Abstract
Oomycete pathogens cause diverse plant diseases. To successfully colonize their hosts, they deliver a suite of effector proteins that can attenuate plant defenses. In the oomycete downy mildews, effectors carry a signal peptide and an RxLR motif. Hyaloperonospora arabidopsidis (Hpa) causes downy mildew on the model plant Arabidopsis thaliana (Arabidopsis). We investigated if candidate effectors predicted in the genome sequence of Hpa isolate Emoy2 (HaRxLs) were able to manipulate host defenses in different Arabidopsis accessions. We developed a rapid and sensitive screening method to test HaRxLs by delivering them via the bacterial type-three secretion system (TTSS) of Pseudomonas syringae pv tomato DC3000-LUX (Pst-LUX) and assessing changes in Pst-LUX growth in planta on 12 Arabidopsis accessions. The majority (∼70%) of the 64 candidates tested positively contributed to Pst-LUX growth on more than one accession indicating that Hpa virulence likely involves multiple effectors with weak accession-specific effects. Further screening with a Pst mutant (ΔCEL) showed that HaRxLs that allow enhanced Pst-LUX growth usually suppress callose deposition, a hallmark of pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI). We found that HaRxLs are rarely strong avirulence determinants. Although some decreased Pst-LUX growth in particular accessions, none activated macroscopic cell death. Fewer HaRxLs conferred enhanced Pst growth on turnip, a non-host for Hpa, while several reduced it, consistent with the idea that turnip's non-host resistance against Hpa could involve a combination of recognized HaRxLs and ineffective HaRxLs. We verified our results by constitutively expressing in Arabidopsis a sub-set of HaRxLs. Several transgenic lines showed increased susceptibility to Hpa and attenuation of Arabidopsis PTI responses, confirming the HaRxLs' role in Hpa virulence. This study shows TTSS screening system provides a useful tool to test whether candidate effectors from eukaryotic pathogens can suppress/trigger plant defense mechanisms and to rank their effectiveness prior to subsequent mechanistic investigation.
Author Summary
Hyaloperonospora arabidopsidis (Hpa) is an obligate biotroph whose population coevolves with its host, Arabidopsis thaliana. The Hpa isolate Emoy2 genome has been sequenced, allowing the discovery of dozens of secreted candidate effectors. We set out to assign functions to these candidate effectors, investigating if they suppress host defenses. We analyzed a sub-set of Hpa candidate effectors (HaRxLs) that carry the RxLR motif, using a bacterial system for in planta delivery. To our surprise, we found that most of the HaRxLs enhanced plant susceptibility on at least some accessions, while few decreased it. These phenotypes were mostly confirmed on Arabidopsis transgenic lines stably expressing HaRxLs that became more susceptible to compatible Hpa isolates. Furthermore, effectors that conferred enhanced virulence generally suppressed callose deposition, a hallmark of plant defense. This indicates that the “effectorome” of Hpa comprises multiple distinct effectors that can attenuate Arabidopsis immunity. We found that many HaRxLs did not confer enhanced virulence on all host accessions, and also that only ∼50% of the effectors that conferred enhanced Pst growth on Arabidopsis, were able to do so on turnip, a non-host for Hpa. Our data reveal interesting HaRxLs for detailed mechanistic investigation in future experiments.
Introduction
Plants face constant attacks by a wide array of microorganisms including bacteria, fungi and oomycetes. Obligate biotrophic pathogens are particularly interesting because they can effectively evade or suppress host recognition, thus thwarting host defenses and enabling pathogen growth and reproduction [1].
In natural environments, plant disease is rare because plants activate a multilayered defense to most potential pathogens [2]. Relatively conserved molecules, called pathogen (or microbe)-associated molecular patterns (PAMPs), are recognized by the plants via pattern recognition receptor proteins (PRRs) [3], [4]. This interaction results in pattern-triggered immunity (PTI). Successful pathogens target effector proteins to the host cell cytoplasm to suppress PTI [5]. To counteract this, plants have evolved a second line of defense comprising resistance (R) proteins that recognize particular effectors either directly or through their activities on plant targets. This recognition leads to effector-triggered immunity (ETI) [2], [5].
It has been proposed that the “effector repertoire” of a given pathogen specifies its ability to infect a given host genotype [6], [7], [8]. Recent publications report many effector candidates predicted in the genomes of filamentous obligate biotrophs [9], [10], [11]. Comparison of effector sets of phylogenetically related species of obligate biotrophs that grow on different hosts reveals little overlap, suggesting host species-specific adaptation [10]. However, there are few studies about the functionality of obligate biotroph effectors on their hosts [12].
The downy mildew Hyaloperonospora arabidopsidis (Hpa) is an obligate biotroph that can only grow on Arabidopsis thaliana [13]. The Hpa-Arabidopsis pathosystem has been used to study host/parasite co-evolution and allowed the identification of cognate avirulent (AVRs) and resistance (R) proteins involved in specific Arabidopsis/Hpa interactions [14], [15]. The sequencing of the Hpa isolate Emoy2 genome revealed its potential to encode at least 134 candidate effectors (HaRxLs) [9]. We report here assessments of the contribution of many of these HaRxLs to Arabidopsis immunity suppression.
Filamentous pathogens likely secrete their effectors from intercellular hyphae or haustoria [16]. Several studies have defined apoplastic and cytoplasmic effectors, based on their target sites in the host [17], [18], [19]. Cytoplasmic effector proteins have been inferred from either their localization inside the host cell (e.g. Uromyces fabae RTP1 protein) [20] or their recognition by host cytoplasmic R proteins; examples include Melampsora lini AVRs (AvrL567, AvrM, AvrP123, AvrP4), Leptosphaeria maculans (AvrLm1) and Blumeria graminis f.sp. hordei (AVRa10, AVRk1) [21], [22], [23]. In oomycetes, the cloning of four AVR genes, Avr1b-1 (Phytophthora sojae), Avr3a (Phytophthora infestans, P.i.), ATR1 and ATR13 (Hpa) ([24], [25], [26], [27]) revealed a common N-terminal organization with signal peptides, enabling secretion from the pathogen, followed by a region that includes the amino acid motifs RxLR (for arginine (Arg), any amino acid, leucine (Leu), Arg) and EER (for glutamine (Glu, Glu, Arg) [28]. Functional analysis of Avr3a demonstrated that it accumulates in and is secreted from P.i. haustoria before its translocation into the host cell and its RxLR and EER motifs are required for delivery [29]. Avr1b requires its RxLR and EER motifs for uptake independently of the presence of the pathogen [30]. Binding of the RxLR EER and RxLR-like motifs of several fungal and oomycete proteins to phosphatidyl-inositol 3-phosphate (PI-3-P) has been proposed to mediate their entry into host cells [31]. In summary, the oomycete and fungal RxLR-like motifs, and the recently described LXLFLAK motif in Crinkler proteins [32] are conserved sequences involved in effector translocation into the host [33], [34]. For Hpa, no apoplastic effectors have been reported and the few effector candidates of Hpa that have LXLFLAK motifs, also carry overlapping RxLR motifs. For that reason we focused our “effectoromics” studies on predicted HaRxL-type effector candidates.
Unlike Phytophthora spp., Hpa is not transformable [35], [36]. Previous reports indicate that the bacterial type-three secretion system (TTSS) can be used to study how non-bacterial effectors can manipulate host cell functions [37], [38]. The phytopathogenic bacterium Pseudomonas syringae possesses a TTSS that translocates effectors to the host cell cytoplasm [39] via signals located on their N-termini [40]. P. syringae pv tomato DC3000 (Pst DC3000) grows on multiple Arabidopsis accessions [41]. Its growth in planta increases in PTI-compromised mutants like fls2, cerk1, sdf2, and crt3 [42], [43], [44], [45], and decreases due to ETI when it delivers bacterial AVRs in plants carrying the cognate R proteins [46], [47], [48], [49]. The Hpa effectors ATR1 and ATR13 can be delivered from P. syringae using fusions to the N-terminus of the bacterial effectors AvrRps4 and AvrRpm1 [37], [50]. This technique has enabled the study of Hpa cytoplasmic effectors by monitoring growth in planta of P. syringae delivering different alleles of ATR1 and ATR13 into Arabidopsis accessions that carry (or not) the cognate R proteins RPP1 and RPP13. Although enhanced pathogen growth due to interference with host defence can be detected, it is likely that effectors whose prime role is to promote the elaboration of haustoria would be missed in this kind of assay.
By genomic and expression analysis of the Hpa isolate Emoy2 we defined 140 HaRxLs that carry a signal peptide and RxLR motif, and ranked them taking into account allelic diversity and expression level. Our aim was to survey a broad set of candidate HaRxLs to investigate if they might play a role in suppressing PTI and/or ETI. For this purpose the Effector Detector Vector (EDV) system [37], with a luciferase-expressing Pst DC3000 strain (Pst-LUX), was used for an initial assessment of whether 64 of these HaRxLs could enhance Pst-LUX growth on at least some Arabidopsis accessions. The majority of HaRxLs were found to increase host susceptibility on multiple accessions revealing a correlation with increased callose suppression. Interestingly, many HaRxLs were not effective on all accessions, implying that host effector targets might evolve to be refractory to effector action. However, although a few HaRxLs reduced bacterial growth on certain accessions, avirulence was rare. Selected HaRxLs were studied in more detail in transgenic plants, confirming their disease-promoting activities. On turnip, a non-host plant for Hpa, fewer HaRxLs enhanced Pst-LUX growth, and more reduced it, providing interesting clues into mechanisms that underpin non-host resistance. In addition to providing novel insights into how parasites impose host susceptibility, these data reveal several high priority HaRxLs for future mechanistic investigations.
Results
Identification of HaRxLs in the Hpa Emoy2 genome
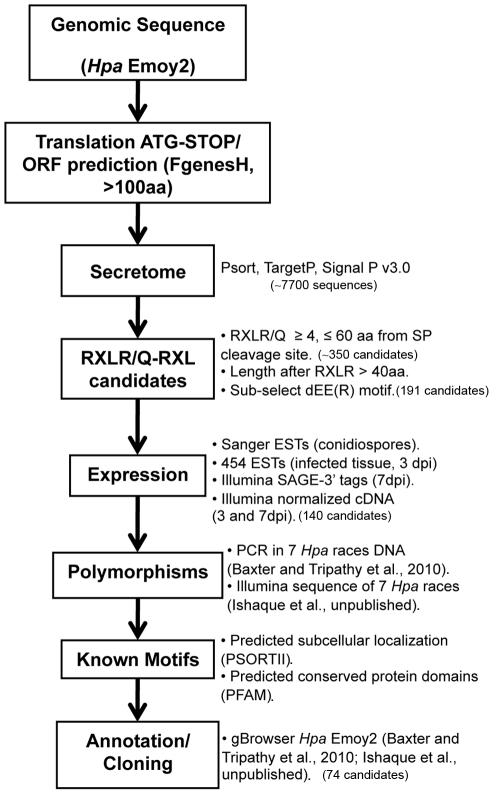
To establish an inventory of the RxLR effector secretome of Hpa, we scanned the draft genome of Emoy2 (http://vmd.vbi.vt.edu) for all possible open reading frames (ORFs) encoding putative proteins longer than 100 amino acids. We then searched these sequences for the presence of signal peptides and from those we extracted gene models carrying RxLR-like motifs (RxLR/Q; RxL) with the sequence and positional constrains defined in Figure 1 (see Materials and Methods). Different sets of HaRxLs were identified depending on the version of the genome used (versions 3.0, 6.0 and 8.3.2). We merged different lists to define a set of 191 HaRxL genes that included the known effector genes ATR1 [26] and ATR13 [27]. Most of the encoded proteins were smaller than 300 amino acids. Approximately 37% had an acidic motif (EE/EER) after the RxLR and ∼6% had a predicted nuclear localization signal (data not shown). This collection included HaRxLs identified by others using similar search algorithms [9], [28].
Figure 1. Bioinformatic pipeline used for the identification of Hyaloperonospora arabidopsidis (Hpa) HaRxLs.
(*) The genome browser is maintained at the Sainsbury Laboratory (gbrowse2.tsl.ac.uk/cgi-bin/gb2/gbrowse/hpa_emoy2_publication).
We next tested which HaRxLs were expressed during the oomycete life cycle and whether these were correctly predicted. A set of Sanger ESTs from germinating Hpa spores ([9] and two different cDNA libraries from infected Arabidopsis plants at 3 and 7 days post inoculation (dpi) were used (see Materials and Methods). We verified expression of 140 of the 191 predicted HaRxLs. Ninety of them were expressed from the asexual spore stage, perhaps ensuring their early availability upon initiation of infection, and remained expressed in planta until 7 dpi. The remaining 51 HaRxLs were expressed either at 3 dpi, 7 dpi or both. Data in column M of Table S1 illustrate the expression pattern of the sub-sets of effectors tested in this work. Of those for which we could confirm expression, the majority (90%) of the HaRxLs was correctly predicted and none had introns (data not shown). Highly expressed and accurately predicted HaRxLs were prioritized for cloning.
We then looked for evidence of polymorphism in effector candidates between seven Hpa isolates (Cala2, Emco5, Emoy2, Hind2, Maks9, Noco2, Waco9). Single Nucleotide Polymorphisms (SNPs) were detected either on PCR products (Baxter et al., unpublished data) or partial assemblies of Illumina short reads (N. Ishaque, unpublished data). Our results indicated that 12% of the HaRxLs were not polymorphic, 56% had between 1 and 10 SNPs, and 31% showed more than 10 and up to 38 SNPs. We classified them as not polymorphic (0 SNPs), low (≥1 SNPs ≤5), medium (≥6 SNPS ≤15) and high polymorphic (>16 SNPs) candidates (Column L, Table S1). For some HaRxLs it was difficult to distinguish heterozygosity from paralogous family members. In consequence, the real level of polymorphism might be underestimated. Recognized Hpa effectors like ATR1 and ATR13 show high levels of polymorphism [26], [27] while we hypothesize that non-recognized virulent effectors, adapted to interact with a specific host target, might have low sequence variability. Hence, candidates belonging to all four above described categories were used in this study.
Construction and verification of an HaRxL library for functional screening using the EDV system
The Effector Detector Vector (EDV) delivers individual effector candidates to host plant cells using the TTSS of Pseudomonas syringae [37]. Seventy-four HaRxLs were cloned into pENTR/pEDV vectors (pEDV-HaRxLs) (Table S1). We obtained 71 fusion proteins (AvrRPS4N1–136–HA tag-HaRxL). Two candidate effectors could not be cloned correctly in pEDV, and another one was truncated and further used as a negative control (NC2, Table S1). Correct in-frame constructs were introduced by conjugation into Pst DC3000 and derivative strains, particularly one expressing the luciferase (luxCDABE) operon of Photorhabdus luminescens (Pst-LUX) [41] (see Materials and Methods for full details). No differences in bacterial growth (either in liquid or solid media) were observed in Pst-LUX clones carrying any of the 71 AvrRPS4N1–136–HA tag-HaRxL fusion proteins regarding the growth of Pst-LUX harbouring AvrRPS4N1–136–HA tag-GFP/AvrRPS4AAAA (data not shown).
We performed in vitro secretion assays to check that the 71 fusion proteins obtained were made in bacteria and secreted to the medium in TTSS-inducing conditions. Secreted protein could be detected as illustrated in Figure S1A. Proteins of the expected size were produced by Pseudomonas for 64 of the pEDV5/6-HaRxLs cloned. No proteins, or protein bands of incorrect size, were observed for the remaining 7 HaRxLs, which were not used in further assays (Table S1, column H). Thus, our library comprised 64 Emoy2 pEDV-HaRxLs.
In Pst DC3000, the TTSS, encoded by the hrp-hrc (hypersensitive response [HR] and pathogenicity-hr conserved) gene cluster, is required for elicitation of HR in non-host plants like tobacco and for full pathogenicity in host plants like tomato [51], [52]. To verify that the HaRxLs proteins did not alter Pseudomonas growth in planta by blocking the TTSS, we performed HR cell death tests in tobacco (Nicotiana tabacum cv. Petit Havana) and disease assays in tomato (Solanum lycopersicum cv. Moneymaker). Of the 64 pEDV-HaRxLs delivered by Pst-LUX in tobacco, only 1 attenuated HR in tobacco while four reduced disease symptoms in tomato. No candidate impaired both activities or completely abolished HR or disease (Table S2 columns C, D and representative examples in Figure S1 B, C). We infer from these results that none of the pEDV-HaRxLs constructs blocked Pst-LUX TTSS translocation of effectors.
The Pst-LUX strain was designed for screening multiple Arabidopsis mutants/accessions that vary in resistance to PstDC3000 [41]. To evaluate the sensitivity of this system, we carefully validated the correlation between the level of bacterial bioluminescence and bacterial growth in planta using ATR1 and ATR13 (Figure S2). ATR1Emoy2/Cala2 and ATR13Emoy2/Emco5 conferred enhanced growth to Pst-LUX in the susceptible genotype Col-0, as did ATR13Emoy2 on Nd-0 plants. This phenotype could be detected as an increase in bioluminescence that correlated with higher numbers of bacteria (colony forming units (cfu)/cm2) (Figure S2). We were also able to detect decreased growth conferred to Pst-LUX by ATR13Emco5 or ATR1Emoy2 in Nd-0 plants (Figure S2).
Several HaRxLs delivered via Pst-LUX increase bacterial growth on multiple Arabidopsis accessions
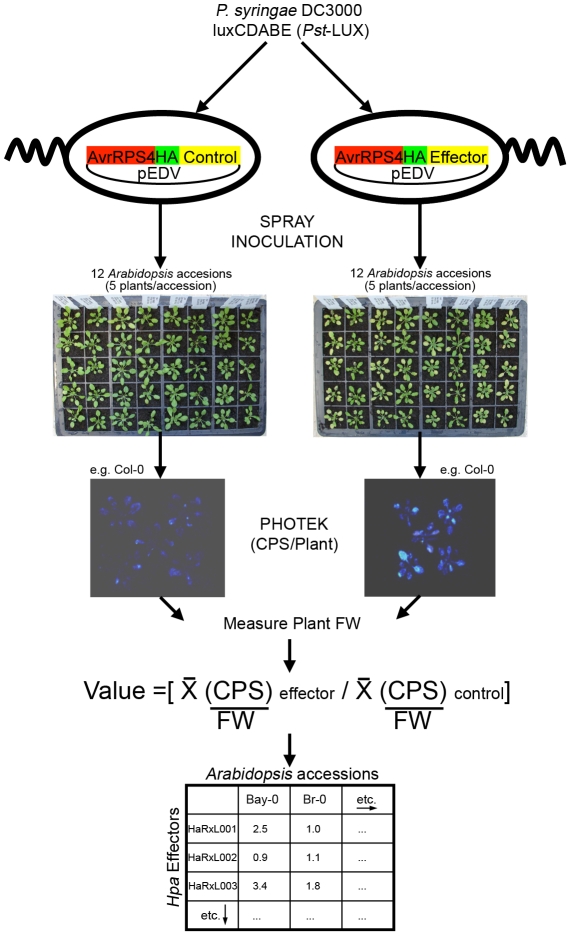
Using spray inoculation, a protocol was developed to assay bacterial growth in a sub-set of the 96 accessions described by Nordborg et al., [53], selected to maximize variability (Bay-0, Br-0, Col-0, Ksk-1, Ler-0, Nd-0, Oy-0, Shakdara, Ts-1, Tsu-0, Wei-0, Ws-0) (Figure 2). Plants were spray-inoculated with Pst-LUX carrying EDV constructs that delivered either a control protein or an HaRxL via TTSS. At 3 dpi, the bioluminescence (photons/fresh weight) emitted by the bacteria was quantified as a measure of bacterial growth (Figure 2, see details in Materials and Methods). The growth of Pst-LUX in planta carrying each HaRxL was compared to a control (see below) and expressed as a ratio. This assay allowed us to establish whether a given HaRxL was able to enhance or decrease Pst-LUX growth, manifested as quantitative differences in bioluminescence, on multiple host accessions in parallel.
Figure 2. Functional screening method.
Hpa effector candidates (HaRxLs) were delivered on 12 Arabidopsis accessions through the bacterial TTSS of the Pst-LUX strain. Levels of bacterial growth were measured quantifying bioluminescence (photon counts) emitted by the bacteria present on whole plants. The ratio of the average photon counts per second (CPS) per gram of fresh weight (FW) emitted by the bacteria delivering a given HaRxL versus the bacteria delivering control proteins was determined per accession. Experiments were repeated at least three times and statistical tests applied. Results and conclusions are shown in Table S2 and Figure 3.
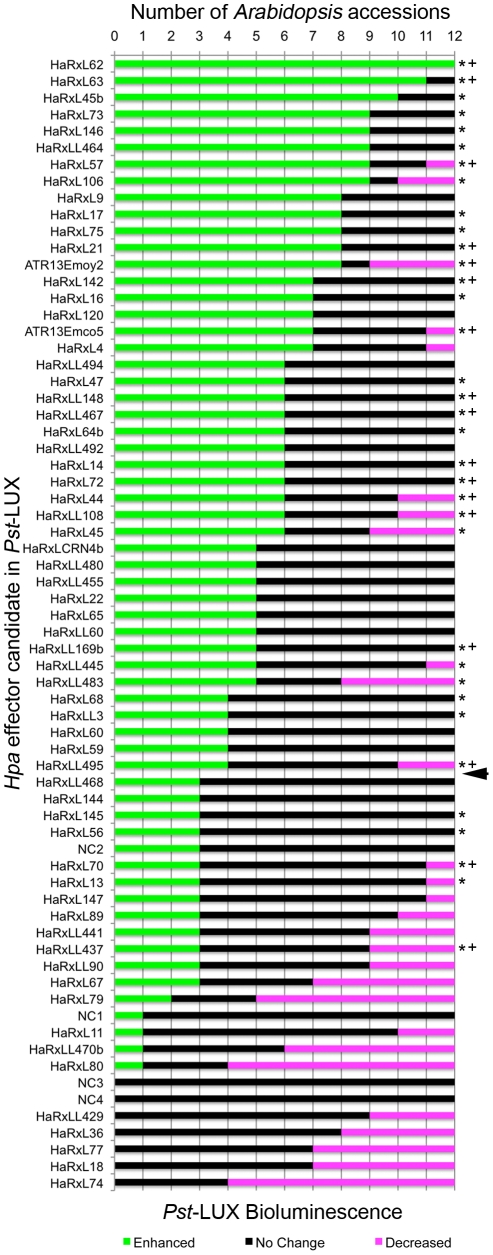
Sixty-four pEDV-HaRxLs and 3 control proteins (EDV5:HA-AvrRPS41–136, EDV6:HA-YFP, EDV6:HA-AvrRps4-AAAA) were delivered via Pst-LUX to 12 different Arabidopsis accessions. At three days post spray-inoculation, the photons/second/g fresh weight (CPS/Fw) were scored for five plants of each accession and averages, standard deviations and errors calculated. The ratio of increase or decrease in the CPS/Fw emitted by a Pst-LUX strain delivering a given pEDV-HaRxL, versus control (in the corresponding EDV5 or EDV6 backbone) was determined, as well as its statistical significance (one tailed T-test, unequal variances assumed) (Figure 2, Table S2). Experiments were repeated at least three times. Given the variability between experiments, the final outcome of each pEDV-HaRxL effect per accession was assessed across experiments and categorized according to the following criteria: i) a reproducible ratio higher or lower than one, showing the same trend on at least two experiments with a minimum statistical significance of p<0.05 on each of them, was considered as either “Enhanced” or “Decreased” growth and labeled with (+) or (−) respectively; ii) a non-reproducible ratio showing opposite statistically significant trends or the same trend but not statistically significant was considered as “No Change” and scored as ( = ) (see Table S2, columns R, S, T). A graphical synopsis of the screening outcome per effector across the 12 host accessions is presented in Figure 3, with the most effective pEDV-HaRxL (HaRxL62) at the top, conferring enhanced Pst growth on all 12 accessions.
Figure 3. Hpa HaRxLs can promote or decrease Pst-LUX growth in different Arabidopsis accessions.
The graph illustrates the outcome of the interaction between 12 Arabidopsis accessions (X axis) and Pst-LUX clones delivering 64 different Hpa effector candidates (HaRxLs, Y axis). Bars indicate the number of host accessions where the delivery of a given Hpa RxLR-like candidate effector by Pst-LUX conferred either enhanced (green bars), decreased (magenta bars) or no change (black bars) in bacterial growth, measured as bioluminescence, compared to the controls. The arrow indicates the threshold set up to consider that a given HaRxL truly enhances Pst-LUX bioluminescence. The asterisks indicate HaRxLs that suppress callose deposition in Col-0 when delivered via Pst-ΔCEL. High suppression levels are marked with (+). For details see Table S2, columns R,S and T. NC 1,2,3,4: negative internal controls.
To distinguish the effector-driven Enhanced/Decreased Pst-LUX growth patterns from the random phenotypes that can be obtained by delivering any given protein into the plant via the EDV system, we included four internal controls (negative controls, NCs). These constructs were truncated versions of HaRxLs (NC2 and NC3), non-secreted proteins with an RxLR-like motif (NC1) or a small bacterial protein with similarity to a xylosidase (NC4, genebank: AP12030.1). NC1 is part of a larger Hpa ORF encoding a putative transposase. NC2 is an early C-terminal truncated version of HaRxL143 (before the RxLR motif), while NC3 is a frame-shift version of HaRxL77 (Table S1). Functional ATR13Emoy2 and ATR13Emco5 alleles were also included. The pattern shown by these internal controls allowed us to establish threshold levels to assess whether a given HaRxL had a credible effect on Pst-LUX growth (Figure 3, Table S2). NC3 and NC4 did not impact Pst-LUX bioluminescence. We attributed the residual effect of NC1 and NC2 on Pst-LUX growth to the random variability of the system (Figure 3) and therefore we set the thresholds as follows: for an effector to be considered as an “enhancer” of Pst-LUX growth it had to show an increased significant change in bioluminescence in four or more accessions. Conversely, as the control ATR13Emco5 was recognized in only 1 accession out of the 12 tested, any effector that decreased the growth of Pst-LUX on one or more accessions was classified as capable of being recognized (Figure 3).
Our results indicate that 43 pEDV-HaRxLs enhanced Pst-LUX growth in planta, while 28 decreased Pst-LUX activity (Figure 3). The majority (72%) of the pEDV-HaRxLs that increased growth in ≥4 accessions did not decrease it in any accession (Green/black bars, Figure 3). This suggests most pEDV-HaRxLs can suppress plant defenses and avoid recognition by the host, although their effectiveness varies between accessions. We found only one pEDV-HaRxL capable of enhancing Pst-LUX growth in all accessions tested (HaRxL62); we infer that its plant target(s) might have little natural variation between accessions and that no R gene(s) recognize it in these accessions. In addition, pEDV-HaRxL9, 17, 21, 45b, 53, 73 and HaRxLL464 were able to increase Pst-LUX growth in 8 or more accessions and had no negative effect on the remaining ones. The pEDV-HaRxLs that decreased Pst-LUX growth did so mainly in ≤3 accessions (68%). This pattern of accession-specific Pst-LUX growth reduction was observed for both alleles of ATR13, and also for pEDV-HaRxL4, 44, 45, 57, 106, 108, HaRxLL445, 483 and 495. Only nine pEDV-HaRxLs conferred decreased bacterial growth in >3 and <8 accessions. Given the lack of accession-specificity of their phenotype we speculate these HaRxLs might affect the virulence activity of Pst effectors (Magenta areas, Figure 3).
Verification via growth curves of HaRxL-induced changes in Pst-LUX growth highlights a subset of effective HaRxLs
We extended the concordance analysis developed with ATR1Emoy2/Cala2 and ATR13Emoy2/Emco5 (Figure S2) to 13 other pEDV-HaRXLs delivered from Pst-LUX by conducting growth curve assays using seven Arabidopsis accessions (Table S3). For this test we selected some HaRXLs representative of the different patterns we observed in Figure 3. Briefly, we tested HaRxL62 because it enhanced Pst-LUX luminescence in all accessions; HaRxL14, HaRxL21, HaRxLL60, HaRxLL464, and HaRxLL492 because they enhanced Pst-LUX luminescence in ≥6, but did not decreased it in any accession. HaRxL44, 45, 57 and 106 also increased bacterial luminescence in ≥6 accessions but decreased it in 1–3 accessions. HaRxL70 was selected among the group of “non-effective” effectors, and HaRxL79 because it reduced Pst-LUX bioluminescence in >3 accessions.
For 32 of 35 combinations (pEDV-HaRXL×accession) we confirmed the correlation between enhanced bioluminescence and increased bacterial growth. These data verified that Pst-LUX bioluminescence reveals the effect of HaRXLs on Pst-LUX growth. We also observed that some HaRXLs have a substantial positive effect on bacterial growth on multiple accessions, and can increase Pst-LUX growth ∼10-fold (Table S3 and Figure S3). In particular, we confirmed that HaRxL62 and HaRxL14 render multiple host accessions more susceptible to bacterial infection (Figure S3). Accession-specific effects were verified for HaRxLL464 and HaRxL21 while putative recognition events, leading to a decrease in bacterial growth, were verified for HaRxL44 in Ler-0, HaRxL57 in Ksk-1 (Figure S3, Table S3), and HaRxL106 in Col-0 (Table S3). No effect was observed for HaRxL70 in Col-0 while the decrease in bacterial growth caused by HaRxL79 was only observed when plants were spray inoculated (Table S3). These data reinforced the usefulness of the EDV Pst-LUX assay for selecting candidates for further work, and confirmed several candidates as a high priority for further investigation.
Host genotypes and levels of HaRxL polymorphism are not correlated with effector-induced changes in Pst-LUX growth
To evaluate if host genotypes influenced the pattern of Arabidopsis responsiveness to the set of HaRxLs tested, the spectrum of effective HaRxLs per accession was analyzed. We found that an average of 42% of the pEDV-HaRxLs enhanced Pst-LUX growth on any given accession, while only ∼11% reduced Pst-LUX growth. Many combinations (46%) did not cause any change in Pst-LUX growth (Figure S4). Enhancement or decrease of susceptibility was not restricted to a particular set of accessions, and did not correlate with those accessions showing resistance or susceptibility to the infection by the Hpa isolate Emoy2 (Figure S4). The only deviations from this pattern were Nd-0, in which most of the pEDV-HaRXLs (73%) increased Pst growth and only ATR13Emco5 was able to decrease it, and Br-0 in which fewer pEDV-HaRXLs in total were effective (31% compared to the average of 42% for all other accessions) (Figure S4). These results are consistent with the idea that some effector targets are widely conserved while others vary between accessions.
The level of polymorphism of HaRxLs did not correlate with the capacity to enhance Pst-LUX growth. Among the 64 candidates tested, 11 were highly polymorphic, 21 had a medium level and 32 showed low polymorphism. HaRxLs categorized in these three groups showed ability to increase bacterial luminescence in an average of 6±2.54, 6±3.15 or 5±2.66 accessions, respectively. For example, HaRxLL464 and HaRxL57 showing low or no polymorphism, and the highly polymorphic HaRxL106 and HaRxL21 were all capable of increasing Pst-LUX growth in 8 or more host accessions.
HaRxLs did not trigger hypersensitive recognition in any Arabidopsis accession after EDV delivery
Isolate Emoy2 is recognized by certain Arabidopsis accessions, indicating effector recognition by R protein(s). In order to identify avirulent HaRxLs in the library, we analysed in detail Pst-LUX growth assays in each of the 12 Arabidopsis accessions (Figure 3, Figure S4). Possible recognition of pEDV-HaRxL strains in our assays was indicated by the decrease in Pst-LUX growth, usually in an accession-specific manner (Figure 3, Table S2). Potentially novel ATR proteins may have been detected in interactions with accessions Col-0, Ler-0, Br-0 and Ksk-1 (Figure S4).
ETI is strongly correlated with HR-like cell death [54], [55] although HR is not always required for resistance [49], [56]. We tested possible recognitions using a weakly virulent Pst DC3000 ΔCEL (Pst-ΔCEL) strain and a modified P fluorescens carrying a functional TTSS (Pf0-1) [57] to deliver potentially recognized HaRxLs to the corresponding “resistant” accessions. We performed localized leaf infiltrations using high doses of bacteria and looked for macroscopic (leaf collapse) and microscopic (dead cells stained with trypan blue) indicators of HR-like cell death 24 h post infiltration.
Surprisingly, no HaRxLs besides the positive control (ATR13Emco5 in Nd-0) provoked clear signs of macroscopic HR. We then stained infiltrated leaves with trypan blue and examined for microscopic lesions. All micro-HR lesions were much smaller and weaker than those triggered by bacterial effectors like AvrRpm1 or AvrRpt2 (data not shown) or by the Hpa effector ATR13Emco5 in Nd-0 (Table S4). In 78 pEDV-HaRXL/accession combinations, we saw micro-HR-like cell death in only 7 interactions, comprising just 4 candidate effectors (HaRxL4, 18, 70 and 80; in bold in Table S4). Similar results were obtained with both Pf0-1 and Pst-ΔCEL strains, except for HaRxL106 where no HR was detected in Col-0 and Ksk-1 when delivered through Pf0-1 (Table S4). Nevertheless, the decrease in bacterial growth observed for Pst-LUX delivering each of these candidate effectors in the corresponding accessions was confirmed by reduction in disease symptoms and bacterial growth using Pst-ΔCEL (data not shown).
None of the mild recognition patterns matched with profiles expected for ATR4, ATR5 and the putative ATR(s) recognized in Ksk-1 and Br-0 (Figure S4). Interestingly, two HaRxLs (HaRxL18 and 70) were weakly recognized in Bay-0, an accession susceptible to isolate Emoy2. These results suggest that the decreases in Pst growth we see in some HaRxL/accession combinations are not due to strong R/AVR interactions. Also, weak recognition of some HaRxLs might not result in HR [49], [56].
Importance of PTI suppression for Hpa infection
Many HaRxLs delivered in planta by Pst-LUX confer increased growth of an already virulent pathogen. Enhanced susceptibility to adapted pathogens is often a result of PTI suppression [42], [43], [44], [45]. PTI- responses, like callose deposition, likely limit the growth of Hpa during infection [58], [59], [60]. Also, ATR13Emoy2/Emco5 can complement HopM1-mediated suppression of callose deposition when delivered by Pst-ΔCEL [37]. Therefore, we investigated if PTI affects Hpa growth, and whether Hpa is able to actively suppress PTI. As no known PAMP has been identified for Hpa, we tested whether Hpa infection alters responses to known PAMPs.
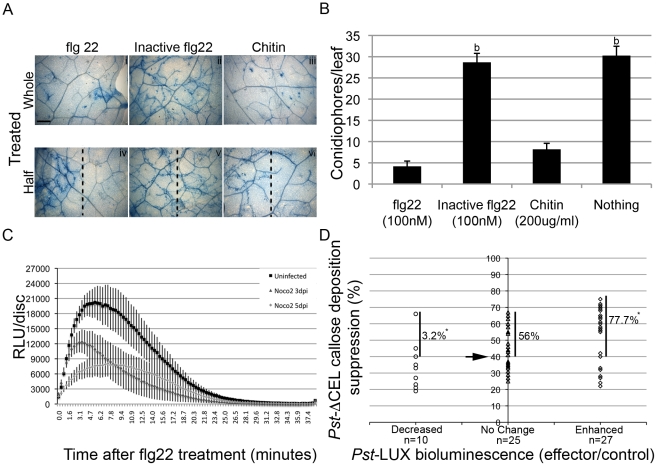
To test if PTI can attenuate Hpa growth during a compatible interaction, we pre-treated young Col-0 plants with flg22 (100 nM), an inactive flg22 (from Agrobacterium tumefaciens), or chitin (200 µg/ml) 24 h before the plants were sprayed with spores of Hpa isolate Noco2. Reduced hyphal growth was observed in the areas where the PAMPs were applied, as assessed by trypan blue staining of the pathogen in the leaves (Figure 4 A). We also noticed a decrease in the rate of Hpa sporulation (Figure 4 B). These phenotypes were not observed with inactive flg22 (Figure 4 A, B), or when the treatment was applied on plants mutant for these PAMP receptors (fls2-1 and cerk1-1, data not shown). These data suggest that the phenomenon is specific for PTI. We also noticed that the “protection” that flg22 and chitin conferred to the plants was higher near the infiltrated site, was dose-dependent, and diminished with the time of pre-infiltration relative to Hpa infection (24 hs>48 hs>72 hs) (Figure 4 A lower panel and data not shown), consistent with the transient nature of the local PTI response [61]. We did not observe extensive local micro-HR in flg22-treated leaves [62]. The “local” effect of flg22 and chitin in restricting Hpa hyphal growth (Figure 4 A) might indicate that either we did not induce systemic acquired resistance (SAR) [63], or we applied Hpa before SAR was established.
Figure 4. Suppression of PTI as a virulence tool for Hpa.
(A) Pre-treatment of Col-0 leaves with flg22 or Chitin reduces Hpa isolate Noco2 hyphal colonization. Leaves of four-week-old Col-0 plants were pre-infiltrated with 100 nM flg22, inactive flg22 (from A. tumefasciens) or Chitin (200 µg/ml) 24 h before inoculation of Hpa Noco 2 (5×104 sp/ml). Pictures show trypan blue stained leaves at 5 days post-Hpa spraying (dps). This experiment was repeated three times with similar results and also for Emoy2 on Oy-0 plants. Panels i, ii and iii: the whole area shown was pre infiltrated. Panels iv, v and vi: only the right side of the picture was infiltrated. Dotted vertical line indicates approximated infiltration boundaries. Bar is 500 µm. (B) Pre-Induced PTI responses reduce Hpa asexual reproduction. Leaves of three-week old Col-0 plants were infiltrated with the indicated solutions 24 h before infection with Noco2 (5×104 sp/ml). Conidiophores per leaf were counted on trypan blue stained leaves excised at 5 dps. Bars represent the average ±2× SE of 40 leaves. This experiment was repeated three times with similar results. (b) p value <0.01, T-test. (C) Hpa infected tissues show reduced ROS response to flg22. Leaf discs from uninfected and infected Col-0 plants were treated with 100 nM flg22, and the level of ROS generated measured with a Luminometer. Values indicated are average of Relative Luminescence Units (RLUs) ± SE of 24 leaf discs. (D) HaRxLs delivered by Pst-ΔCEL in Col-0 plants suppress callose deposition. Effector's impact on the level of Pst-ΔCEL-triggered callose deposition is presented in the Y-axis. The average reduction (in percentage) of callose deposits observed when a given candidate effector was delivered, compared to the number of callose deposits observed when control proteins were delivered by Pst-ΔCEL, is represented by the shapes in the body of the graph. HaRxLs were also categorized according to their phenotype on Pst-LUX bioluminescence in Col-0 (X-axis). The arrow indicates the threshold set up to consider callose deposition as significantly suppressed. The numbers in the body of the graph correspond to the percentage of HaRxLs able to suppress callose deposition among each bioluminescence category. (*) Indicates p<0.05 of Z-test versus random distribution expected for the number (n) of HaRxLs on each group.
One of the earliest PTI responses is the generation of reactive oxygen species (ROS burst) [64]. To determine if Hpa can suppress ROS, we measured flg22-induced ROS in infected leaf tissues. We observed a highly reproducible reduction by ∼50% in ROS accumulation induced by flg22 if leaves were pre-infected with Hpa isolate Noco2 (Figure 4C). Oy-0 plants infected with Emoy2 showed the same pattern (data not shown). Thus, Hpa infection can dampen PTI responses.
Most HaRxLs that increase bacterial growth in Col-0 can suppress Pst-ΔCEL-induced callose deposition
PTI results in callose deposition in the cell wall [65] and microbial effectors that impair PTI also suppress callose deposition [66], [67], [68], [69]. Pst-ΔCEL is unable to suppress callose deposition due to lack of HopM1 and AvrE [70]. We introduced pEDV-HaRxL constructs into Pst-ΔCEL and evaluated if they could restore callose suppression when infiltrated in Col-0.
Sixty-two pEDV-HaRxLs and 2 control proteins were delivered through this system. Due to variability between leaf reactions in the same plant and among experiments, we established a threshold to define significant reductions in callose deposition (see Materials and Methods). Taking into account the maximum levels of random callose suppression observed after delivery of the negative controls (NC1 and NC2), we set up the threshold to 40% of callose suppression because negative controls could reduce up to 29% or 34% of the callose dots found when Pst-ΔCEL delivered the additional controls (EDV5:HA-AvrRPS41–136, EDV6:HA-YFP or EDV6:HA-AvrRps4AAAA). Using this stringent criterion we found that 35 HaRxLs were able to suppress callose deposition by more than 40%, while 27 HaRxLs did not. Those effectors complementing the phenotype of Pst-ΔCEL are indicated with asterisks (*) in Figure 3.
We noticed that most of the HaRxLs able to complement Pst-ΔCEL were also able to enhance Pst-LUX growth in four or more host accessions (Figure 3). To establish the degree of correlation between both phenotypes, the extent of callose suppression was compared with the changes in Pst-LUX growth produced by each effector in the accession Col-0 (Figure 4 D). For this, we classified the effector's conferred phenotype in the host as follows: enhanced susceptibility = 27 HaRxLs (right side of the graph), decreased susceptibilty = 10 HaRxLs (left side of the graph), no change = 25 HaRxLs (axes intersection). When we plotted the percentage of callose suppression of each of the effectors in these groups, we found 77.7% of HaRxLs candidate effectors with a positive effect on Pst-LUX growth in Col-0 were able to suppress callose deposition, while only 3.2% of those decreasing Pst-LUX growth could reduce callose levels (Figure 4 D). These percentages deviate significantly from those expected for a random distribution (Z-test p<0.05). The HaRxLs located at the top right side of Figure 4 D were those that strongly suppressed callose deposition, and are indicated with plus (+) signs in Figure 3. The HaRxLs with no effect on Pst-LUX growth showed no clear trend in ability to suppress callose deposition.
We conclude that most HaRxLs that enhance Pst growth also suppress PTI, increasing host-susceptibility. Based on the data presented in Figure 3, Tables S2 and S3, Figure S3 and Figure 4D, we prioritized HaRxL14, 21, 44, 45/45b, 57, 62, 106, HaRxLL60, 464, 492 and 495 for further detailed studies. HaRxL14, 21, 44, 57 and 62 were chosen because they increased Pst-LUX growth in more than 6 accessions and strongly suppressed callose deposition in Col-0 (>60%) (Figure 3 and top right side on Figure 4D). HaRxLL464 and HaRxL45/45b strongly enhanced Pst-LUX growth in 9 or more accessions, but showed a mild reduction in callose deposits (Figure 3 and Table S2). HaRxL106 conferred enhanced susceptibility to Pst-LUX in several ecotypes, except Col-0, where it nevertheless reduced callose deposits (Figure 3). HaRxLL60 and HaRxLL492 conferred enhanced growth of Pst-LUX in 5–6 accessions, but were unable to complement the Pst-ΔCEL phenotype.
Arabidopsis transgenic lines constitutively expressing HaRxLs show PTI suppression and enhanced susceptibility
To investigate if the phenotypes observed with the Pst-EDV-delivery screenings were also conferred by stably expressing the corresponding HaRxLs directly in the host plant, transgenic Arabidopsis plants were generated initially in the Col-0 background. The following HaRxLs were expressed from the constitutive (CaMV 35S) promoter: HaRxL14, 21, 44, 45/45b, 57, 62, 106, HaRxLL60, 464, 492 and 495. Of these, for three candidates (HaRxL62 and HaRxL45/45b) either we did not obtain transgenic lines or the ones generated showed segregation of pleiotropic effects, and in consequence are not described here. Some plants (lines 35S-HaRxLL464 and 35S-HaRxL44) showed a 20–30% increase in fresh weight and others (line 35S-HaRxLL60) a 30–40% decrease in fresh weight. In some cases (35S-HaRxL106 and 35S-HaRxLL60) the shape of the leaves changed, becoming either elongated and darker, or serrated and smaller, respectively (data not shown). The level of expression of the transgene in each line was verified by semi-quantitative RT-PCR (Figure S5).
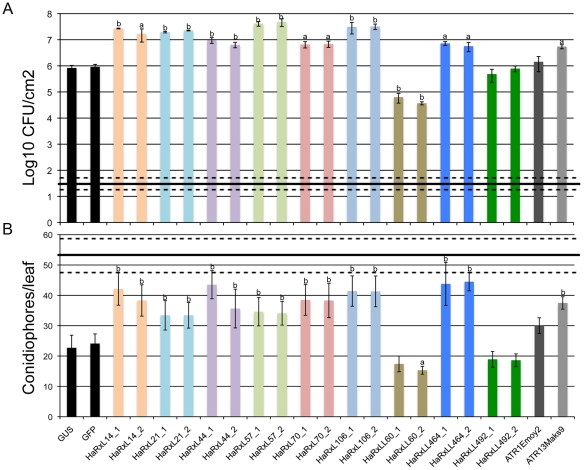
Using three independent lines for the remaining nine candidate effectors, that did not showed perturbed growth, we assessed whether in planta over-expression altered pathogen development, PTI or ETI. We characterized the responses of these transgenic lines to both bacterial (Pst-LUX, Pst ΔavrPto/ΔavrPtoB) and oomycete pathogens (Hpa isolates Noco2 and Emoy2) (Figure 5 and Figure S6). Transgenic lines expressing different HaRXLs showed increased susceptibility to Pst-LUX when spray-inoculated (8 lines, Figure S6), or to Pst ΔavrPto/ΔavrPtoB when infiltrated (7 lines, Figure 5 A). Also, seven lines showed enhanced susceptibility to Hpa isolate Noco2 (Figure 5 B). These phenotypes were observed in at least two out of the three transgenic lines recovered for each effector.
Figure 5. Arabidopsis Col-0 plants expressing constitutively HaRxLs support enhanced growth of P. syringae ΔavrPto/ΔavrPtoB and Hpa isolate Noco2.
(A) Four leaves of three five-week-old plants of two independent transgenic lines per HaRxL were infiltrated with Pst-ΔavrPto/ΔavrPtoB at OD600 = 0.0005. Bacterial growth was determined at 3 dpi by traditional growth curve assays. Bacterial populations immediately after inoculation (3 h; 0 dpi) were averaged among plants and are represented by the solid black horizontal line, with 2× SE represented by the dashed horizontal lines. (a) T-test p value<0.05, (b) T-test p value<0.01.This experiment was repeated two times with similar results. (B) Two-week-old seedlings were spray inoculated with a suspension of 1×104 conidiospores per ml of Hpa isolate Noco2. At 6 dps, whole seedlings were cut and stained with Trypan blue. The number of conidiophores per leaf was counted in 4 leaves per seedling. Ten seedlings were analyzed per transgenic line per HaRxL. The horizontal black and dashed lines represent the average ±2× SE of the number of conidiophores per leaf found in the hyper-susceptible mutant Col-0 eds1-2. (a) T-test p value<0.01, (b) T-test p value<0.05. This experiment was repeated three times with similar results.
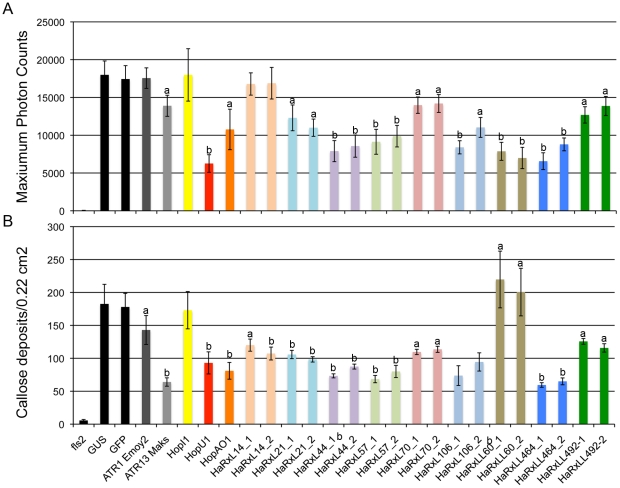
We investigated if the transgenic lines were compromised in ROS burst and callose deposition in response to flg22 (Figure 6). Eight 35S-HaRxLs were able to reduce flg22-triggered ROS accumulation by 22 to 65% compared to controls (Figure 6 A). Also, callose deposition was diminished by an average of 40% compared to controls (Figure 6 B). The ROS and callose suppression in transgenic lines expressing HaRxL 14, 21, 44, 57, 106, HaRXLL 464 was comparable to that observed in plants that express the bacterial effectors HopU1 and HopAO1 (Figure 6 A, B). In summary, six different Hpa HaRxLs, when stably-expressed in planta, displayed a positive correlation between increased susceptibility to Pst and/or Hpa and reduced levels of ROS and callose deposition elicited by flg22 (Figure 7).
Figure 6. Arabidopsis plants expressing constitutively HaRxLs accumulate less ROS and/or callose in response to flg22.
(A) Leaf discs from four-week-old transgenic plants expressing the indicated HaRxL were sampled and floated in water 14 to 16 h prior to flg22 treatment. Photon emission was measured every 100 milliseconds for 40 minutes. Lines and error bars represent the mean of maximum values of photon counts ±2× SE of 24 independent leaf discs. This experiment was repeated four times with similar results. (B) Leaves of four-week-old transgenic lines were hand inoculated with 100 nM of flg22. Twenty-four hours post-inoculation, leaf discs were sampled and stained with aniline blue for visualization of callose dots. The bars represent mean ±2× SE of callose dots per image photographed (field of 0.22 square centimeters). Callose dots were quantified with ImageJ. Twenty leaf discs were analyzed per transgenic line. This experiment was repeated three times with similar results.
Figure 7. Summary of phenotypes observed upon expression of HaRxL effectors in Arabidopsis.
Graphical comparison of the results obtained using the transient EDV assays and stable constitutive expression for nine different HaRxLs. The phenotypes analyzed include bioluminescence of Pst-LUX, suppression of callose deposition triggered by Pst- ΔCEL, growth of Pst- ΔavrPto/ΔavrPtoB, suppression of ion leakage triggered by delivering AvrRPM1 via Pf0-1, growth (conidiation) of Hpa compatible (Noco2) and incompatible (Emoy2) isolates, and suppression of the levels of ROS and callose deposition triggered by flg22 treatments.
To establish if any of the nine HaRxLs could also compromise ETI, we tested transgenic lines for altered resistance to Hpa Emoy2 which is recognized in Col-0 [71]. Two-week-old seedlings were sprayed with Emoy2 conidiospores and trypan blue-stained at 5 dpi. While some restricted hyphal growth was detected, we did not observe asexual or sexual reproduction -in true leaves- in any line (summarized in Figure 7 and data not shown). We then studied the ETI response to AvrRpm1 from P. syringae pv. maculicola [72]. AvrRpm1 was delivered via Pf0-1 in leaves of 4-week-old plants and a macroscopic HR recorded. The onset of HR was delayed but not completely suppressed in four different lines (data not shown). We therefore performed a more sensitive assay by quantifying the levels of ion leakage upon AvrRpm1 detection (Figure S7). Notably, the same four sets of transgenic lines (35S-HaRxL106, 35S-HaRxLL492, 35S-HaRxLL464 and 35S-HaRxLL60), slightly but significantly reduced the levels of ion leakage caused by recognition of AvrRpm1 compared to the control lines (Figure S7 and Figure 7).
Our results suggest that the majority (six out of nine) of Hpa Emoy2 HaRxLs constitutively expressed in planta gave phenotypes (enhanced susceptibility to Pst-LUX and suppression of PAMP-triggered callose deposition) consistent with the results obtained from the Pst-LUX EDV-screening in the Col-0 accession (Figure 7). Two transgenic lines (35S-HaRxL106 and 35S-HaRxLL60) showed phenotypes that contrasted with those observed in the EDV screen on Col-0, and for another line (35S-HaRxL70) no effect was detected with the EDV system in Col-0, while enhanced Pst-LUX growth was observed in the over-expressing plants (Figure 7). When HaRxL106 is delivered by EDV from Pst DC3000 strains, it appears to be recognized by Col-0 and Ksk-1 accessions, but this HR-like cell death is not evident when it is delivered by Pf0-1 (Table S4). Expression of 35S-HaRxLL60 caused the plants to be smaller, accumulate callose constitutively and become resistant to the pathogens tested (Figure 5, Figure 6B, Figure S6). Nevertheless, 35S-HaRxLL60 reduced flg22 ROS production (Figure 6A). The set of six lines with enhanced susceptibility to Pst and/or Hpa also displayed a reduced ROS burst and callose deposition after PAMP treatment (Figure 7). Overall, our analysis points to suppression of PTI-related responses as a predominant mode of action of Hpa candidate effectors in planta.
Emoy2 HaRxL candidate effectors are mostly ineffective or reduce pathogen growth in the Hpa non-host Brassica rapa
Most plants are resistant to most pathogens, and this so called “non-host resistance” (NHR) could be caused by either ineffectiveness of effectors, resulting in failure to suppress PTI, or recognition of effectors, resulting in resistance via ETI [8]. To test these hypotheses we delivered HaRxLs via Pst-LUX in the non-host Brassica rapa cv Just Right (turnip).
Pst-LUX is virulent in turnip and causes disease symptoms at 3 dpi when inoculated at low dose. We tested our collection of HaRxL-carrying Pst-LUX strains in turnip and monitored symptoms and growth. After three rounds of screening we found that 20 effectors can alter Pst-LUX growth in turnip (13 increase, 7 decrease), while the remaining 44 did not cause any changes (Figure S8 and Table S2, column U). We compared these data with the number of “effective” HaRxLs in one given Arabidopsis accession (39 in Col-0) and the average for the 12 accessions we tested (35) (green plus magenta bars vs. black bars in Figure S4). In contrast, only thirteen HaRxLs increased Pst-LUX growth in turnip, while in Arabidopsis accessions an average of 27, and minimum of 18 (in Br-0) increased Pst-LUX growth (see Figure S4 and S8). Those candidates enhancing Pst-LUX growth in turnip also did so in >3 Arabidopsis accessions, implying that their effect on plant immunity is not species-specific and some plant targets might be conserved. Conversely, while similar numbers of HaRxLs decrease Pst-LUX growth in Arabidopsis (8 in average) and turnip (7), we noticed three HaRxLs (HaRxL17, HaRxL47 and HaRxL63) that reduced Pst-LUX growth and disease symptoms in turnip that did not show this phenotype in the 12 Arabidopsis accessions. To assess if these HaRxLs might be specifically recognized in turnip and contributing to NHR against Hpa, we used higher dose inocula to deliver them using Pst-ΔCEL. We did not observe HR-like cell death, but we confirmed the reductions in growth and disease symptoms (data not shown). It remains possible that these HaRxLs might be triggering weak ETI in turnip that does not involve HR-like cell death but still contributes to NHR (see Discussion).
Discussion
Genome sequences of plant pathogens have enabled searches for effectors that might manipulate host cells ([9], [28], [73], [74], [75], this work). Verified effectors provide molecular probes to investigate plant defense mechanisms and better understand pathogen adaptation to hosts. In Hpa, ∼134–140 HaRxLs have been identified [9], [28], this work] and the challenge is to identify those that are functional and then investigate their biological mechanisms.
For effector discovery, we combined the use of a heterologous system for Hpa candidate effector delivery from Pst DC3000, with a rapid and sensitive assay for bacterial growth in planta, and for suppression of callose synthesis. To independently assess the efficacy of the most promising candidates, we also tested the consequences of constitutively expressing selected HaRxLs in planta.
EDV vectors and Pst-LUX strains provide a sensitive assay for effects of HaRxLs on plant immunity
We found that of 64 expressed HaRxLs that could be secreted by the EDV delivery system, 43 (∼67%) could increase the growth of Pst-LUX in several (>4) host accessions. We were surprised that so many candidate effectors can enhance growth of an already virulent pathogen. The false positive rate is likely to be low, while the false negative rate could be high, because we set a stringent threshold to judge a putative effector “effective” compared to four internal controls. Despite the intrinsic variability of Pst-LUX spraying assays, the phenotypes were reproducible, even when the measurable differences were small (Table S2 and S3). We found that detection of LUX activity is more sensitive and less laborious than growth curve assays and noticed that 2- fold differences in LUX emission usually corresponded to differences between 0.3 and 0.6 log in growth. These might be considered small contributions to virulence, but are consistent with previously reported observations for bacterial effectors such as AvrRpm1 AvrRpt2, AvrPtoB, HopF2, HopAO1 and HopU1, where their individual contribution to Pst DC3000 growth is of the same order of magnitude (around 0.4 log) or only detectable using low virulence Pst mutants [47], [69], [76], [77], [78].
A strong advantage of the EDV approach is that one Pst-DC3000 strain can be tested on many different Arabidopsis accessions to reveal accession-specific differences in HaRxL efficacy. This would be extremely laborious by generating stable transgenic lines in multiple accessions. Furthermore, some HaRxLs confer severe pleiotropic defects when expressed directly in planta, hampering efforts to test whether such lines are immunocompromised.
Hpa virulence likely involves multiple effectors with weak accession-specific effects
Since not all HaRxLs are effective in all accessions, it seems likely that each Hpa isolate expresses a repertoire of effectors, each of which may be functional on some but not all host genotypes. The level of infection in Col-0 by compatible Hpa isolates is quite variable, with Waco9 more virulent than Noco2, which is more virulent than Emco5. Such observations are usually interpreted in terms of variation in avirulence gene content. However, variation in host targets as well as in Hpa effector complement may also underpin quantitative differences in host susceptibility. Hpa isolate Emoy2 was reported as having the highest likelihood of producing high levels of sporulation in a study involving 96 Arabidopsis accessions, and isolate Emco5 the lowest [79]. Although Hpa virulence appears to depend on multiple virulence genes with weak effects, rather than a few genes with strong effects, some effectors, such as HaRxL62, 14, 44, 57 and 106, are particularly effective and will repay detailed mechanistic investigation in the future.
HaRxLs that enhance Pst growth usually suppress callose deposition
Significantly, we found that most of the HaRxLs (77%) that increase Pst-LUX growth in Col-0 were also able to suppress Pst-ΔCEL-induced callose deposition. Conversely, those HaRxLs reducing Pst-LUX growth in Col-0 were generally unable to suppress callose deposition. Callose deposition is a late PTI response (though also associated with ETI). We speculate that HaRxLs may enhance Pst-LUX growth via additional PTI suppression, either alone or in conjunction with Pst effectors. The increased susceptibility to Pst-LUX observed using the EDV system was usually consistent with phenotypes of plant lines that constitutively express the corresponding HaRxLs (Figure 7). Moreover, seven of the transgenic lines also showed increased susceptibility to Hpa isolate Noco2. We infer that enhanced susceptibility results from suppression of host mechanisms that are active against diverse pathogens. The fact that they further elevate virulence conditioned by Pst-DC3000 effectors may reflect HaRxLs interference with targets that are not identical to those of Pst-DC3000, resulting in an additive effect.
Infection with Hpa, and many Hpa effectors, suppress PTI
Assays using flg22-induced ROS or callose deposition on stable transgenic lines indicate that the main target of HaRxLs is PTI. All of the transgenic lines tested showed either reduced levels of flg22-induced ROS or callose deposition or both. The PAMP complement in Hpa is unknown, as are their receptors and downstream signal transduction pathways in Arabidopsis. Several molecules have been reported as oomycete PAMPs [17], [80] but their existence in Hpa is not known [9], [81]. We show that pre-elicitation of PTI by bacterial and fungal PAMPs impairs Hpa growth and reproduction, indicating that to infect, Hpa must counteract these host responses. Moreover, in host tissues where high numbers of haustoria are established, PTI responses are attenuated.
PTI involves multiple processes that can be attenuated by diverse pathogen effectors [76], [77], [78], [82]. Our data support the idea that the function of the majority of the effector proteins is to inhibit plant immunity [4], [83], [84]. For Pst, 13 out of 28 active effectors (12 belonging to Pst-DC3000) have been reported to suppress PTI [84], [85], [86]. Thus, ∼50% of this bacterial pathogen's effector repertoire targets PTI in one host. Importantly, this hemibiotroph can infect Solanaceae as well as Brassicaceae, so more effectors might emerge as PTI suppressors when other host species are studied. It has also been observed that 91% of Pst-DC3000 effectors, when delivered at high titers from Pf0-1, are able to suppress the HR induced by the bacterial effector HopA1 (from P. syringae syringae) in tobacco [38]. Since tobacco is a non-host for Pst-DC3000, this study again points to a high functional redundancy between effectors in suppressing HR. In oomycetes, experimental characterization of several RxLR effector genes suggests that many function to suppress host defenses [37], [50], [86]. Also, 3 out of 32 P. infestans RXLR candidate effectors were able to suppress PAMP- triggered programmed cell death (PCD) in N. benthamiana, while another 13 induced either non-specific or R-mediated PCD [87].
HaRxLs are rarely avirulence determinants
We also identified HaRxLs that reduced Pst-LUX growth in the interaction with Arabidopsis accessions, and investigated whether they are new avirulence determinants (ATRs). Surprisingly, we observed that strong incompatibility caused by HaRxLs is rare. None was able to trigger macroscopic ETI when delivered in planta at high titer, as ATR13Emco5 did in Nd-0. Instead, several were identified that can reduce Pst-LUX growth in specific Arabidopsis accessions and four triggered micro HR-like lesions when delivered via Pst-ΔCEL. Since we tested only 64 HaRxLs from just one isolate, on only 12 host accessions, our survey was not exhaustive, and the anticipated ATR4 might not have been in our repertoire. Alternatively, the EDV assay may not be sensitive enough to detect new Hpa ATRs because these ATR-RPP recognitions are weaker than those already described with this system (ATR13-RPP13 or ATR1-RPP1). Conceivably, some ATRs might not carry an RxLR motif and therefore were not identified as candidate effectors in our bioinformatic analysis, as with the recently cloned ATR5 [88]. It also remains possible that either the sub-cellular localization or post-translational modifications of the EDV-delivered HaRxLs are not similar enough to their native form to be able to elicit ETI, although this has not been the case for ATR1 and ATR13 alleles [37], [50].
Non-host resistance could involve a combination of recognized HaRxLs and ineffective HaRxLs
We also tested if the recognition or non-functionality of HaRxLs could be involved in non-host resistance to Hpa in Brassica rapa (turnip). We found that HaRxLs were “less effective” in turnip, but those HaRxLs that enhanced Pst-LUX growth in B. rapa also did so in Arabidopsis, suggesting conservation in their targets. Notably, three HaRxLs conferred reduced Pst-LUX and Pst-ΔCEL growth in B. rapa, but did not reduce growth in any Arabidopsis accession. Therefore, the inability of Hpa to grow in turnip might result not only from reduced “effectiveness” of the effector complement, but also from recognition in the “non-host” of a subset of effectors that are not recognized by most Arabidopsis accessions.
Concluding observations
As with any screening protocol, this heterologous system has some limitations. For example, HaRxLs that require extensive post-translational modifications will not be correctly produced by a prokaryotic system. Also, the co-delivery of an HaRxLs with c.a 30 effectors from Pst DC3000 might alter the outcome of the assay if positive or negative interactions exist between them. This might explain some of the discrepancies we observed between results obtained with the EDV system and those generated by expressing the candidate effectors directly in the plant. A further potential limitation of the EDV system is that effectors required to elaborate a haustorium inside the host cell might not be revealed as promoting Pst growth by the assays we developed. Despite such limitations, most of the phenotypes observed with the EDV system were confirmed in the transgenic lines.
In conclusion, the EDV-based system has enabled the systematic analysis of the biological relevance of effector candidate proteins. The Pst-LUX and Pst-ΔCEL screens allowed the generation of a “ranking of effectors” that permitted the selection of highly interesting candidates as targets for subsequent mechanistic studies. Further detailed investigations of Hpa effectors will help reveal how Hpa alters host cellular processes to promote its growth and reproduction.
Materials and Methods
Bioinformatic identification of HaRxLs
Different versions of the genome of Hpa isolate Emoy2 (http://vmd.vbi.vt.edu; v3.0, v6.0 and v8.3.2) were translated in all 6 reading frames. ORFs from ATG to Stop codon were identified using FgenesH (www.softberry.com) and GETORF (http://emboss.sourceforge.net). Only sequences that encoding ≥100 aminoacids were considered. Secreted proteins were identified using SignalP v3.0 (score cutt-off >0.9), TargetP and PSort (www.psort.org) [89]. Proteins were considered as secreted if two out of three programs called the Signal Peptide as significant. HaRxLs were selected as fulfilling the following criteria: i) Signal Peptide (SP) length <30 amino acids, ii) RxLR-like motif (RxLR/Q, RxL) between 4 and 60 amino acids from the SP cleavage site, iii) predicted protein had >40 amino acids after the RxLR like motif. Redundancy in the ORF dataset was corrected using BlastP. Sequences with 100% identity and E<10−5 were clustered and simplified to the one with highest SP score. A sub-set of RxLR-EE proteins was identified carrying an acidic motif (EE, EER/G/D) [28] between 4 and 30 aminoacids from the RxLR like motif. The expression of HaRxLs was verified using the following resources: i) ESTs generated by Sanger sequencing and 454 sequencing from cDNA extracted from Emoy2 conidiospores [6], ii) Illumina sequence tags (SAGE using 3′ tags from 7 dpi infected tissue), iii) Illumina normalized/concatamerized cDNA (from 3 and 7 dpi infected tissue) (Ishaque et al., unpublished) and iv) RT-PCR with primers designed at 100 bp flanking the ORF sequence (Baxter et al., unpublished). The presence of the predicted and alternative ATGs and Stops Codons, as well as introns, was verified. Nucleotidic sequence polymorphisms on the HaRxLs accross seven Hpa isolates was assessed using either PCR products or in silico assemblies of Illumina short reads (Ishaque et al., unpublished). The HaRxLs were roughly classified as: No polymorphic (0 SNPs), Low (≥1 SNPs ≤5), Medium (≥6 SNPS ≤15) and High Polymorphic (>16 SNPs). To complete the characterization of the Hpa Emoy2 HaRxLs set, its sub-cellular localization (PSORTII) and presence of known conserved protein domains using Coil, Gene3D, HMMPfam, HMMSmart, HMMTigr, PFAM and Prosite was recorded. These data are available in Table S1 for the subset of HaRxLs cloned in this work.
Cloning of HaRxLs
Selected HaRxLs were amplified from genomic DNA extracted from conidiospores of isolate Emoy2 using proofreading polymerase (Accuprime Pfx, Invitrogen) and standard PCR conditions. To generate the HaRxLs collection, the primers were designed to amplify from the signal peptide cleavage site or the RxLR (inclusive) until after the stop codon (3′ untranslated region, UTR). For cloning in pEDV3 or pEDV5 [37], the primers were designed to have SalI/ClaI and BamHI/BglII restriction sites at the 5′ and 3′ ends respectively. For cloning in pEDV6, a Gateway destination version of pEDV3, the sequence CACC was added at the 5′ of the Forward primer. PCR products were gel purified (Qiagen) and ligated (EDV3/5) or recombined in pENTRY-SD-D-TOPO/pDONR221 following the manufacturer's instructions and electroporated in Escherichia coli DH5∝. Gentamycin (EDV3/5) or kanamycin (pENTR/pDONR) resistant colonies were selected on plates and colony PCR performed with M13F and M13R primers. Colonies carrying the right size insert were selected for plasmid purification and sequencing. For EDV6, the correct inserts on pENTRY/pDONR vectors were recombined using Gateway LR clonase or LR clonase II enzyme mix (Invitrogen). The in frame fusion of vector-HA tag-HaRxL sequences were confirmed by sequencing with M13F and M13R primers. Plasmids were mobilized from E.coli DH5∝ to wild-type or mutant Pst strains by standard triparental matings using E. coli HB101 (pRK2013) as a helper strain. Bacterial growth in vitro was controlled at 12, 24, 32 and 44 hs post inoculation of 10 ml Kings B media with a dilution corresponding to 0.00001 OD of an overnight culture of each of the Pst-LUX clones harbouring a different HaRxL or control proteins (GFP, AvrRPS4AAAA). Three colonies per clone were assayed in different experiments. Growth was measured assessing turbidity at OD600 (for liquid cultures) or counting colonies of plated dilutions (in solid media). No significant differences in growth kinetics were observed for the Pst-LUX carrying HaRxLs regarding the clones carrying control proteins or the empty vector pEDV5.
Bacterial strains
Bacterial strains used in this study include E. coli DH5∝, Pseudomonas syringae pv tomato DC3000 carrying the luxCDABE operon from Photorhabdus luminescens (Pst-LUX) [41], Pseudomonas syringae pv tomato DC3000 mutant ΔCEL [90], Pseudomonas syringae pv tomato DC3000 double mutant ΔavrPto/ΔavrPtoB [91], Pseudomonas fluorescens Pf0-1 carrying a functional TTSS [57] and Agrobacterium tumefasciens GV3101 (pMP90 RK). E. coli, and Agrobacterium were grown in low salt Luria-Bertani broth at 37°C (E. coli) or 28°C (Agro) using either liquid media or petri dishes. Pseudomonas strains were grown in either LB or King's B medium at 28°C in liquid media or petri dishes. Antibiotics concentrations (µg/ml) were as follows: Rifampicin 100, Kanamycin 50, Gentamycin 25, Spectinomycin 50, Chloramphenicol 50, Tetracycline 10, Carbenicillin 50.
Plant materials and growth
Arabidopsis accessions used in this study were obtained from NASC. The fls2-1 mutant was obtained from Cyril Zipfel and the cerk1-1 mutant was a kind gift of JP Rathjen. Transgenic lines constitutively expressing HopU1 and HopAO1 were kindly provided by Jim Alfano. Turnip seeds (Brassica rapa cv Just Right) were purchased from Gurney's seeds (http://gurneys.com). Tobacco (Nicotiana tabacum cv petit Havana) and tomato (Solanum lycopersicum cv Moneymaker) seeds were obtained from John Innes Horticultural services. Arabidopsis plants were grown in Scotts and Levington F1 modular compost in controlled environment rooms under short day cycles (10 h/14 h day/night and 150–200 µE/m2s) at 22°C and 60% relative humidity and slightly watered every day from below. Tobacco, tomato and turnip plants were grown under similar conditions as Arabidopsis for 5 weeks post-germination. Plants expressing constitutively HaRxLs were generated by recombining the corresponding ORFs cloned in pDONR221 in the Gateway destination binary vector pB2GW7 [92] under the control of the CaMV 35S promoter. Constructs were transferred to A. tumefaciens strain GV3101 (pMP90 RK) [93] and transformed into Arabidopsis accession Col-0 by the floral dipping method. Primary transformants (T1) were selected on soil containing BASTA (Bayer CropScience, Wolfenbüttel, Germany) and self-pollinated. The progeny of the T2 generation was observed and 3∶1 (BASTA-resistant/BASTA-susceptible) segregating lines were taken further. Homozygous lines were selected by examining the BASTA resistance of T3 seedlings. Three independent transgenic lines per HaRxL (T4s) were analysed and for simplicity we present results for two.
Pathogen growth and inoculations
Primary streaks of Pst-LUX complemented with the controls or HaRxLs were made from isolated colonies onto selective King's B plates and grown overnight at room temperature. Selected individual colonies were then spread with a sterile loop in solid KB plates and incubated overnight at room temperature to produce even bacterial lawns. Cells were scraped from plates with a sterile loop and suspended in 50 to 100 ml of 10 mM MgCl2 to a final OD600 of 1. Dilution series were made from these suspensions to: spray (OD600: 0.2) or infiltrate (OD600 = 0.001) Arabidopsis plants, or to infiltrate tobacco (OD600 = 0.01), tomato (OD600 = 0.001) or turnip (OD600 = 0.001) plants. For tobacco and turnip, leaf panels of the third- to fifth-oldest leaves of were infiltrated by pricking the leaves with a dissecting needle and infiltrating with a blunt syringe. pEDV-HaRxLs were compared with controls on the same leaf. For tomato, leaflets of the third and fourth most recently expanded leaves were used. Concentrations of other bacterial pathogens used in this work are stated on the corresponding figure legends or other sections of M and M. Hpa isolates Emoy2 and Noco2 were maintained in compatible host accessions and inoculated onto 2-week-old plants at 1 or 5×104 conidiospores/ml. After infection, plants were covered with a transparent lid to maintain high humidity (90–100%) conditions in a growth chamber at 16°C for 7 days in short day (10 h/14 h day/night) cycle. To increase the ratio pathogen/host biomass for gene expression analysis, plants were sprayed with the conidiospores solution (or water as control) were kept uncovered in low humidity (60%).
EDV-Pst LUX screening method
Arabidopsis plants of all 12 different accessions were grown in arrays of five individual cells and shuffled randomly before inoculation. Five four-week-old plants of each accession were sprayed with Pst-LUX bacterial suspensions (2×108 cfu/ml, 0.03% (v/v) Silwet L-77) carrying pEDVs encoding for control or HaRxL proteins. Spraying was done using an airbursh system attached to a compressor (GS, model AS18). About 3 mls of bacterial suspension was used per five plants at a pressure of 10–12 psi. Sprayed plants were kept under a transparent lid to keep high humidity conditions. At three days post-spraying, sets of whole five plants were placed in an ultra low light CCD camera (Photek, www.photek.com). Photons emitted per second were scored per plant and referred to the whole plant fresh weight to account for the foliar area. The average level of Photon Counts per Second per gram of Fresh Weight was obtained for each Pst-LUX EDV-delivered protein and the ratio versus the control was scored on each accession (Figure 2). To verify the correspondence between increase on photon emission and bacterial population growth, leaf discs were sampled from the five plants sprayed to generate at least 6 technical replicates. The leaves were surface sterilized (30 s in 70% ethanol, then 30 s sterile distilled water). Traditional growth curve assays were performed as described [94].
Gene expression analysis
Total RNA was extracted from three-week-old HaRxL overexpressing lines (T4 generation) using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). 1 µg DNAse-treated total RNA was reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen). For semi-quantitative RT-PCR 1 µl of cDNA was used per reaction and amplified with an initial denaturation step at 95°C for 3 minutes followed by 23 cycles with the following conditions: 45 s at 94°C, 30 s at 55°C and 30 s at 72°C. In a last cycle a final elongation step at 72°C for 5 min was added. PCR products were separated on 1.5% TAE agarose gels. To control the equal amount of cDNA in every reaction the Actin2 gene (At3g18780) was used. The specificity of the primers for amplification of the HaRxLs transcripts was tested on pDONR221 clones harboring the corresponding HaRxLs. In case of control RT-PCRs on Arabidopsis Col-0 wild type plants cDNA, no unspecific amplification using the HaRxLs' primers was observed. The sequences of primers used in this study are available on request.
Measurement of Reactive Oxygen Species (ROS) generation
ROS released by the leaf tissue in response to flg22 was measured using a chemiluminescent assay [95]. Briefly, leaf discs (0.38 cm2) from Col-0, fls2-1 mutant, transgenic lines expressing a given 35S-HaRxL lines, and control lines were sampled. At least 24 leaf discs from four five-week-old plants per accession/line were sampled and floated for 14–16 h in sterile distilled water in a 96 flat-bootom white multiwell plate (Greiner Bio-One) kept in the dark. ROS production in response to flg22 was measured by replacing the water by 100 ul of a working solution of Luminol (34 µg/ml final concentration, Sigma), Horseradish Peroxidase (20 µg/ml final concentration, Sigma) and flg22 (100 nM final concentration, Peptron, South Korea). The plate was immediately introduced in a Luminometer (Varioskan, Thermo Scientific) and photon counts were recorded every 40 seconds for at least 40 minutes.
Callose and Trypan Blue Stainings and microscopic analysis
Leaves of five-week-old plants were hand-infiltrated at the bottom of the leaf area with 5×107 cfu/ml (OD600 = 0.05) Pst-ΔCEL suspensions. Bacteria complemented with control or HaRxL proteins were infiltrated in different plants of the same set. Twenty leaf samples were taken 12 to 14 h after inoculation at the top of the infiltrated area to avoid visualization of mechanical damage induced callose. Leaf discs were cleared 2 times (1 h) in 96% ethanol and then re-hydrated in ethanol/water series (70%, 50%, 30%) per 30 min each. Leaf disc were floated in water 1 h and then transferred to a solution of 0.01% (w/v) Aniline Blue (Gurr, BHD, England) in K2HPO4 Buffer (150 mM pH 9.5) for 1 h. The samples were then transferred to Glycerol 60% (v/v) and mounted for observation with a Leica DM R fluorescence microscope using UV light and I3 filter (A4-UV). Pictures were taken with a Leica DFC 300 FX Digital Camera. Images were analyzed and callose dots quantified using Image J software and an in house written Macro. The same imaging system was used to visualize Hpa infection structures after staining with lactophenol trypan blue. The number of conidiophores per leaf was scored by manually scanning the abaxial and adaxial surfaces of each leaf on at least four true leaves of five-ten plants per accession/transgenic line analyzed.
Ion leakage measurements
Five-week-old plants were infiltrated with 1×108 cfu/ml (OD600 = 0.1) P. fluorescens Pf0-1 carrying pVSP61-AvrRPM1. To be able to detect HR symptoms the bacteria had to be grown in plates. Leaf discs were collected right after infiltration using a cork borer number 4. Twenty-four leaf discs were collected per transgenic line and shacked in falcon tubes with 45 ml distilled water for 1 h at room temperature. Four leaf discs were transferred multi-well plates with 2 ml distilled water. The level of ion leakage caused by AvrRPM1 recognition by RPM1 in the Col-0 background was detected as an increment in conductivity. This was measured with a Conductivity meter (Horiba Twin B-173) every 60 min over 14 h on at least 6 technical replicates per transgenic line.
Secretion assay and protein analysis
To verify the accumulation proteins corresponding to the AvrRPS4-effector fusions in Pseudomonas and its secretion by the TTSS, the strains complemented with Hpa HaRxLs were grown overnight at 28°C in 25 ml liquid LB media to a final OD600 = 0.3, centrifuged, washed twice with 10 mM MgCl2 and re-suspended in 20 ml minimal media (hrp-inducing minimal medium, MM) [50 mM potassium phosphate buffer, 7.6 mM (NH4) 2SO4, 1.7 mM MgCl2, 1.7 mM NaCl, pH 6.0, with 20 mM glucose added]. The cultures were incubated at 22°C at 200 rpm on a rotator shaker to a final OD600 of 0.5. The pellet and supernatant fractions were separated by centrifugation at 5,200× g for 15 min at 4°C. The pellets were re-suspended in 300 ul of bacterial protein extraction buffer [100 mM NaCl, 25 mM Tris-HCl pH 8, 10% glycerol, 10 mM DTT and 1× protease inhibitors cocktail (Complete EDTA-free tablets, Roche)], sonicated 3 times (10 s), and centrifuged at 12.000 rpm for 5 minutes. Then 100 µl of supernatant were taken and 25 µl of SDS loading buffer were added. Samples were boiled for 5 minutes at 96°C before loading. The supernatant of the 20 ml MM culture was filtered through a 0.2 µm pore filter and concentrated using centricon YM-10 columns (Amicon Bioseparation) by sequential centrifugations of 30 min at 5000 g at 4°C. The process was repeated several times. Proteins in the final 2 ml of concentrated supernatant were precipitated using Strataclean beads (Stratagene). Five to ten µl of beads were used per ml of supernatant; incubated 10 min at 4°C inverting gently, centrifuged at 2000 g for 2 min and re-suspended in 25 ul Laemmli buffer containing 0.1 M NaOH. Samples were boiled for 5 minutes at 96°C and centrifuged at 12.000 rpm for 2 minutes before loading. The pellet fraction (15 µl) and the culture fluid fraction (25 µl) were analyzed by SDS–PAGE, electro blotted onto PVDF membrane (Bio-Rad), and probed with horseradish peroxidase- conjugated high affinity anti–HA antibody (Roche) and re-probed with anti-NPTII antibody (Upstate). Bands were visualized using PICO kit (Thermo Scientific) and imaged with Kodak scientific imaging film.
Supporting Information
Verification of the functionality of the TTSS of Pst -LUX clones expressing HaRxLs. (A) Immunoblot showing the accumulation of AvrRPS4 1–136-HA-HaRxL fusions on the bacterial pellet (left panel) and its secretion into TTSS-inducing minimal media (right panel, supernatant). Approximated molecular weights in KDa are shown on the left of each panel. (B) Four-week old tobacco (N. tabacum cv. petit havana) leaves were infiltrated with Pst-LUX clones carrying either control proteins (YFP, AvrRPS4-AAAA), or HaRxLs at OD600 = 0.01. Pst-hrcC and Pst-hrcC-LUX strains were included as positive controls for TTSS impairment. Symptoms of HR cell death were screened at 2,3 and 4 dpi. Picture was taken at 2 dpi. (C) Five-week old tomato (Solanum lycopersicum cv. moneymaker) leaflets syringe infiltrated with OD600 = 0.001 of different Pst-LUX clones expressing HaRxLs or controls (YFP). Leaflets were detached and imaged at 3 dpi with a Photek camera to detect bioluminescence.
(TIF)
Correlation between Pst -LUX bioluminescence and its growth in planta . (A), (C) Five-week-old plants of the indicated Arabidopsis accessions were spray-inoculated at OD600 = 0.2 with Pst-LUX delivering the indicated Hpa effector or control proteins. At 3 dpi, five whole plants per treatment were imaged using a Photek camera to record photons counts per second. Bars illustrate the average photon counts per gram of plant fresh ± SD. (a) p value of T-test assuming unequal variances <0.05, (b) p<0.01. (B), (D) Twenty-four leaf discs obtained from the above mentioned plants were excised and used to determine the number of bacteria per leaf area, showed in Log10 scale. Bars indicate the average ± SD of six technical replicates. One-way ANOVA test was applied with (a) p<0.05, (b) p<0.01.
(TIF)
Behavior of Pst- LUX delivering HaRxLs assessed via growth curves. Histograms illustrate the changes in growth levels (measured as colony forming units –CFU-) of Pst-LUX strains delivering the indicated HaRxLs, compared to control strains, on different Arabidopsis accessions.
(TIF)
Pattern of HaRxLs induced changes in Pst -LUX virulence plotted per Arabidopsis accession. Bars indicate the number of HaRxLs that enhanced (green), decreased (red) or did not changed (black) the growth of Pst-LUX on each Arabidopsis accession tested. The outcome of the interaction of the Hpa isolate Emoy2 with each accession is indicated as Susceptible (S) or Resistant (R). Known and predicted ATR/RPP interactions are described. (?) indicate putative/unknown ATR/RPP genes.
(TIF)
Semi-quantitative RT-PCR applied to RNA extracted from the stable transgenic lines generated and tested in this paper. Expression levels of each HaRxL were tested in two independent homozygous transgenic lines (1,2). For comparison, the expression of the Arabidopsis housekeeping gene actin (Act2) is shown.
(TIF)
Arabidopsis Col-0 plants expressing constitutively HaRxLs support enhanced growth of Pst -LUX. Five four-week-old plants of two independent transgenic lines expressing the corresponding Hpa candidate effector and one line per control protein, were sprayed at OD600 = 0.2 with Pst-LUX. At 3 dpi, photon counts per plant were measured, as well as plant's fresh weight. Bars represent means of 5 replicates ±2× Standard Errors (SE). (a) p value of T-test assuming unequal variances <0.05; (b) p<0.01. This experiment was repeated three times with similar results.
(TIF)
Constitutive expression of four different HaRxLs caused a mild reduction on the levels of ion leakage triggered by AvrRPM1 recognition. Five-week-old plants were hand infiltrated with Pf0-1 delivering P. maculicola AvrRPM1 at OD600 = 0.1. Twenty-four leaf discs were sampled from four infiltrated transgenic plants per line per HaRxL in each of five different experiments. At least six technical replicates were done per line. Conductivity was measured in the water were discs were floating, as an indication of ion leakage into the media. Measurements were taken every hour until the peak of ion leakage was detected in the wild type and control lines (around 14 hours post-infiltration). Bars correspond to the average ±2× SE of the maximum level of ion leakage observed for each line expressed as a percentage of the value displayed by Col-0 wild type (100%). Values take into account averaged results of 5 different experiments. (a) p value<0.05 of two tailed Z-test.
(TIF)
Fewer HaRxLs can alter the growth of Pst -LUX in the Hpa -non-host Brassica rapa compared to Arabidopsis . Columns show the distribution of the number of candidate effectors that enhance or decrease Pst-LUX virulence in the non-host B. rapa compared to one accession (Col-0) and the average results of the screening in the set of twelve accessions of the host Arabidopsis.
(TIF)
Names and sequences of Hyaloperonospora arabidopsidis isolate Emoy2 HaRxLs candidates cloned. (a) Nucleotide sequence predicted in genome versions of the Hpa Emoy2 race (V3.0 to V6.0) generated previously to the published one (v8.3.2; Baxter and Tripathy et al., 2010). (b) Translation of the predicted nucleotide sequence in Hpa Emoy Genome v8.3.2. (c) Nucleotidic sequence cloned in pENTRY/pDONR vectors. The clones were amplified from the Signal Peptide cleavage until the stop codon, adding a Methionine (M) at the N-terminal, unless stated otherwise. (d) Sequence cloned in pEDV vectors. When pEDV6 was used, sequences were identical to the pENTRY donors. For clones in pEDV3/5 (non-Gateway versions) no pENTRY clones were generated. (e) Comparison of sequence similarity of the pEDV clone with the predicted nucleotide sequence on the Emoy2 genome (Column G vs Column C). (f) Comparison of sequence similarity of the pEDV clone translated aminoacid sequence with the predicted one the Emoy2 genome (Column H vs Column D). (g) Level of Polymorphism of candidate RxLR effectors predicted by “in silico” comparison of the Hpa Emoy2 race genome (v8.3.2, v6.0, v3.0) with the genomes of seven Hpa races sequenced via illumina short reads. The number of SNPs predicted is indicated between brakets. We classified them as No polymorphic (0 SNPs), Low (≥1 SNPs ≤5), Medium (≥6 SNPS ≤15) and High Polymorphic (>16 SNPs). (h) Level of Expression of candidate RxLRs was tested using different expression libraries, and categorized as: 1- Sanger ESTs obtained from Spores, 2- 454 ESTs generated from infected tissue at 3 days post-inoculation (dpi), 3- Illumina short reads generated from infected tissue at 3 and 7 dpi. (i) Predicted subcellular localization using Wolf-PSORT (of sequences after signal peptide cleavage site). The localization stated corresponds to the ones consistently found using both Plant and Fungi databases. The score numbers representing the number of closest neighbours in each alignment are represented in brakets. (+) Cloned from RxLR onwards, without M. (*) Cloned from RxLR onwards, X or L replaced by M. (°) Cloned from SP cleavage site onwards, with M introduced. ND Not determined. (1) There are two sequences highly similar in Emoy2 genome. The paralog cloned here is not assembled in the published Emoy2 genome (v8.3.2). They differ only in three aminoacids (EGMIW×KNMIR). (2) Putative gene family. The paralog cloned is not the one assembled in V8.3.2 of the genome. (3) The cloned sequence not assembled in v8.3.2. It is highly similar except for the C-terminal end. Putative paralogs. (4) Putative gene family. The paralog cloned is not assembled in v8.3.2 of the genome. (5) There are two paralog sequences in Emoy2 genome. Only HaRxL45 is assembled in v8.3.2. (6) These RxLR candidates are not assembled in the V8.3.2 of the genome, but they were predicted in previous versions of it (v3.0 and v6.0). (7) The sequence of this clone has an internal deletion of 20 aminoacids regarding the sequence predicted in v8.3.2 of the genome. Putative paralog. (8) NC1,2,3,4: Negative controls.
(XLS)
Effect of 64 HaRxLs on Pst -LUX bioluminescence tested across 12 Arabidopsis accessions. (a) Pst-LUX complemented with the corresponding candidate HaRxL or control was infiltrated in leaves of Nicotiana tabacum cv. Petit Havana. Development of hypersensitive response (HR) cell death was evaluated at 2 days post-infiltration. Representative picture on Figure S1B. (b) Pst-LUX complemented with the corresponding candidate HaRxL or control was infiltrated in leaflets of Lycopersicum esculentum cv. Moneymaker. Disease sympthoms were determined between 3 and 5 dpi. Presence of the bacteria on the lessions was confirmed imaging Pst-LUX bioluminescence. Representative picture on Figure S1 C. (c) Values in the body of the table correspond to the ratio of Pst-LUX bioluminescence between a Pst-LUX clone delivering a given HaRxL candidate effector and a Pst-LUX clone delivering a control protein. The bioluminescence counts per second (CPS) per mg of plant fresh weight (FW) were averaged for five plants per Arabidopsis accession per experiment. In summary: Value = [X (CPS/FW) effector/X (CPS/FW) control] (d) Summary of the Hpa candidate effector's effect per ecotype. This conclusion is based on the reproducibility of a given trend of change (2 out of 3 experiments at least) and the statistical significance of the change regarding the control on each individual experiment. T-test p values assuming unequal variances are indicated as follows: >0,01 dark grey boxes, >0,05 light grey boxes. Mathematical symbols indicate increased (+), decreased (−) or no change ( = ) in bioluminescence/FW regarding control. (e) Number of Arabidopsis accessions on wich Pst-LUX bioluminescence activity increased (+), decreased (−) or did not change ( = ) when delivering a given HaRxL candidate effector. (f) Scoring the bioluminescence activity of Pst-LUX delivering a given HaRxL candidate effector in Brassica rapa cv Just Right (Turnip) leaves. Symbols as in (e). (g) Negative controls.
(XLS)
Comparison of photon counts versus colony forming units for Pst -LUX. Results obtained delivering 13 Hpa candidate effectors via EDV by Pst-LUX in different Arabidopsis accessions. Numbers in the body of the table correspond to the ratio between Pst-LUX delivering the indicated effector and the control Pst-LUX clone delivering YFP. For the spray inoculation, the quantization of the bacterial bioluminescence (Photon Counts) and colony forming units (CFUs) is displayed for two to 3 replicate experiments. Experiments displaying the results obtained when the bacteria are introduced in the plant by syringe infiltration are shown for comparison. For the spray inoculation experiments, bacterial growth was scored at 3 dpi using five plants to record Pst-LUX bioluminescence. The same plants were ground to determine bacterial growth by plating in selective media and CFUs. Experiments in parallel were done inoculating the bacteria at OD600: 0.001 by syringe infiltration. Numbers highlighted in bold indicate T-test p value<0.05. (*) Result differs from the one obtained with the EDV screen (see Table S2). (a) Values of photon counts correspond to the ratio of counts per second (CPS)/fresh weight (FW) in grams of Pst-LUX clones delivering via EDV the stated Hpa candidate effectors versus CPS/FW of the YFP or AvrRPS4AAAA control. (b) Values of CFUs correspond to the ratio colony forming units (CFUs)/FW of Pst-LUX clones delivering via EDV the stated Hpa candidate effectors versus CFUs/FW of the YFP control or AvrRPS4AAAA control. (c) The concordance between the Photon counts and CFUs ratios is indicated.
(DOC)
Putatively recognized Hpa candidate effectors and HR-like cell death. The table displays the number of leaves showing HR-like micro-lesions when Trypan blue-stained 24 h post-infiltration with Pst-ΔCEL (OD600 = 0.01) or Pf0-1 (OD600 = 0.1) delivering the indicated Hpa RXLR effector candidates in the corresponding Arabidopsis accessions. Numbers in parenthesis correspond to Pf0-1 data. Twenty-four leaves were infiltrated per candidate per bacterium. This experiment was repeated twice for each vehicle bacterium, with similar results. Values significantly different from negative controls (NC1 or NC2) across replicate experiments are indicated in bold (p value T-test<0.05).
(DOC)
Acknowledgments
We thank the JIC horticulturist team, Matthew Smoker, Avril Soards, Tina Payne and Jaqueline Bautor for technical assistance. We thank Craig Brooks-Rooney, Nicolas Battich, Belen Tejada-Romero, Sharon Hall, Peter Bittner-Eddy and Astrild Vaudaine for contributing with the cloning of HaRxLs. We thank Cyril Zipfel, Sophien Kamoun, Ruslan Yatusevik and Adriana Cruz-Cabral for helpful comments on the manuscript. This paper is dedicated to Tiziano and Francesco Lunello-Fabro and to Federico Lunello.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the following grants: The ERA-PG Effectoromics project funded by the British Biotechnological and Biological Sciences Research Council (BBSRC), the German Research Foundation (Deutsche Forschungsgemeinshaft, JEP/DFG) and the Germany-Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NGI/NOW) to GF, JS, MC, RLA, JEP, GV, JB and JDGJ. The HFSP grant RGP0057/20067-C to DG, JLB and JDGJ; The Gatsby Foundation GAT2545 to SJMP, AR, RL, DJS, EK and GF. The BBSRC grants BB/F0161901, BB/E024882/1 to NI and JDGJ; The BBSRC CASE studentship T12144 to NI and a Marie Curie early stage training program fellowship (019727) to SJMP. The EMBO ALTF 614–2009 and Marie Curie FP7-PEOPLE-2009-IEF funded MCC. The BBSRC grants BB/E024815/1, BB/G015066/1, BB/F005806/1 supported JB. The funders had no role in experimental design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Oliver R, Ipcho S. Arabidopsis pathology breathes new life into the necrotrophs-vs.-biotrophs classification of fungal pathogens. Mol Plant Pathol. 2004;5:347–352. doi: 10.1111/j.1364-3703.2004.00228.x. [DOI] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Zipfel C. Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol. 2009;12:414–420. doi: 10.1016/j.pbi.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324:742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Hoorn RA, Kamoun S. From Guard to Decoy: a new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raffaele S, Farrer RA, Cano LM, Studholme DJ, MacLean D, et al. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science. 2010;330:1540–1543. doi: 10.1126/science.1193070. [DOI] [PubMed] [Google Scholar]
- 7.Hajri A, Brin C, Hunault G, Lardeux F, Lemaire C, et al. A “repertoire for repertoire” hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PLoS One. 2009;4:e6632. doi: 10.1371/journal.pone.0006632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipka U, Fuchs R, Lipka V. Arabidopsis non-host resistance to powdery mildews. Curr Opin Plant Biol. 2008;11:404–411. doi: 10.1016/j.pbi.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330:1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330:1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- 11.Schirawski J, Mannhaupt G, Munch K, Brefort T, Schipper K, et al. Pathogenicity determinants in smut fungi revealed by genome comparison. Science. 2010;330:1546–1548. doi: 10.1126/science.1195330. [DOI] [PubMed] [Google Scholar]
- 12.Oliva R, Win J, Raffaele S, Boutemy L, Bozkurt TO, et al. Recent developments in effector biology of filamentous plant pathogens. Cell Microbiol. 2010;12:1015. doi: 10.1111/j.1462-5822.2010.01484.x. [DOI] [PubMed] [Google Scholar]
- 13.Slusarenko AJ, Schlaich NL. Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol Plant Pathol. 2003;4:159–170. doi: 10.1046/j.1364-3703.2003.00166.x. [DOI] [PubMed] [Google Scholar]
- 14.Holub EB. Natural history of Arabidopsis thaliana and oomycete symbioses. Eur J Plant Pathol. 2008;122:91–109. [Google Scholar]
- 15.Coates ME, Beynon JL. Hyaloperonospora arabidopsidis as a pathogen model. Annu Rev Phytopathol. 2010;48:329–345. doi: 10.1146/annurev-phyto-080508-094422. [DOI] [PubMed] [Google Scholar]
- 16.Torto TA, Li SS, Styer A, Huitema E, Testa A, et al. EST Mining and Functional Expression Assays Identify Extracellular Effector Proteins From the Plant Pathogen Phytophthora. Genome Res. 2003;13:1675–1685. doi: 10.1101/gr.910003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev of Phytopathol. 2006;44:41–60. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- 18.Birch PRJ, Rehmany AP, Pritchard L, Kamoun S, Beynon JL. Trafficking arms: oomycete effectors enter host plant cells. Trends Microbiol. 2006;14:8–11. doi: 10.1016/j.tim.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Qutob D, Kemmerling B, Brunner F, Küfner I, Engelhardt S, et al. Phytotoxicity and Innate Immune Responses Induced by Nep1-Like Proteins. Plant Cell. 2006;12:3721–3744. doi: 10.1105/tpc.106.044180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kemen E, Kemen AC, Rafiqi M, Hempel U, Mendgen K, et al. Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol Plant Microbe In. 2005;18:1130–1139. doi: 10.1094/MPMI-18-1130. [DOI] [PubMed] [Google Scholar]
- 21.Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell. 2006;18:243–256. doi: 10.1105/tpc.105.035980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gout L, Fudal I, Kuhn ML, Blaise F, Eckert M, et al. Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol. 2006;60:67–80. doi: 10.1111/j.1365-2958.2006.05076.x. [DOI] [PubMed] [Google Scholar]
- 23.Ridout CJ, Skamnioti P, Porritt O, Sacristan S, Jones JD, et al. Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell. 2006;18:2402–2414. doi: 10.1105/tpc.106.043307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan W, Cao M, Leung D, Tyler BM. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol Plant Microbe In. 2004;17:394–403. doi: 10.1094/MPMI.2004.17.4.394. [DOI] [PubMed] [Google Scholar]
- 25.Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci U S A. 2005;102:7766–7771. doi: 10.1073/pnas.0500113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen RL, Bittner-Eddy PD, Grenvitte-Briggs LJ, Meitz JC, Rehmany AP, et al. Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science. 2004;306:1957–1960. doi: 10.1126/science.1104022. [DOI] [PubMed] [Google Scholar]
- 27.Rehmany AP, Gordon A, Rose LE, Allen RL, Armstrong MR, et al. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell. 2005;17:1839–1850. doi: 10.1105/tpc.105.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Win J, Morgan W, Bos J, Krasileva KV, Cano LM, et al. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell. 2007;19:2349–2369. doi: 10.1105/tpc.107.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 30.Dou D, Kale SD, Wang X, Jiang RH, Bruce NA, et al. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell. 2008;20:1930–1947. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kale SD, Gu B, Capelluto DG, Dou D, Feldman E, et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 2010;142:284–295. doi: 10.1016/j.cell.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Schornack S, van Damme M, Bozkurt TO, Cano LM, Smoker M, et al. Ancient class of translocated oomycete effectors targets the host nucleus. Proc Natl Acad Sci U S A. 2010;107:17421–17426. doi: 10.1073/pnas.1008491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birch PR, Boevink PC, Gilroy EM, Hein I, Pritchard L, et al. Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr Opin Plant Biol. 2008;11:373–379. doi: 10.1016/j.pbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Panstruga R, Dodds PN. Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science. 2009;324:748–750. doi: 10.1126/science.1171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Judelson HS, Tyler BM, Michelmore RW. Transformation of the oomycete pathogen, Phytophthora infestans. Mol Plant Microbe In. 1991;4:602–607. doi: 10.1094/mpmi-4-602. [DOI] [PubMed] [Google Scholar]
- 36.Huitema E, Smoker M, Kamoun S. A straightforward protocol for electro-transformation of Phytophthora capsici zoospores. Methods Mol Biol. 2010;712:129–135. doi: 10.1007/978-1-61737-998-7_11. [DOI] [PubMed] [Google Scholar]
- 37.Sohn KH, Lei R, Nemri A, Jones JDG. The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell. 2007;19:4077–4090. doi: 10.1105/tpc.107.054262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo M, Tian F, Wamboldt Y, Alfano JR. The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe In. 2009;22:1069–1080. doi: 10.1094/MPMI-22-9-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornelis GR. The type III secretion injectisome, a complex nanomachine for intracellular toxin delivery. Biol Chem. 2010;391:745–751. doi: 10.1515/BC.2010.079. [DOI] [PubMed] [Google Scholar]
- 40.Mudgett MB, Chesnokova O, Dahlbeck D, Clark ET, Rossier O, et al. Molecular signals required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc Natl Acad Sci USA. 2000;97:13324–13329. doi: 10.1073/pnas.230450797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan J, Crooks C, Lamb C. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 2008;53:393–399. doi: 10.1111/j.1365-313X.2007.03303.x. [DOI] [PubMed] [Google Scholar]
- 42.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 43.Gimenez-Ibanez S, Ntoukakis V, Rathjen JP. The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal Behav. 2009;4:539–541. doi: 10.4161/psb.4.6.8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. Embo J. 2009;28:3428–3438. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci U S A. 2009;106:15973–15978. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salmeron JM, Staskawicz BJ. Molecular characterization and h r p dependence of the avirulence gene avrPro from Pseudomonas syringae pv. tomato. Mol Gen Genet. 1993;239:6–16. doi: 10.1007/BF00281595. [DOI] [PubMed] [Google Scholar]
- 47.Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell. 2004;16:2822–2835. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 49.Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 50.Rentel MC, Leonelli L, Dahlbeck D, Zhao B, Staskawicz BJ. Recognition of the Hyaloperonospora parasitica effector ATR13 triggers resistance against oomycete, bacterial, and viral pathogens. Proc Natl Acad Sci USA. 2008;105:1091–1096. doi: 10.1073/pnas.0711215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindeberg M, Stavrinides J, Chang JH, Alfano JR, Collmer A, et al. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol Plant Microbe. 2005;In18:275–282. doi: 10.1094/MPMI-18-0275. [DOI] [PubMed] [Google Scholar]
- 52.Lin NC, Martin GB. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol Plant Microbe In. 2005;18:43–51. doi: 10.1094/MPMI-18-0043. [DOI] [PubMed] [Google Scholar]
- 53.Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, et al. The Pattern of Polymorphism in Arabidopsis thaliana. PLoS Biology. 2005;3:1289–1299. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodman RN, Novacky AJ. The Hypersensitive Response in Plants to Pathogens: A Resistance Phenomenon. St. Paul: APS Press; 1994. [Google Scholar]
- 55.Greenberg JT, Yao N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 2004;6:201–211. doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 56.Yu IC, Parker J, Bent AF. Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA. 1998;95:7819–7824. doi: 10.1073/pnas.95.13.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas WJ, Thireault CA, Kimbrel JA, Chang JH. Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J. 2009;60:919–928. doi: 10.1111/j.1365-313X.2009.03998.x. [DOI] [PubMed] [Google Scholar]
- 58.Donofrio NM, Delaney TP. Abnormal callose response phenotype and hypersusceptibility to Peronospoara parasitica in defence-compromised arabidopsis nim1-1 and salicylate hydroxylase-expressing plants. Mol Plant Microbe In. 2001;14:439–450. doi: 10.1094/MPMI.2001.14.4.439. [DOI] [PubMed] [Google Scholar]
- 59.Dong X, Hong Z, Chatterjee J, Kim S, Verma DP. Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta. 2008;229:87–98. doi: 10.1007/s00425-008-0812-3. [DOI] [PubMed] [Google Scholar]
- 60.Meyer D, Pajonk S, Micali C, O'Connell R, Schulze-Lefert P. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 2009;57:986–999. doi: 10.1111/j.1365-313X.2008.03743.x. [DOI] [PubMed] [Google Scholar]
- 61.Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006;20:537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naito K, Taguchi F, Suzuki T, Inagaki Y, Toyoda K, et al. Amino Acid sequence of bacterial microbe-associated molecular pattern flg22 is required for virulence. Mol Plant Microbe In. 2008;9:1165–1174. doi: 10.1094/MPMI-21-9-1165. [DOI] [PubMed] [Google Scholar]
- 63.Mishina T, Zeier J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic aquired resistance in Arabidopsis. Plant J. 2007;50:500–513. doi: 10.1111/j.1365-313X.2007.03067.x. [DOI] [PubMed] [Google Scholar]
- 64.Gomez-Gomez L, Boller T. Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 2002;7:251–256. doi: 10.1016/s1360-1385(02)02261-6. [DOI] [PubMed] [Google Scholar]
- 65.Gomez-Gomez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999;18:277–284. doi: 10.1046/j.1365-313x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- 66.Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci U S A. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J, Shao F, Li Y, Cui H, Chen L, et al. A Pseudomonas syringae Effector Inactivates MAPKs to Suppress PAMP-Induced Immunity in Plants. Cell Host Microbe. 2007;1:175–185. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 69.Underwood W, Zhang S, He SY. The Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J. 2007;52:658–672. doi: 10.1111/j.1365-313X.2007.03262.x. [DOI] [PubMed] [Google Scholar]
- 70.DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci U S A. 2004;101:9927–9932. doi: 10.1073/pnas.0401601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tor M, Holub EB, Brose E, Musker R, Gunn N, et al. Map positions of three loci in Arabidopsis thaliana associated with isolate-specific recognition of Peronospora parasitica. (downy mildew). Mol Plant Microbe In. 1994;7:214–222. [Google Scholar]
- 72.Rohmer L. Nucleothide sequence, functional characterization and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola. Mol Microbiol. 2003;47:1545–1562. doi: 10.1046/j.1365-2958.2003.03402.x. [DOI] [PubMed] [Google Scholar]
- 73.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 74.Jiang RH, Tripathy S, Govers F, Tyler BM. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci U S A. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 76.Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, et al. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–281. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- 77.de Torres M, Mansfield JW, Grabov N, Brown IR, Ammouneh H, et al. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 2006;47:368–382. doi: 10.1111/j.1365-313X.2006.02798.x. [DOI] [PubMed] [Google Scholar]
- 78.Wilton M, Subramaniam R, Elmore J, Felsensteiner C, Coaker G, et al. The type III effector HopF2(Pto) targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc Natl Acad Sci USA. 2010;107:2349–2354. doi: 10.1073/pnas.0904739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nemri A, Atwell S, Tarone AM, Huang YS, Zhao K, et al. Genome-wide survey of Arabidopsis natural variation in downy mildew resistance using combined association and linkage mapping. Proc Natl Acad Sci USA. 2010;107:10302–10307. doi: 10.1073/pnas.0913160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaulin E, Dramé N, Lafitte C, Torto-Alalibo T, Martinez Y, et al. Cellulose Binding Domains of a Phytophthora Cell Wall Protein Are Novel Pathogen-Associated Molecular Patterns. Plant Cell. 2006;18:1766–1777. doi: 10.1105/tpc.105.038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tor M. Pattern-recognition receptors [PRRs] in plants. Jo Leukocyte Biol. 2008;84:A1–A2. [Google Scholar]
- 82.Kvitko BH, Park DH, Velasquez AC, Wei CF, Russell AB, et al. Deletions in the Repertoire of Pseudomonas syringae pv. tomato DC3000 Type III Secretion Effector Genes Reveal Functional Overlap among Effectors. PLoS Pathog. 2009;5:e1000388. doi: 10.1371/journal.ppat.1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Block A, Li GY, Fu ZQ, Alfano JR. Phytopathogen type III effector weaponry and their plant targets. Curr Op Plant Biol. 2008;11:396–403. doi: 10.1016/j.jbi.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cunnac S, Lindeberg M, Collmer A. Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr Op Microbiol. 2009;12:53–60. doi: 10.1016/j.mib.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 85.Hann DR, Rathjen JP. The long and winding road: virulence effector proteins of plant pathogenic bacteria. Cell Mol Life Sci. 2010;67:3425–3434. doi: 10.1007/s00018-010-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dou D, Kale SD, Wang X, Chen Y, Wang Q, et al. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell. 2008;20:1118–1133. doi: 10.1105/tpc.107.057067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oh SK, Young C, Lee M, Oliva R, Bozkurt TO, et al. In Planta Expression Screens of Phytophthora infestans RXLR Effectors Reveal Diverse Phenotypes, Including Activation of the Solanum bulbocastanum Disease Resistance Protein Rpi-blb2. Plant Cell. 2009;21:2928–2947. doi: 10.1105/tpc.109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bailey K, Cevik V, Holton N, Byrne-Richardson J, Sohn K, et al. Molecular Cloning of ATR5Emoy2 from Hyaloperonospora arabidopsidis, an avirulence determinant that triggers RPP5-mediated defense in Arabidopsis. Mol Plant Microbe In. 2011;24:827–838. doi: 10.1094/MPMI-12-10-0278. [DOI] [PubMed] [Google Scholar]
- 89.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 90.Badel JL, Nomura K, Bandyopadhyay S, Shimizu R, Collmer A, et al. Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol Microbiol. 2003;49:1239–1251. doi: 10.1046/j.1365-2958.2003.03647.x. [DOI] [PubMed] [Google Scholar]
- 91.Lin NC, Martin GB. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol Plant Microbe In. 2005;18:43–51. doi: 10.1094/MPMI-18-0043. [DOI] [PubMed] [Google Scholar]
- 92.Karimi M, Inze D, Depicker A. GATEWAY((TM)) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 93.Koncz C, Schell J. The promoter of the TL -DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- 94.Katagiri F, Thilmony R, He SY. The Arabidopsis Thaliana-Pseudomonas Syringae Interaction. Arabidopsis Book. 2002;1:1–35. doi: 10.1199/tab.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Keppler LD, Baker CJ, Atkinson MM. Active oxygen production during a bacteria-induced hypersensitive reaction in tobacco suspension cells. Phytopathology. 1989;79:974–978. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Verification of the functionality of the TTSS of Pst -LUX clones expressing HaRxLs. (A) Immunoblot showing the accumulation of AvrRPS4 1–136-HA-HaRxL fusions on the bacterial pellet (left panel) and its secretion into TTSS-inducing minimal media (right panel, supernatant). Approximated molecular weights in KDa are shown on the left of each panel. (B) Four-week old tobacco (N. tabacum cv. petit havana) leaves were infiltrated with Pst-LUX clones carrying either control proteins (YFP, AvrRPS4-AAAA), or HaRxLs at OD600 = 0.01. Pst-hrcC and Pst-hrcC-LUX strains were included as positive controls for TTSS impairment. Symptoms of HR cell death were screened at 2,3 and 4 dpi. Picture was taken at 2 dpi. (C) Five-week old tomato (Solanum lycopersicum cv. moneymaker) leaflets syringe infiltrated with OD600 = 0.001 of different Pst-LUX clones expressing HaRxLs or controls (YFP). Leaflets were detached and imaged at 3 dpi with a Photek camera to detect bioluminescence.
(TIF)
Correlation between Pst -LUX bioluminescence and its growth in planta . (A), (C) Five-week-old plants of the indicated Arabidopsis accessions were spray-inoculated at OD600 = 0.2 with Pst-LUX delivering the indicated Hpa effector or control proteins. At 3 dpi, five whole plants per treatment were imaged using a Photek camera to record photons counts per second. Bars illustrate the average photon counts per gram of plant fresh ± SD. (a) p value of T-test assuming unequal variances <0.05, (b) p<0.01. (B), (D) Twenty-four leaf discs obtained from the above mentioned plants were excised and used to determine the number of bacteria per leaf area, showed in Log10 scale. Bars indicate the average ± SD of six technical replicates. One-way ANOVA test was applied with (a) p<0.05, (b) p<0.01.
(TIF)
Behavior of Pst- LUX delivering HaRxLs assessed via growth curves. Histograms illustrate the changes in growth levels (measured as colony forming units –CFU-) of Pst-LUX strains delivering the indicated HaRxLs, compared to control strains, on different Arabidopsis accessions.
(TIF)
Pattern of HaRxLs induced changes in Pst -LUX virulence plotted per Arabidopsis accession. Bars indicate the number of HaRxLs that enhanced (green), decreased (red) or did not changed (black) the growth of Pst-LUX on each Arabidopsis accession tested. The outcome of the interaction of the Hpa isolate Emoy2 with each accession is indicated as Susceptible (S) or Resistant (R). Known and predicted ATR/RPP interactions are described. (?) indicate putative/unknown ATR/RPP genes.
(TIF)
Semi-quantitative RT-PCR applied to RNA extracted from the stable transgenic lines generated and tested in this paper. Expression levels of each HaRxL were tested in two independent homozygous transgenic lines (1,2). For comparison, the expression of the Arabidopsis housekeeping gene actin (Act2) is shown.
(TIF)
Arabidopsis Col-0 plants expressing constitutively HaRxLs support enhanced growth of Pst -LUX. Five four-week-old plants of two independent transgenic lines expressing the corresponding Hpa candidate effector and one line per control protein, were sprayed at OD600 = 0.2 with Pst-LUX. At 3 dpi, photon counts per plant were measured, as well as plant's fresh weight. Bars represent means of 5 replicates ±2× Standard Errors (SE). (a) p value of T-test assuming unequal variances <0.05; (b) p<0.01. This experiment was repeated three times with similar results.
(TIF)
Constitutive expression of four different HaRxLs caused a mild reduction on the levels of ion leakage triggered by AvrRPM1 recognition. Five-week-old plants were hand infiltrated with Pf0-1 delivering P. maculicola AvrRPM1 at OD600 = 0.1. Twenty-four leaf discs were sampled from four infiltrated transgenic plants per line per HaRxL in each of five different experiments. At least six technical replicates were done per line. Conductivity was measured in the water were discs were floating, as an indication of ion leakage into the media. Measurements were taken every hour until the peak of ion leakage was detected in the wild type and control lines (around 14 hours post-infiltration). Bars correspond to the average ±2× SE of the maximum level of ion leakage observed for each line expressed as a percentage of the value displayed by Col-0 wild type (100%). Values take into account averaged results of 5 different experiments. (a) p value<0.05 of two tailed Z-test.
(TIF)
Fewer HaRxLs can alter the growth of Pst -LUX in the Hpa -non-host Brassica rapa compared to Arabidopsis . Columns show the distribution of the number of candidate effectors that enhance or decrease Pst-LUX virulence in the non-host B. rapa compared to one accession (Col-0) and the average results of the screening in the set of twelve accessions of the host Arabidopsis.
(TIF)
Names and sequences of Hyaloperonospora arabidopsidis isolate Emoy2 HaRxLs candidates cloned. (a) Nucleotide sequence predicted in genome versions of the Hpa Emoy2 race (V3.0 to V6.0) generated previously to the published one (v8.3.2; Baxter and Tripathy et al., 2010). (b) Translation of the predicted nucleotide sequence in Hpa Emoy Genome v8.3.2. (c) Nucleotidic sequence cloned in pENTRY/pDONR vectors. The clones were amplified from the Signal Peptide cleavage until the stop codon, adding a Methionine (M) at the N-terminal, unless stated otherwise. (d) Sequence cloned in pEDV vectors. When pEDV6 was used, sequences were identical to the pENTRY donors. For clones in pEDV3/5 (non-Gateway versions) no pENTRY clones were generated. (e) Comparison of sequence similarity of the pEDV clone with the predicted nucleotide sequence on the Emoy2 genome (Column G vs Column C). (f) Comparison of sequence similarity of the pEDV clone translated aminoacid sequence with the predicted one the Emoy2 genome (Column H vs Column D). (g) Level of Polymorphism of candidate RxLR effectors predicted by “in silico” comparison of the Hpa Emoy2 race genome (v8.3.2, v6.0, v3.0) with the genomes of seven Hpa races sequenced via illumina short reads. The number of SNPs predicted is indicated between brakets. We classified them as No polymorphic (0 SNPs), Low (≥1 SNPs ≤5), Medium (≥6 SNPS ≤15) and High Polymorphic (>16 SNPs). (h) Level of Expression of candidate RxLRs was tested using different expression libraries, and categorized as: 1- Sanger ESTs obtained from Spores, 2- 454 ESTs generated from infected tissue at 3 days post-inoculation (dpi), 3- Illumina short reads generated from infected tissue at 3 and 7 dpi. (i) Predicted subcellular localization using Wolf-PSORT (of sequences after signal peptide cleavage site). The localization stated corresponds to the ones consistently found using both Plant and Fungi databases. The score numbers representing the number of closest neighbours in each alignment are represented in brakets. (+) Cloned from RxLR onwards, without M. (*) Cloned from RxLR onwards, X or L replaced by M. (°) Cloned from SP cleavage site onwards, with M introduced. ND Not determined. (1) There are two sequences highly similar in Emoy2 genome. The paralog cloned here is not assembled in the published Emoy2 genome (v8.3.2). They differ only in three aminoacids (EGMIW×KNMIR). (2) Putative gene family. The paralog cloned is not the one assembled in V8.3.2 of the genome. (3) The cloned sequence not assembled in v8.3.2. It is highly similar except for the C-terminal end. Putative paralogs. (4) Putative gene family. The paralog cloned is not assembled in v8.3.2 of the genome. (5) There are two paralog sequences in Emoy2 genome. Only HaRxL45 is assembled in v8.3.2. (6) These RxLR candidates are not assembled in the V8.3.2 of the genome, but they were predicted in previous versions of it (v3.0 and v6.0). (7) The sequence of this clone has an internal deletion of 20 aminoacids regarding the sequence predicted in v8.3.2 of the genome. Putative paralog. (8) NC1,2,3,4: Negative controls.
(XLS)
Effect of 64 HaRxLs on Pst -LUX bioluminescence tested across 12 Arabidopsis accessions. (a) Pst-LUX complemented with the corresponding candidate HaRxL or control was infiltrated in leaves of Nicotiana tabacum cv. Petit Havana. Development of hypersensitive response (HR) cell death was evaluated at 2 days post-infiltration. Representative picture on Figure S1B. (b) Pst-LUX complemented with the corresponding candidate HaRxL or control was infiltrated in leaflets of Lycopersicum esculentum cv. Moneymaker. Disease sympthoms were determined between 3 and 5 dpi. Presence of the bacteria on the lessions was confirmed imaging Pst-LUX bioluminescence. Representative picture on Figure S1 C. (c) Values in the body of the table correspond to the ratio of Pst-LUX bioluminescence between a Pst-LUX clone delivering a given HaRxL candidate effector and a Pst-LUX clone delivering a control protein. The bioluminescence counts per second (CPS) per mg of plant fresh weight (FW) were averaged for five plants per Arabidopsis accession per experiment. In summary: Value = [X (CPS/FW) effector/X (CPS/FW) control] (d) Summary of the Hpa candidate effector's effect per ecotype. This conclusion is based on the reproducibility of a given trend of change (2 out of 3 experiments at least) and the statistical significance of the change regarding the control on each individual experiment. T-test p values assuming unequal variances are indicated as follows: >0,01 dark grey boxes, >0,05 light grey boxes. Mathematical symbols indicate increased (+), decreased (−) or no change ( = ) in bioluminescence/FW regarding control. (e) Number of Arabidopsis accessions on wich Pst-LUX bioluminescence activity increased (+), decreased (−) or did not change ( = ) when delivering a given HaRxL candidate effector. (f) Scoring the bioluminescence activity of Pst-LUX delivering a given HaRxL candidate effector in Brassica rapa cv Just Right (Turnip) leaves. Symbols as in (e). (g) Negative controls.
(XLS)
Comparison of photon counts versus colony forming units for Pst -LUX. Results obtained delivering 13 Hpa candidate effectors via EDV by Pst-LUX in different Arabidopsis accessions. Numbers in the body of the table correspond to the ratio between Pst-LUX delivering the indicated effector and the control Pst-LUX clone delivering YFP. For the spray inoculation, the quantization of the bacterial bioluminescence (Photon Counts) and colony forming units (CFUs) is displayed for two to 3 replicate experiments. Experiments displaying the results obtained when the bacteria are introduced in the plant by syringe infiltration are shown for comparison. For the spray inoculation experiments, bacterial growth was scored at 3 dpi using five plants to record Pst-LUX bioluminescence. The same plants were ground to determine bacterial growth by plating in selective media and CFUs. Experiments in parallel were done inoculating the bacteria at OD600: 0.001 by syringe infiltration. Numbers highlighted in bold indicate T-test p value<0.05. (*) Result differs from the one obtained with the EDV screen (see Table S2). (a) Values of photon counts correspond to the ratio of counts per second (CPS)/fresh weight (FW) in grams of Pst-LUX clones delivering via EDV the stated Hpa candidate effectors versus CPS/FW of the YFP or AvrRPS4AAAA control. (b) Values of CFUs correspond to the ratio colony forming units (CFUs)/FW of Pst-LUX clones delivering via EDV the stated Hpa candidate effectors versus CFUs/FW of the YFP control or AvrRPS4AAAA control. (c) The concordance between the Photon counts and CFUs ratios is indicated.
(DOC)
Putatively recognized Hpa candidate effectors and HR-like cell death. The table displays the number of leaves showing HR-like micro-lesions when Trypan blue-stained 24 h post-infiltration with Pst-ΔCEL (OD600 = 0.01) or Pf0-1 (OD600 = 0.1) delivering the indicated Hpa RXLR effector candidates in the corresponding Arabidopsis accessions. Numbers in parenthesis correspond to Pf0-1 data. Twenty-four leaves were infiltrated per candidate per bacterium. This experiment was repeated twice for each vehicle bacterium, with similar results. Values significantly different from negative controls (NC1 or NC2) across replicate experiments are indicated in bold (p value T-test<0.05).
(DOC)