultraspiracle has been shown to mediate maturationally related transcriptional responses to JH in the honey bee (13) (SI Appendix, Table S9). This result further supports our hypothesis that additional factors like hormones may drive global gene expression across the brain. (iv) This TRN linked the TF Deaf1 to bee aggression, as has been shown previously (7), and the predicted targets of Deaf1 overlapped significantly with the targets of Deaf1 in Drosophila. These findings suggest that many modules of the bee brain TRN involve direct physical interactions between TF and target genes and highlight strong evolutionary conservation across bees and flies, despite a ca. 300-Myr divergence.
Conclusions
This bee brain TRN does not encompass all of the layers of complexity inherent in brain function, which includes indirect and combinatorial regulation of gene expression by TFs and regulation via noncoding RNAs, epigenetic mechanisms, and posttranscriptional processes. However, its ability to accurately model a surprisingly high percentage of the transcriptome—absent information on nonlinear interactions between genes, physical interactions, time course, or brain subregion localization, and using only a partial (∼30%) list of TFs (21)—suggests that the relationship between brain gene expression and behavior is both stronger and more predictable than previously imagined.
Methods
Experimental Designs of BeeSpace Experiments.
The integrated nature of the BeeSpace Project enabled us to control and standardize aspects of field, laboratory, and informatics work across a large number of studies. All experiments were performed with bees from a single species, the Western honey bee A. mellifera. Field experiments were performed during the summers of 2007 and 2008, primarily at the Bee Research Facility at the University of Illinois (Urbana, IL). Additional fieldwork was performed at Ixtapan de la Sal, Mexico (Africanized vs. European honey bees) (7), Avignon, France (Northern vs. Southern European subspecies (22), Arizona State University (high vs. low pollen hoarding), and East Tennessee State University (time training).
Hereditary influences were studied by comparing bees from known different genotypic backgrounds, either different subspecies or selected lines. Bees were sampled from colonies derived from populations maintained as described previously (7, 22, 23). Environmental influences were studied by minimizing genetic variation among bees; we used colonies each derived from a queen instrumentally inseminated by semen from a single male. To generate large numbers of bees with similar genotypes, most experiments used sets of colonies, each headed by sister queens instrumentally inseminated with semen from brother drones. We generated three sets of source colonies by this approach, all derived from typical mixed stock of North European honey bee subspecies.
Source colonies were maintained uniformly, according to standard beekeeping practices. Behavioral collections were made directly from these colonies, or bees were introduced into different host colonies according to the various experimental paradigms used in individual experiments. Each study was replicated in at least two independent field trials using bees from different source colonies. Bees were collected directly into liquid nitrogen or dry ice and stored at −80 °C before brain dissection and gene expression analysis.
Global Analysis of Brain Gene Expression and Social Behavior.
Unsupervised hierarchical clustering and MDS were performed using MATLAB. Pearson’s correlation was used as a metric to compute the distances between transcriptomic responses (log-transformed P values) in the dataset and unweighted average distance (UPGMA) (24) was used to construct the dendrogram. To test the accuracy of the clustering, we used the cophenetic correlation metric (25), which measures how faithfully the tree represents the dissimilarities among observations. We found it to be at a very high value of 0.89, suggesting an accurate fit to the data. We also performed additional statistics to verify the significance of the result (SI Appendix, Table S3 and Fig. S1). Both random sampling of various subsets of the transcriptome (SI Appendix, Table S3) and clustering after removing one of the phenotypes (SI Appendix, Fig. S1) revealed that the behavior-specific clusters we obtained were robust. In both the above-mentioned control experiments, the aggression subcluster of transcriptional profiles for environmental factors sometimes grouped into a separate cluster of its own; nevertheless, all its members were retained in the same cluster. We also estimated the global P value of the tree, on the basis of a method suggested in ref. 26. Bootstrapping estimates (based on 1,000 runs) were used to calculate P values using the pvclust (27) package in R. These P values were then used to calculate the global P value, defined as the minimum of the P values for the nodes. We estimated the global P value to be less than 0.001.
Identification of Honey Bee Transcription Factors.
Few TFs have been experimentally identified and characterized in the honey bee. We therefore focused on genes that are robust orthologs of TFs that have been experimentally characterized in Drosophila. We used a list of 256 TFs for which there was little or no change in critical residues between the bee gene and its Drosophila ortholog (28). Of these putative honey bee TFs, 236 were present in the brain gene expression profiles, and 190 had predicted targets in the TRN.
Network Reconstruction.
Our ASTRIX method is built on two well-known algorithms used for network inference and regression—ARACNE (4) and LARS (10)—to infer interactions and predict expression in new conditions. ARACNE is an information–theoretic method for identifying transcriptional interactions from microarrays or other gene expression data. It is specifically designed to scale up to the complexity of regulatory networks in mammalian cells (4). ARACNE has been shown to correctly predict the regulatory network of c-Myc in human B cells (4) and to accurately reconstruct the TRN responsible for epithelial-to-mesenchymal transformation in human brain tumors (3). In ARACNE, a TF and putative target gene are predicted to interact if the mutual information between their expression levels is above some set threshold. An additional feature of the ARACNE algorithm is that it uses the data processing inequality (DPI) technique to eliminate the majority of indirect interactions inferred by coexpression methods. In this way, ARACNE both predicts interactions between TFs and putative target genes and performs variable reduction, retaining only the strongest, direct interactions.
LARS is a regression algorithm used for inferring relationships between a dependent variable (in this case putative target gene expression) and one or more independent variables (predictors; in this case TFs inferred by ARACNE). LARS outputs a quantitative model, which can be used to predict the response variable on the basis of the states of the dependent variables. LARS is a model selection algorithm, which is a less greedy version of the traditional forward selection method. It selects a parsimonious set of predictors from a large collection of possible covariates for the prediction of a response variable (10). Given that there is almost no physical TF–target interaction data available for honey bees, it seemed prudent to emphasize parsimony at every step.
There are several advantages of our approach for network inference over using ARACNE or LARS alone. First, whereas ARACNE gives only the topology of the network, this combination outputs both the topology and also whether each interaction is activating or inhibitory. Second, we can estimate the percentage of the variance in a target gene’s expression, which is explained by each TF, and predict the expression of a target gene in new conditions. Third, by performing variable reduction and removing indirect interactions in ARACNE, we avoid problems faced by LARS with large numbers of interactions and highly correlated variables (10). Most importantly, ASTRIX only selects the subset of TF–gene relationships that can accurately predict the target gene’s expression quantitatively in the training set, and only these interactions are moved forward to the test set for validation.
Cross-Validation and Accuracy of Fit.
The quantitative output of the TRN model enabled us to test the accuracy of the predictions in new conditions. All data used in this procedure were normalized before network inference to have row variances of 1; this normalization means that for the influence of a TF on a given target gene, we can uniformly interpret the magnitude of the coefficients α, β, and σ, and use their magnitudes to rank the individual interactions (29). We quantified the accuracy of the inferred model by measuring the RMSD. RMSD has the same units as variance, thus providing an estimate for the amount of variance of the gene explained by the model. We also quantified the Pearson correlation between the model’s predictions and the actual expression data.
We performed two tests to determine the TRN’s ability to predict the expression of each gene in new states. In 10-fold cross-validation using random subsets of samples as test sets, using the expression levels of the TFs alone, our model made highly accurate gene expression predictions with a mean RMSDtest = 0.35. We selected only the genes that we could predict well in the training set with a RMSD threshold = 0.5, and the resulting mean RMSDtrain was 0.35. We repeated the analysis using correlation as a metric. Correlation also yielded the same result: the test correlation was as good as the correlation in the training set (0.87), suggesting a good fit to the data without overfitting. Second, we performed “leave-one-state-out” cross-validation, in which all of the bees from 1 of the 48 states were removed from the training set; this is a more stringent test in that the model is tested on a “new” behavioral state, i.e., for which it has not been trained. We again found an equally good accuracy in training and test set using a RMSD threshold of 0.5 in training for selecting genes. (RMSDtrain = 0.33, RMSDtest = 0.32; SI Appendix, Fig. S4).
After estimating the accuracy of fit in cross-validation, we then used the entire dataset and inferred the final network using ARACNE and LARS. We then selected genes that could be predicted with correlation >0.8 to be in the final model—exactly as we had done on each training set in the error estimation stage. The mean correlation of fit in the whole set was 0.87. We built the final network on the basis of the LARS model for each gene. We chose the TF–target gene interactions that had a regression coefficient greater than 0.1. The β-values (regression coefficients) give an estimate of the amount of variance explained by each TF. The putative regulators with regression coefficients of less than 0.1 were pruned out and the final network was determined, which consists of 2,176 genes and 190 TFs.
Estimation of Background Accuracy and Controls for Expression Prediction.
We performed three experiments to estimate the model’s accuracy on control conditions. (i) We inputted a random dataset, the same size as our dataset, normalized and preprocessed in the same way, and then tried to infer from it a network model. This control model did not predict any interactions significantly below the required mutual information threshold (P < 1E-6), suggesting that very few, if any, interactions obtained in the experimental bee brain TRN could have occurred by random chance. (ii) We used the honey bee expression data and then randomly shuffled the expression of TFs (but not putative targets) across experiments. Once again, no significant interactions were predicted. Although some interactions had high mutual information, none of the predicted interactions had a training correlation greater than 0.8. The average test set correlation of this control model across all 9,544 genes was only 0.22 in 10-fold cross-validation. These results suggest that the accurate predictions of the bee brain TRN (r > 0.8; mean test set correlation, r = 0.87) were specifically due to interactions between TFs and their predicted target genes rather than to cryptic structure in the dataset or the modeling approach used. (iii) We tested the ability of the bee brain TRN to model gene expression data in a prominent subregion of the brain, the mushroom bodies (SI Appendix, Fig. S9). We again trained the model on all whole-brain data, then used the mushroom body-specific foraging experience dataset (SI Appendix, Table S1) as a test set (r = 0.36). To determine the significance of the model’s performance, we measured the test prediction using the mean expression of the TFs alone instead of using the linear model predicted by our approach; the average test set prediction correlation was only 0.01.
Measuring Relative Gene Shuffling.
We used differential rank conservation (DIRAC) (19) to identify changes in relative gene expression ordering in networks. DIRAC quantifies and assesses expression consistency by ranking the target genes within a selected module on the basis of expression.
Supplementary Material
Acknowledgments
We extend special gratitude to Alissa Eisenstein for her outstanding microarray “spot finding” work throughout the BeeSpace Project. We also thank S. Sinha for the list of bee cis-regulatory elements; J. Drnevich and C. Fields for database construction; R. E. Page and G. V. Amdam for the high- and low-pollen hoarding bees; E. Carefoot for contributions to Fig. 2B; and D. F. Clayton, J. Earls, C. Fields, C. Funk, J. M. George, M. E. Hudson, T. Gernat, C. C. Lutz, S. Ma, Y. Ono, K. Scott, S. Sinha, M. B. Sokolowski, L. J. Stubbs, and S. Zhong for reviewing the manuscript. BeeSpace experiments funded by National Science Foundation Frontiers in Biological Research Award EF25852 (to B.R.S., Principal Investigator). Additional support for transciptional regulatory network (TRN) analysis comes from: National Institutes of Health Director’s Pioneer Award 1DP1OD006416 (to G.E.R.) and National Cancer Institute Howard Temin Pathway to Independence Award, Department of Defense W81XWH-08-1-0420, Grand Duchy of Luxembourg, and the Roy J. Carver Charitable Trust (N.D.P.). Microarray data meet minimum information about microarray experiment (MIAME) standards, see SI Appendix, Table S10 for accession nos. for each experiment (http://www.ebi.ac.uk/microarray-as/ae/). The TRN is publicly available at http://price.systemsbiology.net/downloads/.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 17861.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114093108/-/DCSupplemental.
References
- 1.Robinson GE. Genomics. Beyond nature and nurture. Science. 2004;304:397–399. doi: 10.1126/science.1095766. [DOI] [PubMed] [Google Scholar]
- 2.Robinson GE, Fernald RD, Clayton DF. Genes and social behavior. Science. 2008;322:896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 5.Bonneau R, et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–1365. doi: 10.1016/j.cell.2007.10.053. [DOI] [PubMed] [Google Scholar]
- 6.Luscombe NM, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 7.Alaux C, et al. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc Natl Acad Sci USA. 2009;106:15400–15405. doi: 10.1073/pnas.0907043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winston ML. The Biology of the Honey Bee. Cambridge, MA: Harvard Univ Press; 1987. p. viii, 281. [Google Scholar]
- 9.Margolin AA, et al. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Ann Stat. 2004;32:407–499. [Google Scholar]
- 11.Roy S, et al. modENCODE Consortium. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. doi: 10.1038/sj.npp.1301544. [DOI] [PubMed] [Google Scholar]
- 13.Riddiford LM. Juvenile hormone action: A 2007 perspective. J Insect Physiol. 2008;54:895–901. doi: 10.1016/j.jinsphys.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Robinson GE. Genomics and integrative analyses of division of labor in honeybee colonies. Am Nat. 2002;160(Suppl 6):S160–S172. doi: 10.1086/342901. [DOI] [PubMed] [Google Scholar]
- 15.Koo JW, Russo SJ, Ferguson D, Nestler EJ, Duman RS. Nuclear factor-kappaB is a critical mediator of stress-impaired neurogenesis and depressive behavior. Proc Natl Acad Sci USA. 2010;107:2669–2674. doi: 10.1073/pnas.0910658107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 17.Pfaff DW, Arnold AP, Etgen AM, Rubin RT, Fahrbach SE, editors. Hormones, Brain and Behavior. New York: Elsevier; 2009. [Google Scholar]
- 18.Bauer R, et al. Schlank, a member of the ceramide synthase family controls growth and body fat in Drosophila. EMBO J. 2009;28:3706–3716. doi: 10.1038/emboj.2009.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy JA, Hood L, Price ND, Geman D. Identifying tightly regulated and variably expressed networks by Differential Rank Conservation (DIRAC) PLOS Comput Biol. 2010;6:e1000792. doi: 10.1371/journal.pcbi.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson CM, Ihle KE, Fondrk MK, Page RE, Amdam GV. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 2007;5:e62. doi: 10.1371/journal.pbio.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adryan B, Teichmann SA. FlyTF: A systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics. 2006;22:1532–1533. doi: 10.1093/bioinformatics/btl143. [DOI] [PubMed] [Google Scholar]
- 22.Whitfield CW, et al. Genomic dissection of behavioral maturation in the honey bee. Proc Natl Acad Sci USA. 2006;103:16068–16075. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page RE, Fondrk MK. The effects of colony-level selection on the social organization of honey bee (Apis mellifera L.) colonies: Colony-level components of pollen hoarding. Behav Ecol Sociobiol. 1995;36:135–144. [Google Scholar]
- 24.Gauch HG, Whittaker RH. Hierarchical classification of community data. J Ecol. 1981;69:537–557. [Google Scholar]
- 25.Sokal RR, Rohlf FJ. The comparison of dendrograms by objective methods. Taxon. 1962;11:33–40. [Google Scholar]
- 26.Levenstien MA, Yang Y, Ott J. Statistical significance for hierarchical clustering in genetic association and microarray expression studies. BMC Bioinformatics. 2003;4:62. doi: 10.1186/1471-2105-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki R, Shimodaira H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, et al. Functional characterization of transcription factor motifs using cross-species comparison across large evolutionary distances. PLOS Comput Biol. 2010;6:e1000652. doi: 10.1371/journal.pcbi.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonneau R, et al. The Inferelator: An algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 2006;7:R36. doi: 10.1186/gb-2006-7-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.