Abstract
Manganese superoxide dismutase (MnSOD) is a nuclear encoded and mitochondrial matrix localized redox enzyme that is known to regulate cellular redox environment. Cellular redox environment changes during transitions between quiescent and proliferative cycles. Human MnSOD has two poly(A) sites resulting in two transcripts: 1.5 and 4.2 kb. The present study investigates if the 3'-untranslated region (UTR) of MnSOD regulates its expression during transitions between quiescent and proliferating cycles, and in response to radiation. A preferential increase in the 1.5 kb MnSOD transcript levels was observed in quiescent cells, while the abundance of the longer transcript showed a direct correlation with the percentage of S-phase cells. Log transformed expression ratio of the longer to shorter transcript was also higher in proliferating normal and cancer cells. Deletion and reporter assays showed a significant decrease in reporter activity in constructs carrying multiple AU-rich sequence that are present in the 3'-UTR of the longer MnSOD transcript. Overexpression of the MnSOD 3'-UTR representing the longer transcript enhanced endogenous MnSOD mRNA levels, which was associated with an increase in MnSOD protein levels and a decrease in the percentage of S-phase cells. Irradiation increases the mRNA levels of the 1.5 kb MnSOD transcript, which was consistent with a significant increase in reporter activity of the construct carrying the 3'-UTR of the shorter transcript. We conclude that the 3'-UTR of MnSOD regulates MnSOD expression in response to different growth states and radiation.
Keywords: MnSOD, 3'-Untranslated region, radiation, polyadenylation, quiescence
Introduction
Manganese superoxide dismutase (MnSOD) is a nuclear encoded and mitochondrial matrix localized redox enzyme that converts mitochondrial generated superoxide to hydrogen peroxide (McCord and Fridovich, 1969). Human MnSOD has two poly(A) sites resulting in two transcripts with the same open reading frame: 1.5 and 4.2 kb. The majority of research is focused on transcriptional control of MnSOD expression, while very little is known about the post-transcriptional mechanisms regulating MnSOD expression (Clerch, 2000; St Clair and Oberley, 1991; Wan et al., 1994; Xu et al., 1999).
Post-transcriptional regulation of mRNAs is known to be governed by the non-coding sequence present in the 5'- and 3'-ends of mRNAs (Chen and Shyu, 1995; Goswami et al., 2000; Guo et al., 1994; Mazumder et al., 2003; Rouault et al., 1988; Shaw and Kamen, 1986). A computational analysis suggests that the average length of the 3'-untranslated region (UTR) of mRNAs in humans is approximately four times longer than the average length of the 5'-UTR (Mazumder et al., 2003). It is worth noting that the length of the 5'-UTR remains relatively constant from fungi to plants and from invertebrates to vertebrates, while the length of the 3'-UTR increases during evolution and with species complexity. This ‘evolutionary expansion’ supports the hypothesis that the 3'-UTRs regulate mRNA levels and translation (Mazumder et al., 2003).
The 3'-UTR harbors not only the polyadenylation signal (PAS) for termination of transcription, but also cis-acting elements like the AU-rich elements (AREs) and microRNA binding sites (Chen and Shyu, 1995; Chivukula and Mendell, 2008; Lewis et al., 2005; Wang and Blelloch, 2009). More than half of human genes are known to have multiple PAS (Edwalds-Gilbert et al., 1997; Lutz, 2008; Tian et al., 2005) resulting in multiple transcripts. The presence of multiple transcripts has often been suggested as a possible mechanism of evolution for phenotypic complexity in mammals (Edwalds-Gilbert et al., 1997). Recent studies have identified a preferential accumulation of transcripts correlating with cellular transformation status. Cancer cells express higher levels of oncogenes that were associated with an increase in the abundance of the transcripts carrying the shorter 3'-UTRs compared to non-transformed cells (Mayr and Bartel, 2009; Sandberg et al., 2008). The mRNA levels of oncogenes carrying the shorter 3'-UTRs correlated with a higher abundance of their corresponding protein levels. These previous reports suggest that transcript selection could be a major regulatory pathway influencing the mRNA levels of growth-promoting genes.
MnSOD activity negatively regulates cellular proliferation (Oberley, 2001; Oberley et al., 1995; Sarsour et al., 2005; Sarsour et al., 2008). The present study investigates if a preferential accumulation of MnSOD transcript is associated with differences in cellular growth states and following treatment with ionizing radiation. Cells in quiescent state and growth-arrest induced by ionizing radiation exhibited an increase in the 1.5 kb MnSOD transcript, which correlated with a higher level of MnSOD protein and activity. A direct correlation was observed between the 4.2 kb MnSOD mRNA levels and percentage of S-phase cells. Results from a luciferase reporter assay exhibited a significant decrease in reporter activity in cells transfected with plasmid DNAs carrying the longer 3'-UTR of MnSOD. These results suggest that a preferential selection of the longer MnSOD transcript and subsequently a decrease in MnSOD protein levels and activity could shift the cellular redox environment towards a more oxidizing state favoring proliferation.
Results
Preferential selection of MnSOD transcripts in proliferating human normal and cancer cells
MnSOD protein levels and activity are higher in quiescence (G0) compared to proliferation (Oberley et al., 1989; Oberley et al., 1995; Sarsour et al., 2005; Sarsour et al., 2008). Human MnSOD has two transcripts 1.5 and 4.2 kb carrying the same open reading frame (Figure 1A). The significance of these two transcripts during transitions between quiescence and proliferation is currently unknown. To determine whether growth-state related changes in MnSOD protein levels and activity could be due to a preferential selection of MnSOD transcripts, a quantitative RT-PCR assay was applied to measure MnSOD mRNA levels in quiescent and proliferating normal human fibroblasts (NHFs). Contact inhibited NHFs were replated at a lower cell density and total cellular RNA isolated at different times post-plating. Cells from duplicate plates were analyzed for cell cycle phase distributions by performing flow cytometry measurements of DNA content. MnSOD mRNA levels were significantly higher (3-fold) in cells with a lower percentage of S-phase (Figure 1B). MnSOD mRNA levels decreased as the percentage of S-phase cells increased. Next, we determined if this inverse correlation between MnSOD mRNA levels and the percentage of S-phase cells could be due to a preferential selection of the two transcripts. The quantitative RT-PCR assay was repeated using primer-pairs designed to distinguish between the two MnSOD transcripts (Figure 1A). The abundance of the 4.2 kb MnSOD transcript was evaluated by designing primer-pairs specific to the 3'-UTR of the longer transcript, while an anchored oligo-(dT)15 reverse primer to the proximal PAS was included in the PCR reactions designed to measure the abundance of the 1.5 kb transcript. These results showed a direct correlation between the 4.2 kb MnSOD transcript levels and the percentage of S-phase cells (R2=0.84). Quiescent cells exhibited an increase in the mRNA levels of the shorter MnSOD transcript (Figure 1C).
Figure 1.

A direct correlation between the abundance of the 4.2 kb MnSOD transcript and percentage of S-phase cells. A. An illustration of the MnSOD transcript: CDS, coding sequence; PAS, polyadenylation signal; ARE: AU-rich sequence; arrows indicate primer sequence used in the quantitative PCR assay. B. Normal human fibroblasts (NHFs) were cultured for different days and harvested for flow cytometry analysis of the percentage of S-phase cells. Total cellular RNA isolated from duplicate dishes were analyzed for total MnSOD mRNA levels using a quantitative RT-PCR assay and primers representing the coding sequence. Fold change was calculated by first normalizing to 18S RNA levels and then relative to MnSOD mRNA levels in cells with 30 percent S-phase. Asterisks represent statistical significance compared to cells with 30 percent S-phase; n=3, p<0.05. C. Quantitative RT-PCR assay was repeated using primers designed to distinguish the mRNA levels of the two MnSOD transcripts (see materials and methods section).
The observation of the S-phase-dependent selection of the 4.2 kb MnSOD transcript was also evident from results obtained from a semi-quantitative RT-PCR assay (Figure 2A). Total cellular RNA isolated from quiescent and exponential cultures of MCF-10A human non-malignant mammary epithelial cells were reverse transcribed and PCR amplified simultaneously to measure mRNA levels of MnSOD and GAPDH. MnSOD mRNA levels were higher in quiescent compared to exponential cultures. These results were comparable to results obtained from a quantitative RT-PCR assay (Figure 2B). It is interesting to note that the distributions of the individual MnSOD transcripts was significantly different in quiescent compared to proliferating cells. The abundance of the 1.5 kb MnSOD transcript was significantly higher in quiescent cells, while the abundance of the 4.2 MnSOD transcript was higher in exponential cultures. The higher abundance of the longer MnSOD transcript in proliferating cells was associated with lower MnSOD protein levels (Figure 2C). Cells with lower percentage of S-phase exhibited higher MnSOD protein levels coinciding with an increase in the 1.5 kb MnSOD transcript levels. In contrast, an increase in the percentage of S-phase cells was associated with an increase in the 4.2 kb MnSOD transcript levels, and a decrease in MnSOD protein levels. The preferential selection of MnSOD transcripts and abundance of MnSOD protein levels were associated with a higher MnSOD activity during quiescence (lower S-phase) compared to proliferation (higher S-phase) (Figure 2D).
Figure 2.

An inverse correlation between MnSOD activity and percentage of S-phase cells. A. A semi-quantitative RT-PCR assay was used to measure MnSOD and GAPDH mRNA levels in exponential and quiescent cultures of MCF-10A human non-malignant mammary epithelial cells. Primers were designed from the coding sequence to measure total MnSOD mRNA levels (top panel), while the abundance of the two transcripts were assessed by designing primers specific to the 3'-UTR. The primer pairs for the 4.2 kb MnSOD transcript were designed from the sequence between the two PAS sites. The reverse primer for the 1.5 kb MnSOD transcript was designed to anchor the first PAS. PCR-amplified products were separated by agarose gel electrophoresis and visualized by staining with ethidium bromide. B. MnSOD mRNA levels in the same cDNA pools were further analyzed by using a quantitative RT-PCR assay using primers representing the coding sequence. C. Immunoblot analysis of MnSOD in cells representative of different percentage of S-phase. Blots were scanned and quantitated using AlphaImager 2000 and ImageJ software. Fold change calculated first by normalizing to actin levels in individual samples and then relative to MnSOD protein levels in cells with 15 percent S-phase. D. Total cellular proteins were used for biochemical measurements of MnSOD activity.
To determine the generality of this phenomenon, the abundance of the two MnSOD transcripts was measured in exponential cultures of human oral squamous (Cal27, SQ20B, FaDu), lung (A549, H292), and breast (MB-231, Sum159) cancer cells (Table 1). Expression ratio of the longer (4.2 kb) to shorter (1.5 kb) transcript was calculated following the method described by Mayr and Bartel et al. (Mayr and Bartel, 2009). These results showed that the abundance of the 4.2 kb MnSOD transcript was higher in exponential cultures of non-malignant and malignant cells (Table I). These results (Figures 1 and 2, and Table I) suggest that the higher abundance of the 4.2 kb MnSOD transcript is a general phenomenon associated with cellular proliferation (percent S-phase) rather than the transformation status of cells.
Table 1.
Expression ratio of the longer to shorter MnSOD transcripts in malignant and non-malignant cells
| Tissue types | Cell lines | Log2 (4.2 kb/1.5 kb) |
|---|---|---|
| Malignant | ||
| Head and Neck | Cal 27 | 1.2 |
| Head and Neck | SQ20B | 1.1 |
| Head and Neck | FaDu | 2.3 |
| Lung | A549 | 1.3 |
| Lung | H292 | 1.5 |
| Breast | Sum159 | 1.4 |
| Breast | MDA-MB-231 | 1.2 |
| Non Malignant | ||
| Breast | MCF 10A | 1.3 |
| Fibroblast | NHF | 1.2 |
MnSOD 3'-UTR regulates luciferase activity in a reporter assay
A dual-luciferase assay was used to determine if the 3'-UTR of MnSOD regulates its mRNA and protein levels. 3'-UTR sequence representing the 1.5 kb MnSOD transcript and deletion constructs carrying 2 and 5 AU-rich (AREs) sequence of the 4.2 kb MnSOD transcript were cloned downstream of a renilla luciferase gene in psiCHECK-2 plasmid DNA (Promega). Control and transfected cells were harvested at 48 h post-transfection and luciferase activity was measured. Renilla-luciferase activity decreased approximately 40% in cells transfected with plasmid DNA carrying 2 AREs compared to cells transfected with plasmid DNA carrying 3'-UTR from the 1.5 kb MnSOD transcript (Figure 3A). Cells transfected with plasmid DNA carrying 5 AREs showed approximately 60% decrease in renilla luciferase activity.
Figure 3.
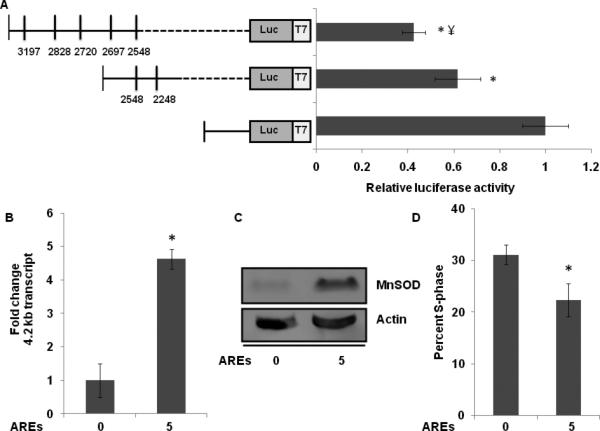
MnSOD 3'-UTR regulates its mRNA levels. A. A RT-PCR assay was used to amplify MnSOD 3'-UTR encompassing 0, 2, and 5 AREs. PCR-amplified cDNAs were directionally cloned into psiCHECK-2 plasmid DNA. MB-231 control and transfected cells were assayed for firefly and renilla luciferase activity. Fold-change was calculated first by normalizing to transfection efficiency followed by normalization to luciferase activity in cells transfected with psiCHECK 2 plasmid DNA without any insert sequence, and then relative to luciferase activity in cells transfected with plasmid DNA carrying MnSOD 3'-UTR sequences without an ARE. Asterisks represent statistical significance relative to cells transfected with MnSOD 3'-UTR sequences without an ARE; ¥ represents statistical significance relative to constructs with two AREs; n=6, p<0.05. B. A quantitative RT-PCR assay was used to measure the mRNA levels of the 4.2 kb MnSOD transcript in MB-231 cells transfected with plasmid DNA carrying MnSOD 3'-UTR with 5 AREs, and without an ARE. Fold change was calculated relative to cells transfected with plasmid DNA carrying MnSOD 3'-UTR without an ARE; n=6, p<0.05. C. Immunoblot analysis of MnSOD protein levels in MB-231 cells transfected with plasmid DNA carrying MnSOD 3'-UTR with 5 AREs, and without an ARE. D. Flow cytometry measurements of the percentage of S-phase cells in MB-231 cells transfected with plasmid DNA carrying MnSOD 3'-UTR with 5 AREs, and without an ARE. Asterisks represent statistical significance relative to cells transfected with plasmid DNA carrying MnSOD 3'-UTR without an ARE; n=3, p<0.05.
Overexpression of the reporter construct carrying 5 AREs of the 4.2 kb MnSOD transcript enhanced endogenous mRNA levels of the longer MnSOD transcript (Figure 3B), suggesting a “decoy” function for the transfected MnSOD 3'-UTR. This increase in MnSOD mRNA levels was also associated with an increase in MnSOD protein levels (Figure 3C). It is worth noting that the “decoy” function for the transfected MnSOD 3'-UTR, and its subsequent enhancement of MnSOD mRNA and protein levels resulted in a significant decrease in the percentage of S-phase cells (Figure 3D). These results suggest that AREs in the 3'-UTR of MnSOD regulate its mRNA levels.
Preferential selection of MnSOD transcripts in irradiated cells
Several studies showed environmental stress agents, e.g. radiation and polychlorinated biphenyls, perturb MnSOD mRNA levels (Akashi et al., 1995; Chaudhuri et al.; Chaudhuri et al.; Oberley et al., 1989). Initially, we determined if ionizing radiation alters MnSOD expression in MB-231 human mammary epithelial cancer cells. Exponential cultures of MB-231 were irradiated with 2-8 Gy and total cellular RNA isolated at 48 h post-irradiation. Results from a quantitative RT-PCR assay showed approximately 2-3 fold increase in MnSOD mRNA levels in 6 and 8 Gy irradiated cells (Figure 4A). MnSOD mRNA levels increased approximately 2-fold at 24 h post-irradiation and 8-fold at 72 h post-irradiation in 8 Gy irradiated MB-231 cells (Figure 4B). Next, we determined if the radiation-induced increase in MnSOD mRNA levels could be due to a preferential selection of MnSOD transcripts. The quantitative PCR assay was repeated using primer-pairs specific to the 1.5 and 4.2 kb MnSOD transcripts. Radiation treatments did not alter the abundance of the 4.2 kb transcript during 24-72 h post-irradiation in 8 Gy irradiated MB-231 cells (Figure 4C). However, the abundance of the 1.5 kb transcript increased approximately 2-fold at 24 h post-irradiation and 6-fold at 72 h post-irradiation (Figure 4D). Radiation-induced increase in the 1.5 kb MnSOD transcript was also associated with approximately 2-fold increase in MnSOD protein levels at 48-72 h post-irradiation (Figure 5A). MnSOD activity increased to 40 U/mg protein at 72 h vs. 18 U/mg protein in un-irradiated cells (Figure 5B).
Figure 4.
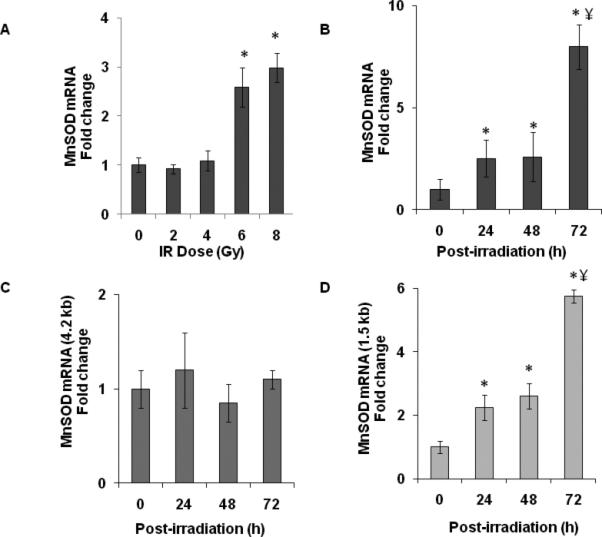
Ionizing radiation selectively enriched the 1.5 kb MnSOD transcript. A. Exponential cultures of MB-231 cells were irradiated with 2-8 Gy and harvested at 48 h post-irradiation for quantitative RT-PCR measurements of MnSOD mRNA levels. Asterisks represent statistical significance relative to MnSOD mRNA levels in unirradiated controls; n=3, p<0.05. B. Control and 8 Gy irradiated MB-231 cells were harvested at indicated times and MnSOD mRNA levels analyzed using a quantitative RT-PCR assay. Fold change calculated relative to un-irradiated control at the time of irradiation (0 h). The quantitative RT-PCR assay was repeated to measure the abundance of the 4.2 kb and 1.5 kb MnSOD transcripts, panels C and D, respectively. Asterisks represent statistical significance relative to 0 h control; ¥ represents statistical significance relative to 24 and 48 h samples; n=3, p<0.05.
Figure 5.
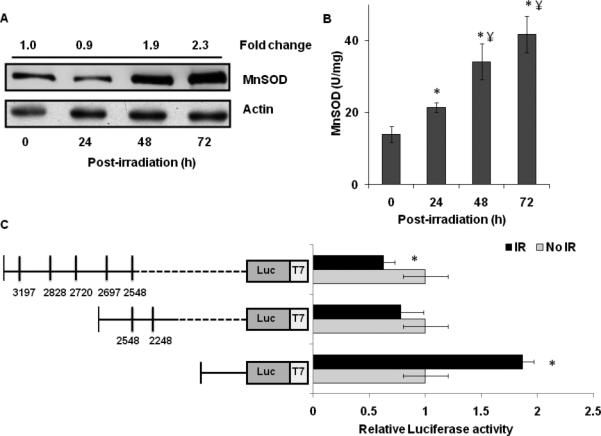
MnSOD 3'-UTR responds to environmental stress. A. Immunoblot analysis of MnSOD protein levels in 8 Gy irradiated MB-231 cells at indicated times post-irradiation. Blots were scanned and quantitated using AlphaImager 2000 and ImageJ software. Fold change calculated first by normalizing to actin levels in individual samples and then relative to MnSOD protein levels in unirradiated control cells at the time of irradiation (0 h). B. Biochemical measurements of MnSOD activity at indicated times post-irradiation. Asterisks represent statistical significance relative to MnSOD activity in unirradiated control cells at the time of irradiation; ¥ represents statistical significance relative to 24 h post-irradiation; n=3, p<0.05. C. Luciferase reporter activity in control and irradiated cells at 48 h post-irradiation. MB-231 cells were transfected with plasmid DNA carrying 0, 2, and 5 AREs of MnSOD 3'-UTR. Control and transfected cells were irradiated with 8 Gy and luciferase activity measured. Fold change was calculated by first normalizing to transfection efficiency and then relative to unirradiated control for each group. Asterisks represent statistical significance compared to unirradiated control for individual group; n=6, p<0.05.
A regulatory role of the 3'-UTR contributing to the preferential selection of MnSOD transcripts in irradiated MB-231 cells was also evident from the results presented in Figure 5C. MB-231 cells were transfected with plasmid DNAs carrying 0, 2, and 5 AREs of MnSOD 3'-UTR. Cells were irradiated with 8 Gy and luciferase activity measured at 48 h post-irradiation. Relative renilla-luciferase activity decreased approximately 50% in irradiated cells expressing 5-AREs, while cells overexpressing 2-AREs showed no significant change in renilla-luciferase activity. The reporter activity of the plasmids carrying the 3'-UTR of the shorter MnSOD transcript without an ARE showed approximately 2-fold increase in renilla-luciferase activity in irradiated vs. un-irradiated control cells. These results showed that irradiation increased the mRNA levels of the 1.5 kb MnSOD transcript, which correlated with an increase in MnSOD protein levels and activity.
Discussion
MnSOD is a redox enzyme that is known to regulate cellular redox environment. MnSOD activity and mitochondrial-generated ROS have been shown to regulate transitions between quiescence and proliferation (Sarsour et al., 2010; Sarsour et al., 2008). The present study investigates whether MnSOD 3'-UTR influences MnSOD expression in quiescent and proliferating cells as well as in response to irradiation. Human MnSOD has two transcripts (1.5 kb and 4.2 kb) with the same open reading frame but different 3'-UTR length. The levels of the shorter transcript increased in quiescent NHFs and MCF-10A cells, while the longer transcript was more abundant in proliferating cells (Figures 1 and 2, Table I). A direct correlation was observed between the levels of the longer MnSOD transcript and percentage of S-phase cells (Figure 1C, R2 = 0.84). The longer MnSOD transcript was more abundant in exponential cultures of malignant and non-malignant human epithelial cells (Table I), suggesting that the selection of the 4.2 kb MnSOD transcript is related to cellular proliferation vs. transformation status. A preferential selection of the longer MnSOD transcript in proliferating cells was associated with a decrease in MnSOD protein levels and activity, while the selection of the shorter transcript during quiescence correlated with an increase in MnSOD protein levels and activity (Figure 2). A decrease in MnSOD activity during proliferation is anticipated to increase the steady-state levels of superoxide facilitating proliferation, while an increase in MnSOD activity is anticipated to decrease the steady-state levels of superoxide supporting a quiescent state. This hypothesis is also consistent with our previously published results (Sarsour et al., 2008).
Recent reports demonstrate a growth-state dependent selection of mRNA isoforms in mouse and human cells (Mayr and Bartel, 2009; Sandberg et al., 2008). Using a genome-wide approach, Sandberg et al. have shown that the abundance of the growth-promoting transcripts truncated at the first PAS increased significantly in proliferating primary murine CD4+ T lymphocytes compared to resting cultures (Sandberg et al., 2008). The increase in the abundance of the shorter transcripts in stimulated cells correlated with a corresponding increase in their protein levels. Mayr et al. have reported similar observation in cancer cells (Mayr and Bartel, 2009). mRNA levels of oncogenes containing multiple PAS (e.g. cyclin D2, FGF2, and IMP-1) are enriched with the shorter transcripts during proliferation coinciding with higher protein levels.
Unlike oncogenes that promote proliferation, MnSOD is a negative regulator of proliferation. MnSOD expression is significantly reduced in proliferating SV40 transformed WI-38 human embryonic lung fibroblasts as well as cancer cells (Church et al., 1993; Oberley, 2001; Oberley et al., 1989; Weydert et al., 2003; Zhong et al., 1997). Ectopic expression of MnSOD has been shown to significantly inhibit cancer cell proliferation both in in vitro cell cultures and in vivo mouse xenograft of human cancers (Church et al., 1993; Weydert et al., 2003; Zhong et al., 1997). Therefore, a preferential selection of the longer transcript is anticipated to decrease MnSOD protein levels and activity facilitating proliferation, while selection of the shorter MnSOD transcript is anticipated to support a quiescent state. Our results that were obtained using semi-quantitative and quantitative PCR assays were also comparable to a previous report where the authors used northern blotting to demonstrate variations in the abundance of the two MnSOD transcripts in normal and SV40 transformed WI38 human lung fibroblasts (St Clair and Oberley, 1991). These authors showed that the 4.2 kb MnSOD transcript was more abundant in SV40 transformed vs. normal WI38 cells. The levels of the shorter transcript was higher in normal vs. SV40 transformed WI38 cells. While the significance of these previous observations was not addressed, our results showed that a higher level of the 4.2 kb MnSOD transcript correlates with proliferation and an increase in the 1.5 kb MnSOD transcript levels correlated with quiescence or slow growth. A preferential selection of the longer transcript of growth inhibitory genes during proliferation is also reported for the Prohibitin gene (Jupe et al., 1996a; Jupe et al., 1996b). The longer (1.9 kb) transcript of Prohibitin was more abundant during proliferation compared to the shorter 1.2 kb transcript. Overexpression of the 3'-UTR of Prohibitin inhibited progression from quiescent to proliferative state in CF3 human fibroblasts. These results suggest that a preferential selection of the shorter transcripts (truncated at the first PAS site) of the growth inhibitory genes would support quiescence (or slowly proliferating cells), while selection of the longer transcripts (truncated at the distal PAS) would facilitate proliferation.
AU-rich (ARE) sequence in the 3'-UTR is known to regulate mRNA levels both positively (e.g. HuR protein binding to AREs) and negatively (e.g. AUF1 protein binding to AREs). Human MnSOD 3'-UTR has multiple AU-rich (AREs) sequence between the two PAS: A(U)3A, A(U)5A, and A(U)6A (Church, 1990). The shorter MnSOD transcript contains only a single A(U)3A motif. Results from a luciferase-reporter assay showed that the reporter activity of cells transfected with plasmid DNA carrying two ARE-motifs of MnSOD 3'-UTR was approximately 40% lower than cells transfected with plasmid DNA carrying MnSOD 3'-UTR sequence without an ARE (Figure 3A). Reporter activity decreased approximately 60% in cells transfected with plasmid DNA carrying five ARE-motifs that are present between the two PAS in MnSOD 3'-UTR (Figure 3A). These results suggest that the AREs between the PAS regulate MnSOD mRNA levels.
Overexpression of the longer MnSOD 3'-UTR significantly increased the endogenous mRNA levels of the 4.2 kb MnSOD transcript, which correlated with a corresponding increase in MnSOD protein levels (Figure 3B and 3C). These results suggest that the exogenous MnSOD 3'-UTR serves as a “decoy” and titrate out trans-factors from binding to the endogenous transcript, which resulted in an increase in the mRNA levels of the 4.2 kb MnSOD transcript. mRNA stabilizing and destabilizing proteins, e.g. HuR and AUF1, have been detected in ribonucleoprotein complexes (Lal et al., 2004). It was suggested that the stability and translation of HuR and AUF1 containing ribonucleoprotein complexes will depend on the target mRNA of interest, abundance of the RNA-binding proteins, subcellular compartment, and cellular environment. Consistent with this hypothesis, we have shown previously that protein binding to Topoisomerase II-alpha 3'-UTR varies during the cell cycle correlating with its changes in mRNA and protein levels (Goswami et al., 2000). Future studies will determine if the abundance of RNA binding proteins in ribonucleoprotein complexes containing MnSOD transcripts varies during quiescence and proliferation, and if such a mechanism could account for the preferential abundance of the two MnSOD transcripts. Furthermore, because MnSOD 3'-UTR also has microRNA target sequence (based on TargetScan bioinformatics evaluation), the decoy function of the exogenous MnSOD 3'-UTR may relieve the 4.2 kb MnSOD transcript from mRNA turnover and/or translation inefficiency leading to increases in its mRNA and protein levels. An additional mechanism for a preferential selection of MnSOD transcripts during quiescence and proliferation could be due to a differential selection of the two polyadenylation sites (PAS). Cell cycle dependent changes in the polyadenylation regulatory pathways, e.g. abundance of the 64 kDa subunit of the cleavage stimulatory factor complex, have been correlated with a preferential selection of PAS in the eIF-2α transcript (Martincic et al., 1998).
Because MnSOD is a negative regulator of proliferation, a decoy-induced increase in MnSOD mRNA and protein levels is anticipated to slow proliferation. In fact, this is what we observed. The percentage of S-phase decreased in cells overexpressing the longer MnSOD 3'-UTR (Figure 3D). Thus, a preferential selection of the shorter MnSOD transcript during quiescence would protect MnSOD mRNA from a post-transcriptional repression, which would increase MnSOD mRNA and protein levels as well as its activity. In contrast, selection of the longer transcript during proliferation would be amenable to post-transcriptional regulation leading to a decrease in MnSOD protein levels and activity.
The preferential selection of the shorter MnSOD transcript was also observed in response to radiation induced growth arrest. MnSOD mRNA levels increased approximately 2- to 3-fold in 6 and 8 Gy irradiated MB-231 cells (Figure 4). While radiation did not change the abundance of the 4.2 kb MnSOD transcript, mRNA levels of the shorter transcript increased approximately 6-fold at 72 h post-irradiation (Figure 4B and 4C). Radiation induced increase in the mRNA levels of the shorter MnSOD transcript was associated with an increase in MnSOD protein levels and activity (Figure 5A and 5B), which corresponds to a delay in cellular proliferation (data not shown). The role of 3'-UTR regulating MnSOD mRNA levels in irradiated cells was further evident from the results presented in Figure 5C. Luciferase activity increased approximately 2-fold in irradiated cells transfected with plasmid DNA carrying the 3'-UTR of the shorter MnSOD transcript compared to un-irradiated transfected cells.
In summary, our results showed a preferential selection of the 1.5 kb MnSOD transcript in quiescent and radiation-induced growth arrested cells. A direct correlation was observed between the abundance of the 4.2 kb MnSOD transcript and percentage of S-phase cells. The increase in the shorter MnSOD transcript levels during quiescence correlated with a higher level of MnSOD protein and activity, while the selection of the longer transcript during proliferation correlated with a lower level of MnSOD protein and activity (Figure 6). The mechanisms regulating MnSOD expression are complex: transcriptional, e.g. AP1, SP1, and NFkB transcription factor binding to MnSOD promoter; post-translational: e.g. phosphorylation; and transcript selection (this study). The role of transcript selection and 3'-UTR adds a new level of complexity to MnSOD expression. We hypothesize that such a complex mode of transcript selection is necessary to fine-tune the cellular redox environment during transition from quiescent through the proliferative cycle.
Figure 6.
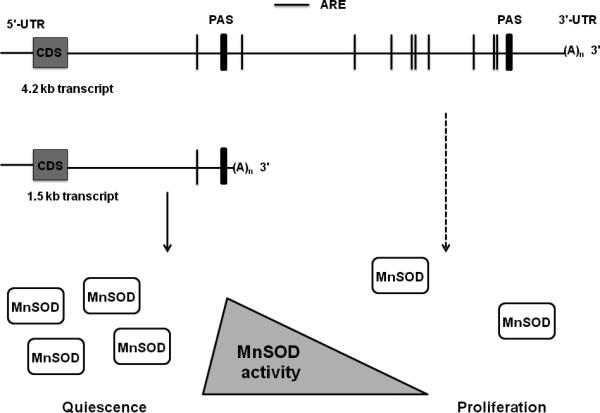
An illustration of the preferential selection of MnSOD transcripts in quiescent and proliferating cells. The abundance of the shorter MnSOD transcript during quiescence correlates with an increase in MnSOD protein levels and activity. The selection of the longer MnSOD transcript during proliferation is associated with a lower level of MnSOD protein and activity. Vertical lines represent sites for AU-rich sequences (ARE); PAS: polyadenylation signal.
Materials and Methods
Cell culture
Normal human fibroblasts (Coriell cell repository), MCF-10A non-malignant and MDA-MB-231 malignant human mammary epithelial cells (ATCC) were cultured following our previously published protocols (Chaudhuri et al., 2010b; Sarsour et al., 2005; Menon et al., 2005). Human oral squamous cell carcinoma (Cal 27, SQ20B, and FaDu), mammary cancer cells (Sum159), and pulmonary carcinoma cells (H292 and A549) were cultured following ATCC protocols. MDA-MB-231 cells were irradiated using a cesium-137 source; dose rate: 0.83 Gy/min.
Flow cytometry measurements of DNA content
Flow cytometry measurements of DNA content and percentage of cells in each phase of the cell cycle were determined following our previously published method (Chaudhuri et al., 2010b; Sarsour et al., 2005).
Antioxidant enzyme activity assay
MnSOD activity was determined by the indirect competitive inhibition assay originally developed by Spitz and Oberley (Spitz and Oberley, 1989).
Immunoblotting assay
Equal amounts of total cellular proteins were separated by 12.5% SDS-PAGE and transferred to nitrocellulose membrane. Blots were incubated with antibody to MnSOD (PharMingen, San Diego, California). Immunoreactive polypeptide was visualized using horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence detection reagents (GE Healthcare, Waukesha, Wisconsin) following manufacturer supplied protocol. Blots were re-probed with antibodies to actin (Santa Cruz Biotechnology, Santa Cruz, California) for comparison of results. Results were quantitated using AlphaImager 2000 (Alpha Innotech, San Leandro, California) and ImageJ software following our previously published protocol (Chaudhuri et al., 2010b).
Quantitative Real Time RT-PCR assay
Trizol reagent (Invitrogen, Eugene, Oregon) was used to isolate total cellular RNA. One microgram of RNA was reverse transcribed using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, California). The cDNA was subjected to Real Time PCR assay using primers specific to MnSOD open reading frame, forward primer: 5'-GGCCTACGTGAACAACCTGAA-3', reverse primer: 5'-CTGTAACATCTCCCTTGGCCA-3', amplicon size, 70 bp. Primer-pairs specific to the 3'-UTR were used to measure the 1.5 and 4.2 kb MnSOD transcript levels; MnSOD 4.2 kb transcript, forward primer: 5'-GCTTTGGTGGTGGATTGAAAC-3', reverse primer: 5'-CATCCCTACAAGTCCCCAAAGT-3', amplicon size, 187 bp; MnSOD 1.5 kb transcript, forward primer: 5'-TAATGATCCCAGCAAGATAA-3', reverse primer (anchored to the first PAS): 5'-TTTTTTTTTTTTTTTGATGGTTG-3', amplicon size, 184 bp. 18S mRNA levels were used as control. The real time PCR assay was carried out with the 2x Power SYBR Green real time master mix (Applied Biosystems) using the following cycle parameters (ABI 7000 Sequence Detection System, Applied Biosystems): inactivation of reverse transcriptase at 95°C for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. A threshold of amplification in the linear range of each sample was selected to calculate the cycle threshold value. The relative mRNA levels were calculated as previously described (Chaudhuri et al., 2010b). Log2 transformed values (Mayr and Bartel, 2009; Sandberg et al., 2008) were used to calculate the expression ratio of the longer to shorter MnSOD mRNA transcript.
Semi-quantitative RT-PCR assay
A semi-quantitative RT-PCR assay was used to simultaneously visualize the abundance of MnSOD and GAPDH mRNA levels in quiescent and exponential cultures of MCF-10A cells. Primer pairs were designed as follows: MnSOD coding sequence, forward primer 5'- CCCTGGAACCTCACATCAAC- 3', reverse primer 5'-CGTGGTTTACTTTTTGCAAGC -3', amplicon size, 566 bp; GAPDH, forward primer 5'-TGAGAAGTATGACAACAGCCTCA -3', reverse primer 5'-CTGTTGAAGTCAGAGGAGACCAC -3', amplicon size, 453 bp. Primer pairs used to selectively amplify the two MnSOD transcripts were: 4.2 kb MnSOD transcript, forward primer 5'-AGGCAGCTGGCTCCGGTTTT- 3', reverse primer 5'-GGCATCCCTACAAGTCCCCAAA -3', amplicon size, 954 bp; 1.5 kb MnSOD transcript, forward primer 5'-AGGCAGCTGGCTCCGGTTTT- 3', reverse primer 5'-CATCAATCCCCAGCAGTGGAATAA- 3', amplicon size, 520 bp. PCR amplified products were resolved by agarose gel electrophoresis and visualized by staining with ethidium bromide.
Dual luciferase reporter assay
Total cellular RNA isolated from MDA-MB-231 cells was reverse transcribed, and cDNA pool was used to PCR-amplify 3'-UTRs specific for the short and long MnSOD transcripts. Primers were designed for directional cloning incorporating NotI and XhoI restriction enzyme sites (underlined). The primers used to amplify the 3'-UTR of the 1.5 MnSOD transcript were: forward primer, 5'-TTTTCTCGAGGCACTGAAGTTCAATGGTGGTGGT-3', and reverse primer, 5'-AAAAGCGGCCGCCCAGGACCTTATAGGGTTTTCAGTATGTACC-3', amplicon size 520 bp. Two separate PCR amplifications were performed to amplify the 3'-UTR of the 4.2 kb MnSOD transcript. The first PCR amplification was performed to amplify the 985 bp of the proximal sequence following the first PAS (Church, 1990). Primer sequences to amplify this sequence region were: 5'-TTTTCTCGAGGCTCATGCTTGAGACCCAAT-3', and 5'-AAAAGCGGCCGCGCTGAGGTGGGACAATCACT-3'. The PCR amplified fragment contains 2 AREs. A second PCR amplification was performed to amplify 1284 bp of the 4.2 kb MnSOD transcript that contains 5 AREs (Church, 1990). The designed primer-pairs were: 5'-TTTTCTCGAGTCTAGGTGACTCTAACTTCCCTGGC-3', and 5'-AAAAGCGGCCGCCTCTCACCCAGAAAGCCAAAGCA-3'. PCR amplified products were cloned into the multiple cloning site (NotI and XhoI) of the psiCHECK-2 reporter vector (Promega). All insert sequences were verified by restriction enzyme digestions and sequence analysis.
MDA-MB-231 cells were seeded at a density of 1 × 105 cells per well of a 24-well dish. Cells were transfected with Lipofectamine 2000 and 100 ng of psiCHECK 2 plasmid DNAs containing MnSOD 3'-UTR sequence specific to the 1.5 and 4.2 kb transcripts. Cells transfected with psiCHECK 2 plasmid DNA without any insert sequence were included as controls. Forty eight hours post-transfection, luciferase activity was measured using dual luciferase reporter assay system following manufacturer supplied protocol (Promega). Firefly-luciferase activity was measured on a Moon Light Luminometer (Pharmingen). One hundred microliters of the Stop and Glo reagent was added, and Renilla luciferase was measured. Firefly luciferase activity was used to normalize for variations in transfection efficiency. Fold-change was calculated first by normalizing to transfection efficiency followed by normalization to luciferase activity in cells transfected with psiCHECK 2 plasmid DNA without any insert sequence, and then relative to luciferase activity in cells transfected with plasmid DNA carrying MnSOD 3'-UTR sequences without an ARE.
Statistics
Statistical significance was determined by one and two-way analysis of variance with Tukey's post hoc test and Student t-tests using GraphPad Prism, version 4 and SPSS (IBM). Results are presented as mean ± standard deviation with results from n ≥ 3 and p < 0.05 considered significant.
Acknowledgements
We thank Dr. Ehab H. Sarsour and Maneesh G. Kumar at the Free Radical and Radiation Biology Division, and Amy Dubinsky and Dr. Beverly L. Davidson at the department of Internal Medicine for technical assistance with the UTR cloning and luciferase reporter assay, Drs. Ann Simons and Melissa Fath for human oral squamous and lung cancer cells, Mr. Gareth Smith with editorial assistance, and the staff at the Flow Cytometry and Radiation and Free Radical Research Core Facilities. Funding from NIH CA111365 and NIEHS P42 ES 013661 supported this work.
Footnotes
Conflict of Interest: There is no conflict of interest.
References
- Akashi M, Hachiya M, Paquette RL, Osawa Y, Shimizu S, Suzuki G. Irradiation increases manganese superoxide dismutase mRNA levels in human fibroblasts. Possible mechanisms for its accumulation. J Biol Chem. 1995;270:15864–9. doi: 10.1074/jbc.270.26.15864. [DOI] [PubMed] [Google Scholar]
- Chaudhuri L, Sarsour EH, Goswami PC. 2-(4-Chlorophenyl)benzo-1,4-quinone induced ROS-signaling inhibits proliferation in human non-malignant prostate epithelial cells. Environ Int. 2010a;36:924–30. doi: 10.1016/j.envint.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri L, Sarsour EH, Kalen AL, Aykin-Burns N, Spitz DR, Goswami PC. Polychlorinated biphenyl induced ROS signaling delays the entry of quiescent human breast epithelial cells into the proliferative cycle. Free Radic Biol Med. 2010b;49:40–9. doi: 10.1016/j.freeradbiomed.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–70. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chivukula RR, Mendell JT. Circular reasoning: microRNAs and cell-cycle control. Trends Biochem Sci. 2008;33:474–81. doi: 10.1016/j.tibs.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church SL. Manganese superoxide dismutase: nucleotide and deduced amino acid sequence of a cDNA encoding a new human transcript. Biochim Biophys Acta. 1990;1087:250–2. doi: 10.1016/0167-4781(90)90213-l. [DOI] [PubMed] [Google Scholar]
- Church SL, Grant JW, Ridnour LA, Oberley LW, Swanson PE, Meltzer PS, et al. Increased manganese superoxide dismutase expression suppresses the malignant phenotype of human melanoma cells. Proc Natl Acad Sci U S A. 1993;90:3113–7. doi: 10.1073/pnas.90.7.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerch LB. Post-transcriptional regulation of lung antioxidant enzyme gene expression. Ann N Y Acad Sci. 2000;899:103–11. doi: 10.1111/j.1749-6632.2000.tb06179.x. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–61. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami PC, Sheren J, Albee LD, Parsian A, Sim JE, Ridnour LA, et al. Cell cycle-coupled variation in topoisomerase IIalpha mRNA is regulated by the 3'-untranslated region. Possible role of redox-sensitive protein binding in mRNA accumulation. J Biol Chem. 2000;275:38384–92. doi: 10.1074/jbc.M005298200. [DOI] [PubMed] [Google Scholar]
- Guo B, Yu Y, Leibold EA. Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J Biol Chem. 1994;269:24252–60. [PubMed] [Google Scholar]
- Jupe ER, Liu XT, Kiehlbauch JL, McClung JK, Dell'Orco RT. The 3' untranslated region of prohibitin and cellular immortalization. Exp Cell Res. 1996a;224:128–35. doi: 10.1006/excr.1996.0120. [DOI] [PubMed] [Google Scholar]
- Jupe ER, Liu XT, Kiehlbauch JL, McClung JK, Dell'Orco RT. Prohibitin in breast cancer cell lines: loss of antiproliferative activity is linked to 3' untranslated region mutations. Cell Growth Differ. 1996b;7:871–8. [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lutz CS. Alternative polyadenylation: a twist on mRNA 3' end formation. ACS Chem Biol. 2008;3:609–17. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- Martincic K, Campbell R, Edwalds-Gilbert G, Souan L, Lotze MT, Milcarek C. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc Natl Acad Sci U S A. 1998;95:11095–100. doi: 10.1073/pnas.95.19.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Fox PL. Translational control by the 3'-UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–8. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem. 1969;244:6049–55. [PubMed] [Google Scholar]
- Menon SG, Coleman MC, Walsh SA, Spitz DR, Goswami PC. Differential susceptibility of nonmalignant human breast epithelial cells and breast cancer cells to thiol antioxidant-induced G(1)-delay. Antioxid Redox Signal. 2005;7:711–8. doi: 10.1089/ars.2005.7.711. [DOI] [PubMed] [Google Scholar]
- Oberley LW. Anticancer therapy by overexpression of superoxide dismutase. Antioxid Redox Signal. 2001;3:461–72. doi: 10.1089/15230860152409095. [DOI] [PubMed] [Google Scholar]
- Oberley LW, McCormick ML, Sierra-Rivera E, Kasemset-St Clair D. Manganese superoxide dismutase in normal and transformed human embryonic lung fibroblasts. Free Radic Biol Med. 1989;6:379–84. doi: 10.1016/0891-5849(89)90083-x. [DOI] [PubMed] [Google Scholar]
- Oberley TD, Schultz JL, Li N, Oberley LW. Antioxidant enzyme levels as a function of growth state in cell culture. Free Radic Biol Med. 1995;19:53–65. doi: 10.1016/0891-5849(95)00012-m. [DOI] [PubMed] [Google Scholar]
- Rouault TA, Hentze MW, Caughman SW, Harford JB, Klausner RD. Binding of a cytosolic protein to the iron-responsive element of human ferritin messenger RNA. Science. 1988;241:1207–10. doi: 10.1126/science.3413484. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–7. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarsour EH, Agarwal M, Pandita TK, Oberley LW, Goswami PC. Manganese superoxide dismutase protects the proliferative capacity of confluent normal human fibroblasts. J Biol Chem. 2005;280:18033–41. doi: 10.1074/jbc.M501939200. [DOI] [PubMed] [Google Scholar]
- Sarsour EH, Goswami M, Kalen AL, Goswami PC. MnSOD activity protects mitochondrial morphology of quiescent fibroblasts from age associated abnormalities. Mitochondrion. 2010;10:342–9. doi: 10.1016/j.mito.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarsour EH, Venkataraman S, Kalen AL, Oberley LW, Goswami PC. Manganese superoxide dismutase activity regulates transitions between quiescent and proliferative growth. Aging Cell. 2008;7:405–17. doi: 10.1111/j.1474-9726.2008.00384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–67. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Spitz DR, Oberley LW. An assay for superoxide dismutase activity in mammalian tissue homogenates. Anal Biochem. 1989;179:8–18. doi: 10.1016/0003-2697(89)90192-9. [DOI] [PubMed] [Google Scholar]
- St Clair DK, Oberley LW. Manganese superoxide dismutase expression in human cancer cells: a possible role of mRNA processing. Free Radic Res Commun. 1991;12-13(Pt 2):771–8. doi: 10.3109/10715769109145858. [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–12. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XS, Devalaraja MN, St Clair DK. Molecular structure and organization of the human manganese superoxide dismutase gene. DNA Cell Biol. 1994;13:1127–36. doi: 10.1089/dna.1994.13.1127. [DOI] [PubMed] [Google Scholar]
- Wang Y, Blelloch R. Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 2009;69:4093–6. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weydert C, Roling B, Liu J, Hinkhouse MM, Ritchie JM, Oberley LW, et al. Suppression of the malignant phenotype in human pancreatic cancer cells by the overexpression of manganese superoxide dismutase. Mol Cancer Ther. 2003;2:361–9. [PubMed] [Google Scholar]
- Xu Y, Kiningham KK, Devalaraja MN, Yeh CC, Majima H, Kasarskis EJ, et al. An intronic NF-kappaB element is essential for induction of the human manganese superoxide dismutase gene by tumor necrosis factor-alpha and interleukin-1beta. DNA Cell Biol. 1999;18:709–22. doi: 10.1089/104454999314999. [DOI] [PubMed] [Google Scholar]
- Zhong W, Oberley LW, Oberley TD, St Clair DK. Suppression of the malignant phenotype of human glioma cells by overexpression of manganese superoxide dismutase. Oncogene. 1997;14:481–90. doi: 10.1038/sj.onc.1200852. [DOI] [PubMed] [Google Scholar]


