Abstract
The self-incompatibility (SI) system of the Brassicaceae is based on allele-specific interactions among haplotypes of the S locus. In all tested self-incompatible Brassicaceae, the S haplotype encompasses two linked genes, one encoding the S-locus receptor kinase (SRK), a transmembrane kinase displayed at the surface of stigma epidermal cells, and the other encoding its ligand, the S-locus cysteine-rich (SCR) protein, which is localized in the pollen coat. Transfer of the two genes to self-fertile Arabidopsis thaliana allowed the establishment of robust SI in several accessions, indicating that the signaling cascade triggered by this receptor–ligand interaction and the resulting inhibition of “self” pollen by the stigma have been maintained in extant A. thaliana. Based on studies in Brassica species, the membrane-tethered kinase MLPK, the ARM repeat-containing U-box protein ARC1, and the exocyst subunit Exo70A1 have been proposed to function as components of an SI signaling cascade. Here we tested the role of these molecules in the SI response of A. thaliana SRK-SCR plants. We show that the A. thaliana ARC1 ortholog is a highly decayed pseudogene. We also show that, unlike reports in Brassica, inactivation of the MLPK ortholog AtAPK1b and overexpression of Exo70A1 neither abolish nor weaken SI in A. thaliana SRK-SCR plants. These results do not support a role for these molecules in the SI response of A. thaliana.
Keywords: receptor signaling, S locus receptor kinase
In the self-incompatibility (SI) system of the Brassicaceae (crucifer) family, the ability of cells of the stigma epidermis to discriminate between “self” and “nonself” pollen grains and to inhibit “self” pollen is based on allele-specific interactions between two highly polymorphic proteins encoded by the S-locus haplotype: the S-locus receptor kinase (SRK), a single-pass transmembrane serine/threonine kinase displayed at the surface of stigma epidermal cells (1), and the S-locus cysteine-rich (SCR) protein, a small secreted protein located in the pollen coat (2, 3), which is the ligand for SRK (4, 5). The binding of SCR to the extracellular domain of its cognate SRK activates the receptor kinase (4–6) and is thought to trigger a response within the stigma epidermal cell that culminates in the inhibition of pollen tube development.
The molecular events precipitated by activation of SRK are currently poorly understood, and the immediate cause of self-pollen inhibition is not known. Nevertheless, a few candidate molecules have been proposed to function as downstream effectors in the SRK signaling pathway in Brassica species. The M-locus protein kinase (MLPK), identified by a map-based cloning approach (7), is thought to correspond to the gene disrupted by the mod, or m, mutation, which causes complete loss of SI in the stigmas of the Brassica rapa variety Yellow Sarson (8). Although a stable complementation experiment has not been reported, particle bombardment-mediated transient expression of MLPK in the stigma epidermal cells of mod plants was reported to complement the self-fertile phenotype of these cells (7). Because the two isoforms produced by the MLPK gene colocalize to the plasma membrane and interact with SRK, they are thought to function in SRK-mediated signaling (9).
Another candidate effector of SRK-mediated signaling is the ARM-repeat containing 1 (ARC1) protein, which was identified in a yeast two-hybrid screen as a protein that interacts with, and is phosphorylated by, the kinase domain of a Brassica napus SRK protein (10). Antisense down-regulation of ARC1 transcripts in transgenic B. napus has been associated with partial breakdown of SI (11). Furthermore, ARC1, like other members of the plant U-box (PUB) proteins, is an E3 ubiquitin ligase that localizes to the proteasome and COP9 signalosome in an SRK-dependent manner (12). Recently, ARC1 was found to interact with Exo70A1 (13), a putative component of the exocyst complex, which functions in polarized secretion in yeast and animals (14, 15). Overexpression of Exo70A1 in stigma epidermal cells has been reported to cause partial breakdown of SI (13). Previous studies showed that Exo70A1 is required for compatible pollen-stigma interactions in Arabidopsis thaliana (16), suggesting that the exocyst is involved in the secretion of “compatibility factors” to the stigma surface (13). Together, these results suggest a model of SRK signaling in which the activated SRK, possibly together with MLPK, results in recruitment and phosphorylation of ARC1, which then targets Exo70A1 for ubiquitination and degradation, thereby precluding the secretion of compatibility factors and causing inhibition of pollen hydration, germination, and tube growth (13). Because a stigma epidermal cell can inhibit a self pollen grain while allowing the growth of a nonself pollen tube, this model requires that ARC1-mediated degradation of Exo70A1 and the resulting depletion of compatibility factors be confined to the region subtending the site of pollen–stigma contact, a process that has yet to be demonstrated.
The self-fertile model plant A. thaliana lacks functional S haplotypes (17–20); however, it can be made to express SI on transformation with any of several SRK-SCR gene pairs isolated from its close self-incompatible relatives Arabidopsis lyrata and Capsella grandiflora (21–23). SRK-SCR transformants of some A. thaliana accessions, such as Col-0, exhibit transient SI (i.e., their stigmas express a robust SI response during a narrow developmental window, but subsequently lose their ability to inhibit self pollen) (22, 23) due to the presence in these accessions of a hypomorphic allele of a modifier locus that causes reduced levels of SRK at late stages of stigma development (24). In contrast, SRK-SCR transformants of other accessions, such as C24 and Cvi-0, express developmentally stable SI (17, 23). Notably, the SI response of A. thaliana SRK-SCR transformants, whether stable or transient, is indistinguishable from the SI response of naturally self-incompatible A. lyrata, C. grandiflora, and Brassica species; an A. thaliana stigma that expresses adequate levels of SRK will inhibit the germination and tube growth of self pollen and typically will allow the growth of up to five self pollen tubes, while supporting abundant WT pollen tube growth. The fact that A. thaliana can express a robust SI response on transformation with just the SRK and SCR gene pairs demonstrates that all other factors required for SI, including components of the SI signaling pathway, have been maintained in the species. Thus, the A. thaliana SRK-SCR self-incompatible model is an excellent platform for analysis of the SRK-mediated signaling that underlies crucifer SI.
Highly polymorphic and physically linked SRK and SCR genes have been identified in all self-incompatible members of the Brassicaceae analyzed to date, including A. lyrata, Arabidopsis halleri, and C. grandiflora (21–23). It thus stands to reason that the SRK-mediated signaling pathway would be conserved across the Brassicaceae, and that orthologs of any downstream effectors of SRK signaling identified in Brassica species also would function in the SI response of other self-incompatible species of the Brassicaceae. Here we report on our use of A. thaliana SRK-SCR plants to test the aforementioned model of SI signaling, and in particular the involvement in SI of the A. thaliana orthologs of Brassica MLPK, ARC1, and Exo70A1. Our results do not support a role for these genes in A. thaliana SI. We discuss the implications of these results for our understanding of the SI signaling pathway.
Results and Discussion
Inactivation or Down-Regulation of AtAPK1b and AtAPK1a Does Not Disrupt the SI Response in A. thaliana SRKb-SCRb Plants.
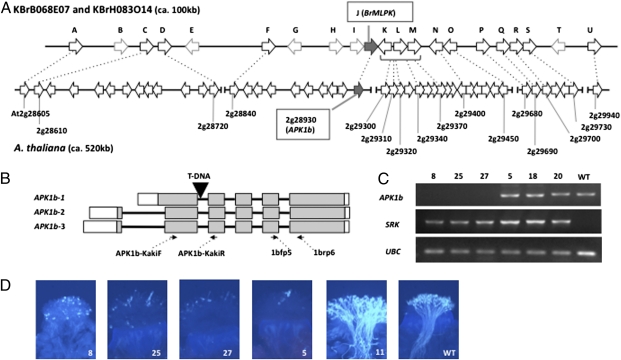
The AtAPK1b (At2g28930) gene is considered to be the ortholog of the B. rapa MLPK (BrMLPK) gene. Both genes belong to the RLCK VII subfamily of protein kinases, and of all A. thaliana members of this subfamily, AtAPK1b shares the greatest sequence identity with BrMLPK (ca. 76% at the amino acid level) (Table S1). Furthermore, the A. thaliana chromosomal region containing this gene shows high synteny with the Brassica chromosomal region that encompasses BrMLPK (Fig. 1A). Also like BrMLPK (9), AtAPK1b exhibits alternative use of the first exon and is predicted to produce transcripts encoding two distinct protein products (Fig. 1B; http://www.arabidopsis.org/servlets/TairObject?id=33491&type=locus).
Fig. 1.
Functional analysis of a T-DNA insertion allele of AtAPK1b. (A) Synteny of the Brassica and A. thaliana chromosomal regions containing MLPK and APK1b, respectively. E-values are listed in Table S5. (B) Structure of the AtAPK1b gene showing the two alternative transcript forms produced by the gene and the location of the T-DNA insertion in the SALK_055314 strain. The boxes indicate exons; shaded boxes represent protein-coding regions, and white boxes designate untranslated regions. The arrows below the diagram indicate the location of the primers (F and R) used for RT-PCR. (C) RT-PCR analysis of stigma RNA isolated from F2 plants derived from a cross of the SALK_055314 T-DNA insertion strain and a Col-0[SRKb-SCRb] plant. Plants 8, 25, and 27 are homozygous for the T-DNA allele and do not express AtAPK1b transcripts, whereas plants 5, 18, and 20 lack the T-DNA insertion and express AtAPK1b transcripts at levels equivalent to those in WT Col-0. All of these F2 plants express SRKb transcripts. UBC transcripts were used as a control. (D) SI phenotype of F2 plants. Stigmas from stage 13 floral buds were pollinated with SCRb-expressing pollen. Plants 8, 25, and 27, which are homozygous for the T-DNA insertion in AtAPK1b and contain SRKb-SCRb, exhibit as robust an SI response as that observed in plant 5, which produces AtAPK1b transcripts and contains SRKb-SCRb. In contrast, plant 11, which is homozygous for the T-DNA allele but lacks SRKb-SCRb, did not exhibit SI, similar to WT Col-0.
To test the involvement of AtAPK1b in SI, we obtained a Col-0 strain carrying a T-DNA insertion in the gene. This strain (Salk_055314) produces no detectable AtAPK1b transcripts in either leaves or stigmas (9, 25). This strain was crossed with a Col-0 plant homozygous for the A. lyrata-derived SRKb-SCRb transgenes (23), which expresses an intense SI reaction in stigmas from stage-13 and early stage-14 floral buds (22). F2 plants were generated, and the accumulation of AtAPK1b transcripts in stigmas was analyzed. As shown in Fig. 1C and Table S2, AtAPK1b transcripts were not detected in plants that inherited the SRKb-SCRb transgenes and were homozygous for the T-DNA insertion, based on segregation analysis in F3 progenies (e.g., plants 8, 25, and 27). In contrast, AtAPK1b transcripts were readily detected in plants containing the SRKb-SCRb transgenes and lacking the AtAPK1b T-DNA insertion (e.g., plants 5, 18, and 20). Pollination assays showed that all of these plants expressed robust SI in stage-13 stigmas (the stage at which the SI response of Col-0[SRK-SCR] plants is most strongly expressed), and no attenuation of SI was observed in plants homozygous for the T-DNA insertion relative to sibs lacking the insertion allele (Fig. 1D). This result indicates that AtAPK1b is not required for the SI response of A. thaliana.
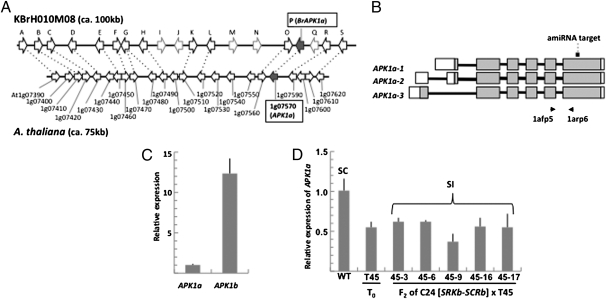
The A. thaliana genome contains a gene, AtAPK1a (At1g07570), which exhibits 82.3% amino acid sequence identity to AtAPK1b. AtAPK1a is unlinked to AtAPK1b and is located on chromosome 1 (Fig. 2A), and it produces three transcript forms predicted to encode distinct protein products (Fig. 2B; www.arabidopsis.org/servlets/TairObject?id=29045&type=locus). However, unlike BrMLPK and AtAPK1b, AtAPK1a is not preferentially expressed in stigmas (9), and it is poorly expressed in these structures; AtAPK1a (APK1a-1, -2, and -3) transcripts levels are 12.3-fold lower than AtAPK1b (APK1b-1, -2, and -3) transcripts (Fig. 2C). Furthermore, B. rapa contains a transcribed gene that exhibits 86.1% amino acid sequence identity to AtAPK1a. This gene is located on BAC AC172873 (KBrH010M08), which is derived from a chromosomal region showing extensive synteny with the AtAPK1a-spanning region of the A. thaliana genome (Fig. 2A), and it clearly does not function redundantly with BrMLPK, because it cannot overcome the self-fertility phenotype effected by homozygosity for the mod mutation. Together, these features indicate that AtAPK1a is not the ortholog of BrMLPK.
Fig. 2.
Expression analysis of AtAPK1a-suppressed plants. (A) Synteny of APK1a-containing chromosomal regions in Brassica and A. thaliana. E-values are listed in Table S5. (B) Structure of the AtAPK1a gene showing its three alternative transcript forms and the location of the sequence targeted by the artificial microRNA (amiRNA target) designed to down-regulate the gene. The arrows below the diagram indicate the location of the primer pairs used for transcript quantitation. (C) Quantitative real-time PCR analysis of AtAPK1b and AtAPK1a transcripts in the stigmas of WT C24 stigmas. (D) Quantitative real-time PCR analysis of AtAPK1a transcripts in the stigmas of WT C24, the T45 AtAPK1a-suppressed primary transformant, and F2 plants derived from crossing T45 with C24[SRKb-SCRb]. All of the F2 plants analyzed contain the SRKb-SCRb transgenes but express AtAPK1a transcripts at lower levels than WT. For all real-time PCR experiments, relative gene expression levels were generated by normalizing the signal using the ubiquitin-conjugating (UBC) gene (At5g25760). SDs from triplicate experiments are indicated by error bars.
Nevertheless, we explored the possibility that AtAPK1a might have assumed the function of AtAPK1b in A. thaliana SI. Several T-DNA strains annotated as containing insertions in AtAPK1a (Salk_56259, Salk_96327, and Salk_22547) were obtained, but all of these strains still expressed AtAPK1a transcripts at appreciable levels. Therefore, we attempted to suppress expression of AtAPK1a in stigmas of the C24 accession using artificial miRNA (26) driven by the promoter of the AtS1 gene, which is active specifically in stigma epidermal cells (27). Of several transformants analyzed, a transgenic plant (T45) showed significant reduction in the already-low levels of stigma AtAPK1a transcripts (Fig. 2D). This plant was crossed to a C24 plant homozygous for the SRKb-SCRb transgenes, which exhibits strong and developmentally stable SI in stigmas starting from stage-13 floral buds and into late-stage flowers (23). Among F2 progenies derived from this cross, several plants that inherited the SRKb-SCRb transgenes showed reduced AtAPK1a transcript levels relative to WT, with reduction levels ranging from 60% to 40% (Fig. 2D). However, all of these AtAPK1a-suppressed plants retained the strong and developmentally stable SI phenotype characteristic of C24[SRKb-SCRb] plants (Table S2).
A. thaliana Ortholog of Brassica ARC1 Is a Fragmented Pseudogene Lacking Most of the Coding Region.
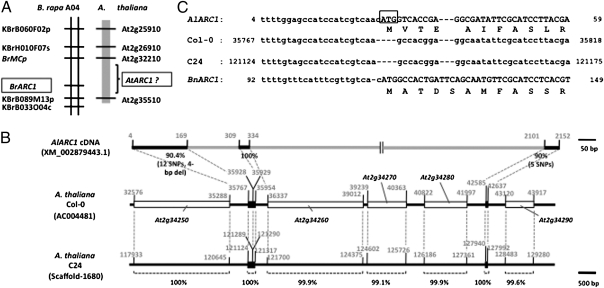
We identified the A. thaliana ortholog of Brassica ARC1 by comparative analysis of A. thaliana, A. lyrata, and B. rapa genomic sequences. The region of A. thaliana chromosome 2 between At2g25910 and At2g35510 was found to share extensive synteny with the ARC1-containing region of the B. rapa A04 linkage group (28, 29) (Fig. 3A). A BLAST search of this region with the B. napus BnARC1 (AF024625.1) did not retrieve any hits; however, a BLAST search with the AlARC1 cDNA (XM_002879443.1, which is 73% identical to BnARC1), resulted in a hit in the At2g34250-At2g34290 region of the A. thaliana Col-0 genome (Fig. 3B). Alignment of the Col-0 ARC1-related sequences with the AlARC1 cDNA sequence identified only a highly degraded ARC1 sequence lacking the initiating methionine codon (Fig. 3C) and consisting of three small fragments interspersed with other genes (Fig. 3B). These fragments exhibited 90%–100% nucleotide sequence identity with regions spanning nucleotides 4–169, 309–334, and 2101–2152 in AlARC1 cDNA.
Fig. 3.
The A. thaliana ortholog of Brassica ARC1. (A) Synteny of the BrARC1-containing region of B. rapa A04 linkage group with the region of A. thaliana chromosome 2 flanked by At2g25910 and At2g35510. (B) Structure of A. thaliana ARC1 sequences in the Col-0 and C24 accessions compared with A. lyrata ARC1 cDNA. Note the highly degraded structure of AtARC1 in both Col-0 and C24. (C) Alignment of the 5′ end of the AlARC1 cDNA sequence with the corresponding region of AtARC1 from Col-0 and C24, and of BnARC1. Note the absence of the initiating methionine codon in the AtARC1 sequences.
A BLAST search of the C24 genome sequence, which covers 96% of the Col-0 reference genome (ref. 30; available at www.1001genomes.org) also identified a fragmented ARC1 sequence identical to the Col-0 ARC1 sequence within Scaffold-1680 (Fig. 3 B and C). Thus, the ARC1 gene of A. thaliana is a highly degraded pseudogene even in C24, an accession in which SRK-SCR transformants express a robust and developmentally stable SI. It is also unlikely that the A. thaliana genome contains another ARC1-like gene that functions in SI. Among the apparently intact PUB genes in A. thaliana, AtPUB17 (At1g29340) is the most similar to BnARC1 (58% amino acid sequence identity; Table S1). However, this gene shares 96.6% and 85.9% amino acid sequence identity, respectively, with sequences that are the presumed orthologs of AtPUB17: an A. lyrata sequence (XM_002890762.1) and a B. rapa sequence within BAC AC189240 (KBrB016E20), both of which are located in chromosomal regions syntenous with each other and with the AtPUB17 region of A. thaliana, but not with the ARC1 regions of A. thaliana, A. lyrata, and B. rapa (Fig. S1). Furthermore, we had previously assayed a T-DNA knockout allele of AtPUB17 and found that this gene has no role in SI (25). Thus, we found no evidence for the involvement of an ARC1-like gene in the A. thaliana SI response.
Overexpression of AtExo70A1 Does Not Weaken SI in SRKb-SCRb Plants.
Despite the lack of a functional ARC1 gene in A. thaliana, we tested the potential involvement of AtExo70A1 in the SI response of C24 plants expressing SRKb and SCRb transgenes. We transformed C24 plants with a construct, designated AtS1pr::AtExo70A1, consisting of the AtS1 promoter fused to the transcriptional unit of AtExo70A1, which is the likely A. thaliana ortholog of Brassica Exo70A1 based on synteny and sequence identity (Fig. S2 and Table S1). Several transformants were generated and crossed to C24 plants carrying SRKb and SCRb transgenes. Because Exo70A1, as a component of the exocyst, functions in secretion, we thought that modifying its expression in stigmas might possibly exert its reported weakening of SI by affecting SRK protein accumulation or targeting either directly or indirectly. Therefore, to allow detection of SRKb protein, we used an SRK-SCR construct designed to express an SRKb molecule containing an N-terminal YFP tag from the AtS1 promoter. The YFP:SRKb fusion produced by the AtS1pr::YFP:SRKb/SCRb transgene is functional, as demonstrated by the SI response observed in pollination assays (Table S3). To test the involvement of AtExo70A1 in SI, we focused on an AtS1pr::AtExo70A1 transformant that carried a single integration of the AtS1pr::AtExo70A1 transgene and exhibited an ∼15-fold increase in stigma AtExo70A1 transcripts relative to WT. This strain was crossed with a plant homozygous for AtS1pr:: YFP:SRKb/SCRb transgenes and to a plant homozygous for the SRKb-SCRb transgenes, which consist of native promoters driving intact coding regions. Although the levels of BnExo70A1 transcripts reported to cause weakening of SI in B. napus have not been given (13), we hypothesized that a more than 10-fold increase in AtExo70A1 transcript levels should provide an adequate test of the role of AtExo70A1 in the SI response of A. thaliana SRK-expressing plants.
We analyzed 64 F2 plants derived from the AtS1pr::YFP:SRKb/SCRb × AtS1pr::AtExo70A1 cross and 56 F2 plants derived from the SRKb-SCRb × AtS1pr::AtExo70A1 cross. Among hygromycin-resistant F2 progenies, the numbers of plants containing the SRKb/SCRb and AtS1pr::AtExo70A1 transgenes, SRKb/SCRb alone, and AtS1pr::AtExo70A1 alone were 27:12:25 in the former cross and 27:10:19 in the latter cross. In both crosses, F1 plants and all F2 plants that inherited the SRKb transgenes, regardless of whether or not they contained the AtS1pr::AtExo70A1 transgene, exhibited an intense SI response on pollination assays (Table S3), and we did not observe any promotion of pollen hydration and pollen tube penetration into the stigma epidermal cell wall in these assays.
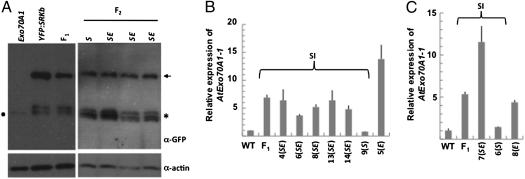
As expected based on the intense SI response observed in these plants, AtExo70A1 overexpression had no significant effect on SRKb protein levels (Fig. 4A). However, the stigmas of these plants exhibited significantly increased AtExo70A1 transcript levels (Fig. 4 B and C). In stigmas of F1 plants derived from both crosses, AtExo70A1 transcript levels were more than fivefold higher than in the stigmas of WT untransformed control plants. Among F2 plants, the stigmas of plants that inherited only the SRKb transgene and lacked AtS1pr::AtExo70A1 (e.g., plant 9 in Fig. 4B and plant 6 in Fig. 4C) accumulated Exo70A1 transcripts at levels equivalent to the low levels observed in untransformed WT control plants, as expected. On the other hand, plants that inherited the AtS1pr::AtExo70A1 transgene alone (e.g., plant 5 in Fig. 4B and plant 8 in Fig. 4C) or both the SRKb transgene and AtS1pr::AtExo70A1 (e.g., plants 4, 6, 8, 13, and 14 in Fig. 4B and plant 7 in Fig. 4C) exhibited increased AtExo70A1 transcript levels relative to plants lacking this transgene, ranging from ∼5-fold to >10-fold. This variability in transcript levels reflects the dosage of the AtS1pr::AtExo70A1 transgene and possibly differences in the developmental age of stigmas used for transcript quantitation. The AtS1 promoter is developmentally regulated during stigma development, and stigmas at somewhat less advanced stages of maturity might have been included in some of the 25 stigma samples used in our analysis, which might have caused us to underestimate AtExo70A1 transcript levels in some plants. In any case, our results indicate that overexpression of AtExo70A1 transcripts in SRKb-expressing stigmas, even at >10-fold higher levels than those effected by the endogenous AtExo70A1 transgene, has no significant effect on the SI response.
Fig. 4.
Expression analysis of stigmas from progenies of the AtS1pr::YFP:SRKb/SCRb × AtS1pr::AtExo70A1 and SRKb-SCRb × AtS1pr::AtExo70A1 crosses. (A and B) Analysis of the AtS1pr::YFP:SRKb/SCRb × AtS1pr::AtExo70A1 cross. (A) Stigma protein gel blot analysis of YFP:SRKb using anti-GFP monoclonal antibodies and anti-actin antibodies to detect actin as a loading control. The arrow indicates the full-length YFP:SRKb protein, and the asterisk indicates the smaller protein species that correspond to the extracellular domain of SRKb and are typically produced by SRK genes. The dot indicates a background protein detected in A. thaliana stigmas by the anti-GFP monoclonal antibody. (B) Quantitative real-time PCR analysis of AtExo70A1 transcripts in stigmas from WT C24, an F1 plant, and representative F2 plants. For each F2 plant, the presence or absence of each transgene is indicated: SE, plants containing both AtS1pr::YFP:SRKb/SCRb and AtS1pr::AtExo70A; S, plants containing only AtS1pr::YFP:SRKb/SCRb; E, plants containing only AtS1pr::AtExo70A. The variation in the relative levels of AtExo70A1 transcripts among F2 plants is due primarily to differences in gene dosage. Of all F2 plants shown, only plant 5 was homozygous for the AtS1pr::AtExo70A transgene. Other sources of variability are stochastic variation in transgene expression between plants and differences in the developmental age of stigmas included in each sample. (C) Quantitative real-time PCR of AtExo70A in progenies of the SRKb-SCRb × AtS1pr::AtExo70A1 cross. Relative expression levels in stigmas from WT C24, an F1 plant, and representative F2 plants are shown. The presence or absence of the transgenes in each F2 plant are as shown in B. Normalization of signals and SDs are as shown in Fig. 2.
Conclusions
SRK-SCR transformants of A. thaliana express an SI response that is as strong as that observed in naturally self-incompatible A. lyrata or Brassica species, either throughout stigma development, as in the C24 accession, or during a narrow window of stigma development, as in the Col-0 accession. This fact implies that the SI signaling pathway triggered by the SRK–SCR interaction, which culminates in the inhibition of self pollen, has been maintained in this species since the switch to self-fertility occurred. Thus, transgenic A. thaliana SRK-SCR plants are a suitable platform for assaying the involvement of specific molecules in the SI response. This study demonstrates that none of the A. thaliana orthologs (or closest paralogs) of the genes that have been implicated in the SI response of Brassica species function in the SI response of A. thaliana SRK-SCR plants. First, a null allele of AtAPK1b does not cause loss or weakening of SI in SRKb-expressing stigmas, unlike the reported loss of SI resulting from a mutation in the B. rapa MLPK gene. Second, the A. thaliana ortholog of the B. napus ARC1 gene is clearly a pseudogene, and a null allele of its closest paralog, PUB17, does not disrupt SI in SRK-expressing stigmas (25). Finally, the observation that overexpression of AtExo70A1 in SRK-expressing stigmas did not produce the weakening of SI reported on overexpression of BnExo70A1 in B. napus indicates that the ARC1/Exo70A1-based model of SI signaling proposed for Brassica does not apply to A. thaliana SRK-SCR plants.
The discrepancies observed between Brassica and A. thaliana might suggest that the SRK–SCR interaction triggers different signaling cascades in the two taxa. Alternatively, the inhibition of self pollen in SI might be the cumulative outcome of multiple SRK-mediated signaling pathways (31) that are differentially utilized in different species of the Brassicaceae. Neither of these scenarios seems probable, however. Signaling pathways that underlie the same process are typically highly conserved over long evolutionary time spans (>100 million years), often across kingdoms (32–34). Indeed, it is precisely this conservation that justifies the use of model systems, such as A. thaliana, to understand biological processes in less tractable nonmodel organisms. It therefore seems unlikely that SRK-mediated signaling was drastically modified in the ∼14–20 million years that separate Brassica and Arabidopsis (35, 36).
A possible explanation for the observed discrepancies is that other genes in the 46-member RLCK VII family and 42-member PUB family of A. thaliana (Table S1) might have assumed the role proposed for MLPK and ARC1 (although the putative ARC1-like protein would have to act on a target other than Exo70A1). Additional data are needed to confirm the roles of ARC1 and MLPK in Brassica SI. At present, the possibility exists that the weakening of SI observed in B. napus plants transformed with the antisense ARC1 transgene (11) might have been caused by down-regulation not of BnARC1 but of a related gene. Supporting this possibility, the stigmas of a B. rapa strain homozygous for a hypomorphic ARC1 allele exhibit no weakening of SI despite expressing 10-fold less ARC1 transcripts than other self-incompatible B. rapa strains (25). In the case of MLPK, it is important to prove that this gene does indeed function in Brassica SI through stable complementation of the mod mutation, rather than through the less robust method of transient expression via particle bombardment of stigmas (7). Resolution of the discrepancies revealed by the results reported here clearly requires an evaluation of the roles in SI of additional A. thaliana MLPK- and ARC1-like genes and reexamination of the roles of MLPK, ARC1, and Exo70A1 in Brassica species.
Materials and Methods
The experiments described herein used A. thaliana Col-0 or C24 plants transformed with the SRKb-SCRb genes isolated from the A. lyrata Sb haplotype (18). Because T-DNA insertional mutants are typically available in the Col-0 accession, we used Col-0[SRKb-SCRb] transformants to assess the effect of T-DNA insertional mutations on SI. However, for experiments involving transgenic down-regulation or overexpression of candidate genes, we used SRKb-SCRb transformants of the C24 accession, which allowed us to monitor the strength of SI by both microscopic examination of pollen tube growth and the amount of seed set.
The SRK-SCR transgenes and T-DNA mutant strains used in this study, along with the constructs designed to manipulate candidate gene expression, are described in SI Materials and Methods. The methods used for plant transformation, pollination assays, DNA gel blot analysis, immunoblot analysis, and expression analysis are also discussed in SI Materials and Methods. The primers used for genotyping by PCR and for expression analysis are listed in Table S4.
Supplementary Material
Acknowledgments
We thank Tiffany Crispell for technical assistance and the reviewers for their constructive comments. This material is based on work supported by National Science Foundation Grant IOS-0744579.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115283108/-/DCSupplemental.
References
- 1.Stein JC, Howlett B, Boyes DC, Nasrallah ME, Nasrallah JB. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc Natl Acad Sci USA. 1991;88:8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schopfer CR, Nasrallah ME, Nasrallah JB. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- 3.Takayama S, et al. The pollen determinant of self-incompatibility in Brassica campestris. Proc Natl Acad Sci USA. 2000;97:1920–1925. doi: 10.1073/pnas.040556397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kachroo A, Schopfer CR, Nasrallah ME, Nasrallah JB. Allele-specific receptor–ligand interactions in Brassica self-incompatibility. Science. 2001;293:1824–1826. doi: 10.1126/science.1062509. [DOI] [PubMed] [Google Scholar]
- 5.Takayama S, et al. Direct ligand–receptor complex interaction controls Brassica self-incompatibility. Nature. 2001;413:534–538. doi: 10.1038/35097104. [DOI] [PubMed] [Google Scholar]
- 6.Shimosato H, et al. Characterization of the SP11/SCR high-affinity binding site involved in self/nonself recognition in Brassica self-incompatibility. Plant Cell. 2007;19:107–117. doi: 10.1105/tpc.105.038869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murase K, et al. A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science. 2004;303:1516–1519. doi: 10.1126/science.1093586. [DOI] [PubMed] [Google Scholar]
- 8.Hinata K, Okazaki K, Nishio T. Proceedings of the 6th International Rapeseed Conference Paris, France, May 17–19, 1983. Vol 1. Paris: Groupe Consultatif International de Recherche sur le Colza; 1983. Gene analysis of self-incompatibility in Brassica campestris var. yellow sarson (a case of recessive epistatic modifier) pp. 354–359. [Google Scholar]
- 9.Kakita M, et al. Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell. 2007;19:3961–3973. doi: 10.1105/tpc.106.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci USA. 1998;95:382–387. doi: 10.1073/pnas.95.1.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone SL, Arnoldo MA, Goring DR. A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science. 1999;286:1729–1731. doi: 10.1126/science.286.5445.1729. [DOI] [PubMed] [Google Scholar]
- 12.Stone SL, Anderson EM, Mullen RT, Goring DR. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell. 2003;15:885–898. doi: 10.1105/tpc.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samuel MA, et al. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell. 2009;21:2655–2671. doi: 10.1105/tpc.109.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu SC, TerBush D, Abraham M, Guo W. The exocyst complex in polarized exocytosis. Int Rev Cytol. 2004;233:243–265. doi: 10.1016/S0074-7696(04)33006-8. [DOI] [PubMed] [Google Scholar]
- 15.Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol. 2006;13:577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- 16.Synek L, et al. AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J. 2006;48:54–72. doi: 10.1111/j.1365-313X.2006.02854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boggs NA, Nasrallah JB, Nasrallah ME. Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000426. doi: 10.1371/journal.pgen.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusaba M, et al. Self-incompatibility in the genus Arabidopsis: Characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell. 2001;13:627–643. [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman-Broyles S, et al. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell. 2007;19:94–106. doi: 10.1105/tpc.106.048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu KK, Shimizu-Inatsugi R, Tsuchimatsu T, Purugganan MD. Independent origins of self-compatibility in Arabidopsis thaliana. Mol Ecol. 2008;17:704–714. doi: 10.1111/j.1365-294X.2007.03605.x. [DOI] [PubMed] [Google Scholar]
- 21.Boggs NA, et al. Expression of distinct self-incompatibility specificities in Arabidopsis thaliana. Genetics. 2009;182:1313–1321. doi: 10.1534/genetics.109.102442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasrallah ME, Liu P, Nasrallah JB. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science. 2002;297:247–249. doi: 10.1126/science.1072205. [DOI] [PubMed] [Google Scholar]
- 23.Nasrallah ME, Liu P, Sherman-Broyles S, Boggs NA, Nasrallah JB. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: Implications for the evolution of selfing. Proc Natl Acad Sci USA. 2004;101:16070–16074. doi: 10.1073/pnas.0406970101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P, Sherman-Broyles S, Nasrallah ME, Nasrallah JB. A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr Biol. 2007;17:734–740. doi: 10.1016/j.cub.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rea AC, Liu P, Nasrallah JB. A transgenic self-incompatible Arabidopsis thaliana model for evolutionary and mechanistic studies of crucifer self-incompatibility. J Exp Bot. 2010;61:1897–1906. doi: 10.1093/jxb/erp393. [DOI] [PubMed] [Google Scholar]
- 26.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dwyer KG, et al. A superfamily of S locus-related sequences in Arabidopsis: Diverse structures and expression patterns. Plant Cell. 1994;6:1829–1843. doi: 10.1105/tpc.6.12.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F, Kitashiba H, Inaba K, Nishio T. A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 2009;16:311–323. doi: 10.1093/dnares/dsp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Udagawa H, et al. Genetic analysis of interspecific incompatibility in Brassica rapa. Theor Appl Genet. 2010;121:689–696. doi: 10.1007/s00122-010-1340-7. [DOI] [PubMed] [Google Scholar]
- 30.Schneeberger K, et al. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci USA. 2011;108:10249–10254. doi: 10.1073/pnas.1107739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tantikanjana T, Nasrallah ME, Nasrallah JB. Complex networks of self-incompatibility signaling in the Brassicaceae. Curr Opin Plant Biol. 2010;13:1–7. doi: 10.1016/j.pbi.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Antunes MS, et al. Engineering key components in a synthetic eukaryotic signal transduction pathway. Mol Syst Biol. 2009;5:270. doi: 10.1038/msb.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: Induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 34.Lam SH, et al. Molecular conservation of estrogen-response associated with cell cycle regulation, hormonal carcinogenesis and cancer in zebrafish and human cancer cell lines. BMC Med Genomics. 2011;4:41. doi: 10.1186/1755-8794-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch MA, Haubold B, Mitchell-Olds T. Molecular systematics of the Brassicaceae: Evidence from coding plastidic matK and nuclear Chs sequences. Am J Bot. 2001;88:534–544. [PubMed] [Google Scholar]
- 36.Yang Y-W, Lai KN, Tai P-Y, Li W-H. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J Mol Evol. 1999;48:597–604. doi: 10.1007/pl00006502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.