This study identifies upregulated proteins in the glaucomatous human retina exhibiting links to TNF-α/TNFR1 signaling, highlights various signaling molecules and regulators of cell death and immune response pathways, links proteomic and epigenetic alterations, and provides a motivating framework.
Abstract
Purpose.
This study aimed to determine retinal proteomic alterations in human glaucoma, with particular focus on links to TNF-α/TNFR1 signaling.
Methods.
Human retinal protein samples were obtained from 20 donors with (n = 10) or without (n = 10) glaucoma. Alterations in protein expression were individually analyzed by quantitative LC-MS/MS. Quantitative Western blot analysis with cleavage or phosphorylation site-specific antibodies was used for data validation, and cellular localization of selected proteins was determined by immunohistochemical analysis of the retina in an additional group of glaucomatous human donor eyes (n = 38) and nonglaucomatous controls (n = 30).
Results.
Upregulated retinal proteins in human glaucoma included a number of downstream adaptor/interacting proteins and protein kinases involved in TNF-α/TNFR1 signaling. Bioinformatic analysis of the high-throughput data established extended networks of diverse functional interactions with death-promoting and survival-promoting pathways and mediation of immune response. Upregulated pathways included death receptor-mediated caspase cascade, mitochondrial dysfunction, endoplasmic reticulum stress, calpains leading to apoptotic cell death, NF-κB and JAK/STAT pathways, and inflammasome-assembly mediating inflammation. Interestingly, retinal expression pattern of a regulator molecule, TNFAIP3, exhibited prominent variability between individual samples, and methylation of cytosine nucleotides in the TNFAIP3 promoter was found to be correlated with this variability among glaucomatous donors.
Conclusions.
Findings of this study reveal a number of proteins upregulated in the glaucomatous human retina that exhibit many links to TNF-α/TNFR1 signaling. By highlighting various signaling molecules and regulators involved in cell death and immune response pathways and by correlating proteomic findings with epigenetic alterations, these findings provide a framework motivating further research.
The prevailing view is that glaucoma pathogenesis is multifactorial, with a complex interplay of elevated intraocular pressure-induced events and genetic/epigenetic/aging-related susceptibility factors contributing to neurodegeneration. Glial activation response and secondary inflammatory/autoimmune processes are also regarded as continuous components of glaucomatous neurodegeneration. It is widely accepted that chronic activation of glial cells and accompanying increases in the production of proinflammatory cytokines, primarily including TNF-α, are hallmarks of inflammation/parainflammation in glaucomatous tissue, although a cause-effect relationship remains to be validated.1,2 TNF-α, with beneficial and neurotoxic effects in the central nervous system (CNS) along with key physiological functions in the maintenance of immune homeostasis, has been implicated in the pathogenesis of a wide spectrum of human neurodegenerative diseases. It is also increasingly evident that TNF-α through the binding of TNFR1, a death receptor, exhibits important links to glial activation response, mediation of retinal ganglion cell (RGC) death, and inflammatory processes during the neurodegenerative injury in glaucoma.3
Despite growing evidence that supports important roles of TNF-α in glaucomatous neurodegeneration, opposing consequences of TNF-α signaling make it difficult to exploit for neuroprotective strategies. Respecting the diverse bioactivities of this multifunctional cytokine, molecular dissection of specific signaling components can provide the possibility to specifically inhibit RGC death or modulate immune response without compromising survival-promoting signals. To better understand molecular components of the neurodegenerative signaling in human glaucoma, this study analyzed retinal protein samples obtained from donor eyes with or without glaucoma. Findings of this comparative analysis supported a prominent upregulation of TNF-α/TNFR1 signaling in the glaucomatous human retina. By highlighting various signaling molecules and regulators involved in cell death and immune response pathways in human glaucoma, these findings provide framework information and motivate further research.
Materials and Methods
Donor Eyes
Retinal protein samples obtained from 10 human donor eyes with glaucoma (age, 84.7 ± 8) and 10 eyes without glaucoma (age, 83.7 ± 7) were individually analyzed by capillary liquid chromatography coupled with linear ion trap mass spectrometry (LC-MS/MS). As previously described,4,5 retinal tissue punches were collected within <6 hours after death, and glaucomatous eyes were well documented.
In addition, cellular localization of selected proteins was determined by immunohistochemical analysis of retinal tissue sections obtained from an additional group of glaucomatous and nonglaucomatous human donor eyes. This group included 38 donor eyes with a diagnosis of glaucoma (age, 76.8 ± 11) and 30 eyes without glaucoma (age, 71.0 ± 15), all fixed within 12 hours after death. Detailed information on donor demographics and clinical data has been previously published.6 All the human donor eyes were handled according to the tenets of the Declaration of Helsinki.
Proteomic Analysis
Protein samples prepared with a lysis buffer containing 50 mM Hepes-KOH pH 8.0, 100 mM KCl, 2 mM EDTA, 0.10% NP-40, 2 mM dithiothreitol, 10% glycerol, and protease and phosphatase inhibitors were analyzed by label-free quantitative LC-MS/MS, as previously described.7 Briefly, trypsin-digested samples were loaded onto an analytical 2D capillary chromatography column packed with strong cation exchange (SCX) and C18 reversed-phase (RP) resin (Phenomenex, Torrance, CA). This biphasic column was attached to an analytical RP chromatography column with an integrated, laser-pulled emitter tip. Peptides were eluted from SCX with seven-step gradients of 5%, 10%, 15%, 30%, 50%, 70%, and 100% of 500 mM ammonium acetate and eluted into a linear ion trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA) according to a linear HPLC gradient (20-minute 0% B, 80-minute 40% B, and 90-minute 60% B at a flow rate of 200 nL/min with mobile phase-A 5% acetonitrile/0.1% formic acid and mobile phase-B 80% acetonitrile/0.1% formic acid). Protein identification from MS/MS spectra was performed with proteomics analysis software (Sequest Sorcerer; Sage-N Research, San Jose, CA), which was set up to search a FASTA formatted human protein database (Human RefSeq) with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 1.2 Da. Meta-analysis software (Scaffold; Proteome Software Inc., Portland, OR) was used to validate peptide and protein identifications based on the criteria of 95.0% and 99.0% probability and at least two peptides as specified by the Prophet peptide8 and protein algorithms.9 The abundance of each identified protein was determined by normalizing the number of unique spectral counts matching to the protein by its predicted molecular weight.10 The Mann-Whitney rank sum test was used to determine the significant difference in protein expression between glaucomatous and control samples.
As in previous studies,4,5,11 we used pathway analysis software (Ingenuity Pathways Analysis [IPA]; Ingenuity Systems, Mountain View, CA) for bioinformatic analysis. Our high-throughput dataset and the corresponding expression values uploaded into the application were analyzed to define functional patterns and generate extended interaction networks using the database (Ingenuity Pathway Knowledge Base; Ingenuity Systems). Canonical pathway analysis identified the pathways from the IPA library that were most significant to the dataset by the right-tailed Fisher's exact test.
Western Blot Analysis
Immunoblotting used primary antibodies to cleaved caspase 1 (1:1000; Millipore, Billerica, MA), caspases 3 and 8 (1:1000; Cell Signaling, Danvers, MA), caspases 9 and 12 (1:500; Abcam, Cambridge, MA), nuclear factor kappa B (NF-κB) subunits, p50 [phospho-Ser932] (1:500; GenWay, San Diego, CA) and p65 [phospho-Ser276] (1:1000; Cell Signaling), signal transducers and activators of transcription (STAT)1 [phospho-Tyr701], STAT2 [phospho-Tyr690], STAT3 [phospho-Ser727], STAT4 [phospho-Tyr693], STAT5 [phospho-Tyr694], STAT6 [phospho-Tyr641], (1:1000; Cell Signaling), and TNF-α–induced protein 3 (TNFAIP3; 1:1000; Epitomics, Burlingame, CA). In addition, a β-actin antibody (Sigma-Aldrich, St. Louis, MO) was used to reprobe immunoblots for loading and transfer control. The secondary antibody incubation used a specific IgG conjugated with horseradish peroxidase (1:2000; Sigma-Aldrich). The primary antibody was omitted to provide negative control. After normalization to β-actin, the average band intensity value obtained from nonglaucomatous samples was used to calculate the fold change in protein expression in glaucomatous samples.
Morphologic Analysis
Double immunofluorescence labeling used the same primary antibodies described for Western blot analysis (1:100). In addition, antibodies against Brn-3 (not subtype selective; 1:200) or glial fibrillary acidic protein (GFAP; 1:200), both from Santa Cruz Biotechnology (Santa Cruz, CA), were used to identify RGCs and astrocytes. A mixture of Alexa Fluor 488- or 568-conjugated species-specific IgGs (1:400; Molecular Probes, Eugene, OR) was used for the secondary antibody incubation. Negative controls were performed by replacing the primary antibody with serum or by using an inappropriate secondary antibody to determine species specificity.
SNP Analysis
Genomic DNA was isolated from retinal tissue samples of 5 donors with glaucoma using a purification kit (DNeasy Blood & Tissue Kit; Qiagen, Hilden, Germany). All fragments were amplified using polymerase (Gotaq Flexi; Promega, Madison, WI) and were sequenced (DNA Analyzer; Applied Biosystems, Foster City, CA). Primer sequences used for amplification and sequencing are provided in Table 1A.
Table 1.
Primer Sequences Used for Amplification and Sequencing
| A. SNP Analysis | ||
| rs112229105 | F | GAAAGCTGTGAAGATACGGGAGAGA |
| R | CCCCGAATAGAGATTCTATATAAAGGTCTC | |
| rs5029941/rs2230926 | F | GCTGTCATCATCTTGTGAAATATCAGTTTG |
| R | GGAGGTTTCTGGTGTTTTCCATTGAG | |
| rs61756235 | F | CAGGTAACAGAGTTCAATGGAATTTGATGA |
| R | CTCTTACTAACCAAGCAAGTCACAGAAC | |
| rs34935799 | F | GGGTGATCATTTGAATGATGGTTTCATG |
| R | ATTCCAAACTTCTTAGCATTTTGTCTGTTC | |
| rs3734553/rs5029957 | F | GTCCCAACAGAAGAGAGCCAGG |
| R | CTCCTGCTCAGACACCCTTAAGC | |
| B. Methylation Sequencing | ||
| Outer primers for first round | F | GATTTAGAGAGTTACGTGATTTTGGAAAG |
| R | TCCAACAAACTCCCAATCCAAAAAC | |
| Inner primers for second round | F | GTATATAATTGAAACGGGGTAAAGTAGATTG |
| R | ATACCAACCCGAAAATCGCTACCCAACATACACC |
F, forward; R, reverse.
Methylation Sequencing
Genomic DNA extracted from retinal tissue samples of 5 donors with glaucoma was subjected to bisulfite treatment (BisulFlash DNA Modification Kit; Epigentek, Farmingdale, NY). After conversion, the promoter region was amplified by nested PCR using DNA polymerase (Platinum Taq Hi-Fi; Invitrogen, Carlsbad, CA) and was sequenced as described. Primer pairs surrounding the CpG island within the TNFAIP3 promoter were designed using the MethPrimer online tool.12 Primer sequences used for amplification and sequencing are provided in Table 1B.
Results
Quantitative LC-MS/MS analysis of human retinal protein samples resulted in the identification of hundreds of proteins with high confidence that exhibited upregulated or downregulated expression in glaucomatous samples. Bioinformatic analysis identified the pathways from the IPA library that were most significantly associated with our high-throughput data. Top canonical pathways most significant to our dataset included death receptor signaling pathway (right-tailed Fisher's exact test; P < 0.05). Here, we present the upregulated proteins exhibiting links to TNF-α/TNFR1 signaling.
As listed in Table 2, upregulated retinal proteins in human glaucoma included TNFR1 and a number of downstream adaptor/interacting proteins, such as TNFR1-associated death domain protein (TRADD), mitogen-activated protein kinase (MAPK)-activating death domain-containing protein, different members of the TNFR-associated factor (TRAF) family, and NF-κB. Identified proteins also included various regulator molecules involved in TNFR signaling, such as caspase 8 and FADD-like apoptosis regulator (CFLAR, also called FLICE-inhibitory protein) and optineurin. Another regulator protein we detected was TNFAIP3, also known as A20, which is a potent inhibitor of NF-κB activation and a negative regulator of TNF-α signaling leading to apoptosis and inflammation. Despite an overall prominent difference between glaucomatous and nonglaucomatous samples, glaucomatous samples exhibited individual differences in increased expression of different proteins. However, the presented data were consistent in at least 6 of 10 glaucomatous samples for each of the listed proteins, except for the regulator proteins, mainly including TNFAIP3. Interestingly, the expression of this protein exhibited prominent individual differences.
Table 2.
Analysis of Retinal Protein Expression in Human Glaucoma: Proteins Involved in the TNFR Signaling
| Symbol | Protein Name | RefSeq | Fold Change |
|---|---|---|---|
| TNF-α | Tumor necrosis factor-alpha | NP_000585 | 3.1* |
| TNFRSF1A (TNFR1) | Tumor necrosis factor receptor superfamily, member 1A | NP_001056 | 2.1* |
| TRADD | TNFRSF1A-associated death domain-containing protein | NP_003780 | 2.3* |
| MADD | MAP kinase-activating death domain-containing protein isoform a | NP_569826 | 1.9* |
| MADD | MAP kinase-activating death domain-containing protein isoform g | NP_569831 | 1.1 |
| CFLAR (FLIP) | Caspase 8 and FADD-like apoptosis regulator | NP_003870 | 1.2 |
| TRAF1 | TNFR-associated factor 1 | NP_005649 | 2.4* |
| TRAF2 | TNFR-associated factor 2 | NP_066961 | 3.6* |
| TRAF3IP1 | TNFR-associated factor 3 interacting protein 1 | NP_056465 | 1.3 |
| TRAF4 | TNFR-associated factor 4 | NP_004286 | 1.5 |
| TRAF4 variant | TNFR-associated factor 4 isoform 2 | NP_665694 | 1.9* |
| TFAF2 (SNX6) | TNFR-associated factor 4-associated factor 2 | NP_067072 | 1.8* |
| TRAF5 | TNFR-associated factor 5 | NP_004610 | 1.4 |
| TRAF6 | TNFR-associated factor 6 | NP_665802 | 2.8* |
| TRAP2 | TNFR-associated protein 2 | NP_002799 | 1.1 |
| TRIP | TNFR-associated factor interacting protein | NP_005870 | 1.2 |
| TIFA | TRAF-interacting protein with FHA domain-containing protein A | NP_443096 | 1.6 |
| KCTD13 | TNF-α-induced protein 1-like adaptor protein | NP_849194 | 2.1* |
| NF-κB | Nuclear factor-kappa B subunit p105 (NFKB1/p50) | NP_003989 | 1.9* |
| IκBE | Nuclear factor-kappa B inhibitor epsilon | NP_004547 | 1.4 |
| TNFAIP3 (A20) | TNF-α-induced protein 3 | NP_006281 | 1.2 |
| TNIP1 | TNF-α-induced protein 3-interacting protein 1 | NP_006049 | 2.5* |
| TNIP2 | TNF-α-induced protein 3-interacting protein 2 | NP_077285 | 1.3 |
| OPTN | NEMO-Related Protein (NRP), optineurin | NP_068815 | 1.2 |
Retinal protein samples obtained from human donor eyes with (n = 10) or without (n = 10) glaucoma were individually analyzed by quantitative LC-MS/MS. All listed proteins were identified with high confidence (greater than 99.0% probability assigned by the Protein Prophet algorithm) based on at least two identified peptides.
Significant difference in protein expression between glaucomatous and control samples (Mann-Whitney rank sum test; P < 0.05).
As shown in Table 3, we detected the upregulation of a number of protein kinases specific to TNFR signaling, such as receptor-interacting serine-threonine kinase 1 (RIPK), NF-κB–inducing kinase (NIK), and inhibitory kappa B (IκB) kinases (IκKs) leading to NF-κB activation. In addition, we detected the upregulation of various other protein kinases in glaucomatous samples that are also linked to TNFR1 signaling. As listed in Table 3, these kinases included numerous members of MAPKs (including the apoptotic c-Jun N-terminal kinase [JNK]) and janus kinases (JAKs).
Table 3.
Analysis of Retinal Protein Expression in Human Glaucoma: Selected Protein Kinases
| Symbol | Protein Name | RefSeq | Fold Change |
|---|---|---|---|
| IKKE | Inhibitor of kappa-B kinase epsilon | NP_054721 | 1.9* |
| IKKG (NEMO) | NF-κB essential modulator, inhibitor of kappa-B kinase gamma | NP_003630 | 3.4* |
| RIPK1 | Receptor (TNFRSF)-interacting serine/threonine kinase 1 | NP_003795 | 2.2* |
| RIPK2 (CARD3) | Receptor-interacting serine/threonine kinase 2 | NP_003812 | 1.1 |
| ROCK1 | Rho-associated protein kinase 1 | NP_005397 | 2.3* |
| RAF1 (c-Raf) | Raf proto-oncogene serine/threonine-protein kinase 1 | NP_002871 | 1.4 |
| STK17A | Serine/threonine kinase 17a (apoptosis-inducing) | NP_004751 | 1.9* |
| STK25 | Serine/threonine kinase 25 (oxidant stress response kinase 1) | NP_006365 | 1.6 |
| MAP4K1 | Mitogen-activated protein kinase kinase kinase kinase 1 isoform 2 | NP_009112 | 1.2 |
| MAP4K2 (GCK) | Mitogen-activated protein kinase kinase kinase kinase 2 | NP_004570 | 6.4* |
| MAP4K4 | Mitogen-activated protein kinase kinase kinase kinase 4 | NP_004825 | 1.1 |
| MAP3K3 (MEKK3) | Mitogen-activated protein kinase kinase kinase 3 | NP_002392 | 1.2 |
| MAP3K4 (MEKK4) | Mitogen-activated protein kinase kinase kinase 4 | NP_005913 | 1.1 |
| MAP3K5 (MEKK5, ASK1) | Mitogen-activated protein kinase kinase kinase 5 | NP_005914 | 2.2* |
| MAP3K6 (MAKK6, ASK2) | Mitogen-activated protein kinase kinase kinase 6 | NP_004663 | 1.3 |
| MAP3K7 (MEKK7) | Mitogen-activated protein kinase kinase kinase 7 | NP_663304 | 1.7 |
| MAP3K14 (NIK) | Mitogen-activated protein kinase kinase kinase 14 | NP_003945 | 2.3* |
| MAP3K18 (TAOK3) | Mitogen-activated protein kinase kinase kinase 18 | NP_057365 | 2.4* |
| MAP2K2 (MKK2, MEK2) | Mitogen-activated protein kinase kinase 2 | NP_109587 | 1.5 |
| MAP2K6 (MKK6, MEK6) | Mitogen-activated protein kinase kinase 6 | NP_002749 | 2.8* |
| MAP2K7 (MKK7) | Mitogen-activated protein kinase kinase 7 | NP_660186 | 1.4 |
| MAPK1 (ERK2) | Mitogen-activated protein kinase 1 (variant 1) | NP_002736 | 4.4* |
| MAPK1 (ERK2) | Mitogen-activated protein kinase 1 (variant 2) | NP_620407 | 1.3 |
| MAPK3 (ERK1) | Mitogen-activated protein kinase 3 | NP_002737 | 3.0* |
| MAPK7 | Mitogen-activated protein kinase 7 | NP_002740 | 1.5 |
| MAPK8 (JNK1) | Mitogen-activated protein kinase 8 | NP_620634 | 2.9* |
| JIP2 | Mitogen-activated protein kinase 8 interacting protein 2 | NP_036456 | 1.1 |
| MAPK10 (JNK3) | Mitogen-activated protein kinase 10 isoform 3 | NP_620446 | 1.1 |
| MAPK13 (p38d) | Mitogen-activated protein kinase 13 | NP_002745 | 1.3 |
| MAPKAPK5 | Mitogen-activated protein kinase-activated protein kinase 5 | NP_003659 | 1.2 |
| PAK2 | P21-activated kinase 2 | NP_002568 | 2.6* |
| PAK 5/7 | P21-activated kinase 5/7 | NP_065074 | 1.8 |
| JAK2 | Tyrosine-protein kinase Janus kinase 2 | NP_004963 | 1.5 |
| JAK3 | Tyrosine-protein kinase Janus kinase 3 | NP_000206 | 1.7 |
Retinal protein samples obtained from human donor eyes with (n = 10) or without (n = 10) glaucoma were individually analyzed by quantitative LC-MS/MS. All listed proteins were identified with high confidence (greater than 99.0% probability assigned by the Protein Prophet algorithm) based on at least two identified peptides.
Significant difference in protein expression between glaucomatous and control samples (Mann-Whitney rank sum test; P < 0.05).
Table 4 shows the proteins linked to apoptosis signaling in the glaucomatous human retina, which included caspases, such as caspase 8 (an early caspase in receptor-mediated pathway) and caspase 9 (related to mitochondrial pathway). Our data also supported the increased expression of various members of the Bcl-2 family controlling the mitochondrial cell death pathway. These included the upregulation of proapoptotic Bax (along with Bid and Bim, which were detectable only in glaucomatous samples) and antiapoptotic Bcl-XL. In addition, we detected the upregulation of various mitochondrial proteins related to the mitochondrial pathway of apoptosis, such as apoptosis-inducing factor and endonuclease G (Table 4). Also detectable was a prominent upregulation of various calcium-dependent cysteine proteases (calpains), another group of cell death mediators, in glaucomatous samples. Caspase recruitment domain-containing proteins that were also detectable in the human retina function in the regulation of both apoptosis and inflammatory responses. Many endoplasmic reticulum (ER)-resident proteins were upregulated in glaucomatous samples (Table 4). These included activating transcription factor 6, 78-kDa glucose-regulated protein, and serine/threonine-protein kinase/endoribonuclease inositol-requiring 1.
Table 4.
Analysis of Retinal Protein Expression in Human Glaucoma: Proteins Linked to Apoptosis
| Symbol | Protein Name | RefSeq | Fold Change |
|---|---|---|---|
| BCL(X)L | Bcl2-like 1 | NP_612815 | 1.9* |
| BAX | Bcl2-associated × protein | NP_620116 | 4.3* |
| BCL2L2 | Bcl2-like 2 | NP_004041 | 1.6 |
| BCL2L10 | Bcl2-like 10 | NP_065129 | 1.4 |
| BIM | Bcl2-like 11 | NP_619527 | † |
| BCL2L13 | Bcl2-like 13 | NP_056182 | 1.1 |
| BID | BH3 interacting domain-containing death agonist | NP_932070 | † |
| ASPP1 | Apoptosis-stimulating protein of p53, 1 | NP_056131 | 1.2 |
| TP53BP1 | P53-binding protein 1 | NP_005648 | 1.3 |
| CYTC | Cytochrome c, somatic | NP_061820 | 1.2 |
| AIFM1 | Apoptosis-inducing factor 1 | NP_665811 | 2.9* |
| AIFM2 | Apoptosis-inducing factor 2 | NP_116186 | 1.6 |
| ENDOG | Endonuclease G | NP_004426 | 3.0* |
| APAF1 | Apoptotic peptidase activating factor 1 | NP_863651 | 1.3 |
| CASP8 | Caspase 8 | NP_001219 | 1.7 |
| CASP9 | Caspase 9 | NP_001220 | 2.1* |
| CASP10 | Caspase 10 | NP_001221 | 1.4 |
| BCL10 | Caspase recruitment domain-containing apoptotic signaling protein | NP_003912 | 2.1* |
| CARD7 (NALP1) | Caspase recruitment domain-containing family member 7 | NP_127497 | 1.1 |
| CARD8 | Caspase recruitment domain-containing family member 8 | NP_055774 | 1.6 |
| CARD9 | Caspase recruitment domain-containing protein member 9 | NP_434700 | 1.4 |
| CARD10 (BIMP1) | Caspase recruitment domain-containing family member 10 | NP_055365 | 1.7 |
| CARD11 | Caspase recruitment domain-containing family member 11 | NP_115791 | 1.1 |
| CARD12 (IPAF) (NLRC4) | Caspase recruitment domain-containing protein member 12 | NP_067032 | 1.2 |
| PDCD10 | Programmed cell death 10; apoptosis-related protein 15 | NP_009148 | 2.7* |
| THAP1 | THAP domain-containing nuclear proapoptotic factor | NP_060575 | 2.0* |
| PAR4 | Prostate apoptosis response protein 4 | NP_002574 | 2.2* |
| ACIN1 | Apoptotic chromatin condensation inducer 1 | NP_055792 | 1.3 |
| DFFA (ICAD) | DNA fragmentation factor alpha | NP_004392 | 2.7* |
| CAPNS1 | Calpain small subunit 1 | NP_001740 | 3.8* |
| CAPN2 | Calpain 2 catalytic subunit | NP_001739 | 1.7 |
| CAPN6 | Calpain 6 | NP_055104 | 1.8* |
| CAPN9 | Calpain 9 | NP_006606 | 1.5 |
| CAPN10 | Calpain 10 | NP_075571 | 2.0* |
| CAST | Calpastatin isoform c, calpain inhibitor | NP_775084 | 2.3* |
| GRP78 (BIP) | 78 kDa glucose-regulated protein, heat shock 70kDa protein 5 | NP_005338 | 5.4* |
| DNAJC10 | DnaJ (Hsp40) homolog C 10 | NP_061854 | 2.2* |
| ERO1LB | Endoplasmic reticulum oxidoreductin 1-like beta | NP_063944 | 2.8* |
| PDI | Protein disulfide isomerase A1 | NP_000909 | 1.4 |
| PDIA6 | Protein disulfide isomerase A6 (ERP5) | NP_005733 | 3.6* |
| ERP29 | Endoplasmic reticulum protein 29 | NP_006808 | 3.3* |
| ERN1 | Serine/threonine-protein kinase/endoribonuclease IRE1 | NP_001424 | 2.5* |
| ATF6 | Activating transcription factor 6 | NP_031374 | 2.7* |
Retinal protein samples obtained from human donor eyes with (n = 10) or without (n = 10) glaucoma were individually analyzed by quantitative LC-MS/MS. All listed proteins were identified with high confidence (greater than 99.0% probability assigned by the Protein Prophet algorithm) based on at least two identified peptides.
Significant difference in protein expression between glaucomatous and control samples (Mann-Whitney rank sum test; P < 0.05).
Proteins detectable only in glaucomatous samples.
In addition to many cell death-promoting proteins, our proteomic data supported a prominent upregulation of various proteins involved in intrinsic adaptive/protective mechanisms, such as inhibitor-of-apoptosis proteins (IAPs), heat shock proteins (HSPs), and a number of antioxidants (data not shown).
Besides increased protein expression supporting NF-κB activation in the glaucomatous human retina, many other proteins linked to inflammatory pathways were upregulated in glaucomatous samples. These included various inflammasome components, including caspase 1, an inflammatory caspase (Table 5).
Table 5.
Analysis of Retinal Protein Expression in Human Glaucoma: Proteins Involved in the Inflammasome
| Symbol | Protein Name | RefSeq | Fold Change |
|---|---|---|---|
| NOD1 (CARD4) | Nucleotide-binding oligomerization domain-containing protein 1 | NP_006083 | 2.1* |
| NOD2 (CARD15) | Nucleotide-binding oligomerization domain-containing protein 2 | NP_071445 | 1.7 |
| NLRC5 | Nucleotide-binding oligomerization domain-containing protein 27 | NP_115582 | 1.4 |
| NLRP3 (NALP3) | NACHT, LRR and PYD domains-containing protein 3 isoform b | NP_899632 | 2.2* |
| CASP1 | Caspase 1 isoform alpha | NP_150634 | 3.1* |
Retinal protein samples obtained from human donor eyes with (n = 10) or without glaucoma (n = 10) were individually analyzed by quantitative LC-MS/MS. All listed proteins were identified with high confidence (greater than 99.0% probability assigned by the Protein Prophet algorithm) based on at least two identified peptides.
Significant difference in protein expression between glaucomatous and control samples (Mann-Whitney rank sum test; P < 0.05).
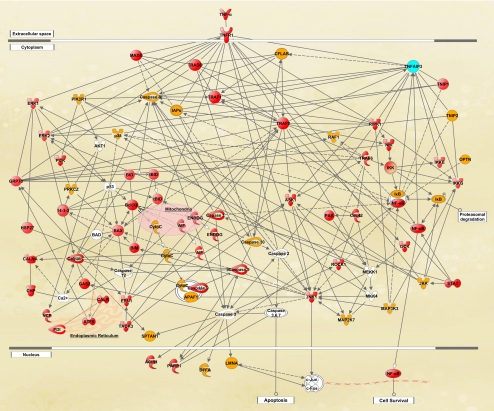
Bioinformatic analysis of the quantitative data established extended functional networks of the identified proteins with links to death-promoting and survival-promoting pathways of the TNF-α/TNFR1 signaling. Figure 1 shows a simplified version of the protein interaction network generated by IPA.
Figure 1.
Bioinformatic analysis of the comparative proteomic data. IPA established extended functional networks of the identified proteins with links to death-promoting and survival-promoting pathways of the TNF-α/TNFR1 signaling. Shown is a simplified version of the protein interactions network, in which proteins shown in red exhibited significantly increased expression and proteins shown in yellow exhibited no significantly increased expression in glaucomatous samples compared with nonglaucomatous control samples. TNFAIP3 expression, shown in blue, exhibited prominent individual differences. Proteins shown in white were not detectable by quantitative LC-MS/MS analysis (detailed information for abbreviated proteins is available at www.ncbi.nlm.nih.gov/protein).
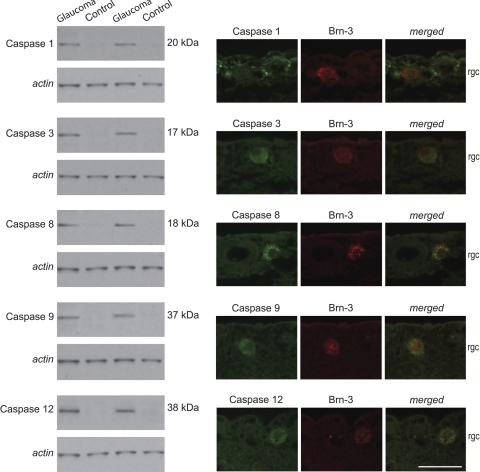
Western blot analysis and immunohistochemistry using specific antibodies to selected proteins validated increased protein expression/activation and cellular localization. To validate caspase activation during glaucomatous neurodegeneration in human eyes, we subjected our retinal protein samples to Western blot analysis and analyzed tissue sections by immunohistochemistry using cleavage site-specific antibodies. Western blot analysis supported caspase activation by protein cleavage in glaucomatous samples (Fig. 2). Based on immunohistochemical analysis, active caspases were detectable only in the inner retina of glaucomatous eyes. As shown in Figure 2, double immunofluorescence labeling of the glaucomatous human retina demonstrated prominent localization of cleaved caspases in RGCs.
Figure 2.
Caspase activation in the glaucomatous human retina. Western blot analysis of retinal protein samples using cleavage site-specific antibodies detected cleaved caspases only in glaucomatous samples. Double immunofluorescence labeling indicated prominent localization of cleaved caspases 3, 8, 9, and 12 (green) in Brn-3–positive RGCs (red) in the glaucomatous human retina, whereas cleaved caspase 1 immunolabeling (green) was more prominent in Brn-3–negative cells corresponding to glia in the inner retina. rgc, retinal ganglion cell layer. Scale bar, 50 μm.
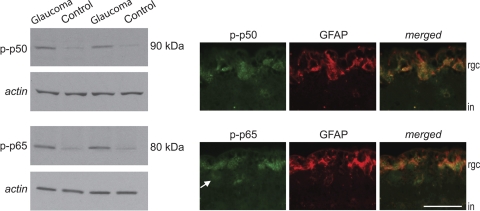
Quantitative LC-MS/MS analysis detected the upregulation of NF-κB subunit p50. Western blot analysis using phosphorylation site-specific antibodies showed significantly increased phosphorylation of this subunit and of another subunit (p65) in glaucomatous samples. Based on double-immunofluorescence labeling, phospho-p50 and phospho-p65 were predominantly localized to GFAP-positive astrocytes in the glaucomatous human retina (Fig. 3).
Figure 3.
NF-κB activation in the glaucomatous human retina. Quantitative Western blot analysis used phosphorylation site-specific antibodies to validate NF-κB activation. Based on β-actin–normalized intensity values, glaucomatous samples exhibited a prominent increase in phosphorylated p50 and p65 compared with nonglaucomatous control samples (Mann-Whitney rank sum test; P < 0.01). Double-immunofluorescence labeling of the glaucomatous human retina indicated localization of these active subunits (green) mainly in GFAP-positive astrocytes (red) in the inner retina. However, some GFAP-negative cells likely corresponding to RGCs (arrow) also exhibited weaker immunolabeling for these phospho-proteins. rgc, retinal ganglion cell; in, inner nuclear layer. Scale bar, 50 μm.
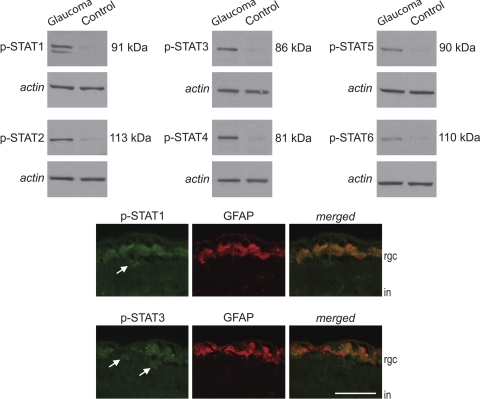
Western blot analysis using phosphorylation site-specific antibodies also supported a significant increase in STAT activation by phosphorylation in glaucomatous samples, and these signaling molecules were localized to RGCs and astrocytes (Fig. 4). We also detected some negative regulators of JAK/STAT signaling in glaucomatous samples, such as feedback inhibitor suppressor of cytokine signaling and protein inhibitors of activated STAT (data not shown).
Figure 4.
STAT activation in the glaucomatous human retina. Quantitative Western blot analysis using phosphorylation site-specific antibodies and β-actin–normalized intensity values demonstrated increased STAT phosphorylation in glaucomatous samples compared with nonglaucomatous control samples. (Mann-Whitney rank sum test; P < 0.01). Double-immunofluorescence labeling indicated localization of these phosphorylated proteins (green) in GFAP-positive astrocytes (red) and also some GFAP-negative cells, likely corresponding to RGCs (arrow) in the inner retina. rgc, retinal ganglion cell; in, inner nuclear layer. Scale bar, 50 μm.
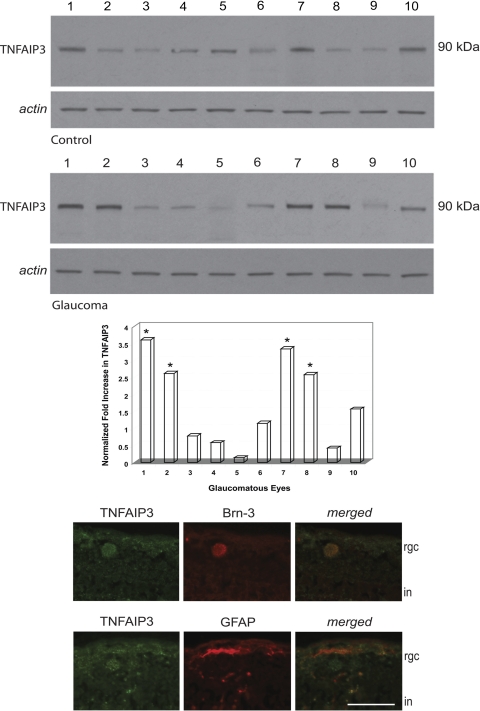
Western blot analysis also aimed to further validate individual differences in the expression of regulator molecules. However, because of the paucity of our glaucomatous donor tissues, comparative analysis of glaucomatous and nonglaucomatous human samples determined only the expression of TNFAIP3 among various regulator proteins. This analysis revealed a greater than two-fold increase in TNFAIP3 expression in 4 of 10 glaucomatous samples; however, TNFAIP3 expression was unchanged or decreased in the rest of the glaucomatous samples (Fig. 5). As shown in Figure 5, immunohistochemical analysis of the human retina demonstrated localization of this regulator protein in both Brn-3-positive RGCs and GFAP-positive astroglia. Individual differences were detectable in retinal TNFAIP3 immunolabeling among glaucomatous donors. However, unlike quantitative LC/MS/MS and Western blot analyses, immunohistochemical analysis primarily determined the cellular localization of selected proteins rather than the quantification of protein extend.
Figure 5.
TNFAIP3 expression in the glaucomatous human retina. Quantitative Western blot analysis using a specific antibody revealed prominent individual differences in the expression level of TNFAIP3. When the average β-actin–normalized intensity value obtained from control samples was used to calculate the fold change in TNFAIP3 immunolabeling, we detected a greater than two-fold increased expression in 4 of 10 glaucomatous samples. *P < 0.01, statistically significant difference (Mann-Whitney rank sum test). However, TNFAIP3 expression was unchanged or decreased in the rest of the glaucomatous samples. Immunolabeling of human tissues demonstrated TNFAIP3 (green) localization in both Brn-3–positive RGCs and GFAP-positive astrocytes (red) in the inner retina. rgc, retinal ganglion cell; in, inner nuclear layer. Scale bar, 50 μm.
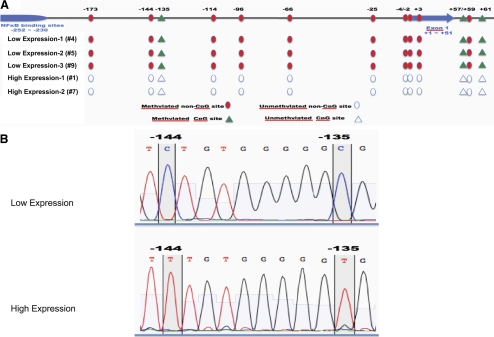
To determine whether genomic variation between donors is correlated with the observed differences in protein expression, we investigated the genomic sequence of TNFAIP3 by direct sequencing of PCR-amplified exon sequences, with particular attention directed toward several known SNPs (rs112229105, rs5029941, rs2230926, rs61756235, rs3734553, rs5029957, rs34935799). No variation was found in these sites between the donors demonstrating high or low expression levels of TNFAIP3 (data not shown). We also investigated the methylation pattern in the promoter region of TNFAIP3 using bisulfite sequence analysis. DNA was extracted from the retinal tissue obtained from 3 glaucomatous donors with low protein expression and 2 glaucomatous donors with high protein expression. As shown in Figure 6, our data indicated that one CpG site (-135) within the promoter region and two CpG sites in the first intron were consistently methylated in donors with low TNFAIP3 expression, whereas methylation was not observed in donors with high expression. Furthermore, we detected a very consistent methylation pattern of non-CpG sites. Cytosine residues at positions -173, -144, -114, - 96, -66, -4, and -2 were consistently methylated in donors with low TNFAIP3 expression (Fig. 6A). Interestingly, our data did not indicate partial conversion of cytosine residues, but all cytosine residues in these positions were either completely methylated or nonmethylated, suggesting that the methylation pattern was consistent across all retinal cell types (Fig. 6B).
Figure 6.
Cytosine nucleotide methylation differences in the TNFAIP3 promoter among glaucomatous human donors. (A) Schematic representation of human TNFAIP3 promoter methylation and location of the detected methylation sites. In glaucomatous retinas with low TNFAIP3 expression (corresponding to glaucomatous donors 4, 5, and 9 in Fig. 5), three CpG and ten non-CpG methylation sites were found. However, in glaucomatous retinas exhibiting high expression of TNFAIP3 (corresponding to glaucomatous donors 1 and 7 in Fig. 5), these sites remained unmethylated. Circles: non-CpG methylation sites; triangles: CpG methylation sites; filled symbols: methylated sites. (B) Examples of sequencing results of the TNFAIP3 promoter after bisulfite conversion in donors exhibiting low (top) or high (bottom) level of TNFAIP3 expression. In donors with high TNFAIP3 expression, the conversion of cytosine residues was complete, suggesting that the vast majority of these residues were unprotected.
Discussion
Cell Death Pathways in Glaucoma
Our high-throughput comparative data obtained by quantitative LC-MS/MS analysis of retinal protein samples, along with the findings of quantitative Western blot analysis and tissue immunolabeling, reflected a prominent upregulation of apoptosis-related pathways and markers of inflammation in human glaucoma with many links to TNF-α/TNFR1 signaling.
We used a label-free approach for quantitative analysis of protein expression by LC-MS/MS. Because of the substantial cost of stable-isotope labeling along with the complexity of sample processing and data interpretation, label-free methods have become more widely used. Although the quantitative assessment of spectral counts presents advantages, a potential caveat is the individual analysis of samples as opposed to mixing the comparative samples before analysis, which is a primary advantage of labeling to eliminate experimental variability. However, previous studies4,5,7 using the label-free approach demonstrated a high level of reproducibility validated by immunoblotting and immunohistochemistry. Our findings here similarly support the usefulness of this method to identify valid expression changes even in relatively low-abundant proteins in glaucomatous tissues. Similar to LC-MS/MS, Western blot analysis-based quantification may be challenging primarily because of variability in transfer and amplification steps. With regard for the preparation-related variability in sample conditions, transfer efficiency, and backgrounds, we reprobed immunoblots with a β-actin antibody and repeated the analysis at least three times by running glaucomatous or nonglaucomatous samples on the same gel and in different combinations with similar results.
Retinal proteins exhibiting increased expression in human glaucoma included TNF-α, TNFR1, and various downstream adaptor/interacting proteins and protein kinases known to regulate diverse consequences of TNF-α/TNFR1 signaling. For example, a proteolytic caspase cascade leads to apoptosis after TNFR1 binding; however, the signaling cascade activating NF-κB primarily promotes cell survival and regulates the expression of a wide variety of proteins that control innate and adaptive immunity.13 One of the specific molecules we detected was TRADD. This signal transducer protein is a component of the multiprotein signaling complex formed after TNFR1 binding, which recruits various proteins including members of the TRAF family. A number of proteins associated with the death receptor-mediated caspase cascade and NF-κB activation appear to bifurcate at TRADD.14
In addition to data supporting the receptor-mediated caspase cascade, cell death signaling in the glaucomatous human retina exhibited links to the mitochondrial pathway.1 Among various proapoptotic members of the Bcl-2 family regulating this pathway, we detected the upregulation of Bax, which is a principal regulator of RGC death.15 We also detected (BH3)-only proapoptotic members of the family in glaucomatous samples, including Bid and Bim. With particular relevance to TNFR signaling, Bid participates in the activation of the mitochondrial cell death pathway on cleavage by caspase 8, a proximal caspase activated after TNFR1 binding.16 Previous studies have implicated Bid in RGC apoptosis in experimental glaucoma17 and Bim in RGC death after optic nerve axotomy.18,19 It has become clear that even with the lack of detectable change in their expression in animal models of glaucoma, (BH3)-only proteins potentiate Bax-mediated cell death by neutralizing antiapoptotic proteins such as Bcl-XL.20
We also detected the increased expression of various ER-resident proteins, including stress-regulated chaperones that catalyze protein folding and function as sensors detecting unfolded protein response (UPR).21 Although UPR is an adaptive response to preserve cell function and survival, its persistence initiates apoptotic cascades, and has been implicated in the pathogenesis of multiple human diseases as in experimental glaucoma.22 In addition to UPR, disturbances in ER calcium homeostasis and redox changes may have important links to ER stress and communications with mitochondria.23 By providing a unique oxidizing environment for disulfide bond formation during protein folding, ER may significantly contribute to mitochondria-generated oxidative stress.24,25 There appears to be a vicious relationship between ER stress and oxidative stress that is likely to play a role in increasing cellular susceptibility to neurodegenerative injury in glaucoma.
Our data also supported the increased expression of calpains in the glaucomatous human retina, which have been shown to contribute to neuronal death in ocular hypertensive rats.26 Besides caspase-independent proteolytic activities, calpains cleave and activate an ER protein, caspase 12, thereby providing a link to the caspase-mediated apoptosis pathway.27
Thus, our data support the coactivation of different apoptotic pathways in the glaucomatous human retina, including caspase- and calpain-mediated pathways, mitochondrial dysfunction, and ER stress. Cross-talk between these pathways may reinforce each other during the apoptotic process in human glaucoma.
Immune Response Pathways in Glaucoma
Opposing the mediation of cell death, we also detected NF-κB activation in the glaucomatous human retina, which plays an essential role as a key regulator of neuronal survival programs induced by TNF-α signaling.28 NF-κB may promote cell survival by inhibiting JNK and inducing antiapoptotic Bcl-2 members, IAPS, and HSPs.29,30 In addition, by controlling the transcription of immune mediators, NF-κB regulates various aspects of innate and adaptive immune responses.31 In glaucomatous samples, we detected the phosphorylation of NF-κB1-p105/p50 and p65 (RelA), two of five distinct but structurally related subunits with specific signaling functions (the others are NF-κB2-p100/52, RelB, and c-Rel). We also detected the increased expression of related kinases, such as RIPK, NIK, and IκK, including a master regulator, IκKγ (NF-κB essential modulator [NEMO]). The activation of NF-κB primarily occurs through activation of the IκK, composed of a heterodimer of the catalytic IκKα and IκKβ subunits and NEMO. Both p50 and p52 subunits participate in target gene transactivation by forming heterodimers with RelA, RelB, or c-Rel. In contrast to the canonical signaling that relies on NEMO-IκK–mediated degradation of IκB followed by RelA/p50 signaling, noncanonical signaling critically depends on the NIK-mediated processing of p100 into p52 and RelB/p52 signaling.13,32 Diverse functions of NF-κB as a master regulator of inflammatory responses and secondary injury processes in the CNS may depend on cell-specific factors and unbalanced activation of different subunit complexes.33
In addition to various other kinase pathways, our proteomic data indicated JAK/STAT signaling in the glaucomatous human retina. This signaling pathway can be triggered in response to multiple stimuli, including TNFR1 binding, and mediates cytokine-mediated inflammatory responses in the CNS.34,35 Recent studies have documented that various components of the JAK/STAT signaling pathway are upregulated in the retina36 and optic nerve37 of ocular hypertensive rats.
Besides the caspases leading to apoptotic cell death, we detected caspase 1 activation in our glaucomatous samples. Stress-induced activation of caspase 1 is recognized as an essential regulator of inflammatory responses through its capacity to process and activate proinflammatory cytokines; therefore, this caspase is considered an inflammatory caspase. In addition to previous evidence supporting TLR signaling5 and complement activation,4 our new data support inflammasome assembly leading to caspase 1 activation in the glaucomatous human retina. These cytosolic multiprotein complexes are assembled as an early innate response to cell stress, and activated caspase 1 initiates an inflammatory cascade by promoting the proteolytic activation of pro-interleukins and the secretion of mature cytokines.38,39 TNF-α signaling has been shown to promote inflammasome activation mediating sterile inflammation,40,41 and the NLRP3 inflammasome has been implicated in the development of neuroinflammation in the CNS.42,43
Thus, presented data support that in addition to NF-κB–mediated inflammation signaling, JAK/STAT signaling and inflammasome may be involved in immune response pathways activated in the glaucomatous human retina and may represent promising targets for immunomodulatory treatment strategies.
Regulation of TNF-α Signaling in Glaucoma
Our proteomic data indicated some specific regulator molecules. One of these molecules was CFLAR, a protease-deficient caspase homolog protein widely regarded as an apoptosis inhibitor.44,45
We also detected optineurin in the human retinal proteome. Based on gene mutations detected among glaucoma patients, optineurin is proposed to be associated with TNF-α–mediated RGC death.46 This TNF-α–inducible protein expressed by RGCs47 appears to constitute a cellular stress sensor mechanism transmitting survival signals.48 A more recent study using microRNA silencing has shown that optineurin inhibits TNF-α–induced NF-κB activation by competitively antagonizing NEMO for RIPK binding.49 It is tempting to further determine whether gene mutations may affect the physiologic function of optineurin to dampen TNF-α signaling, thereby increasing neuronal susceptibility to glaucomatous injury.
Another important regulator molecule we detected was TNFAIP3; however, as verified by quantitative Western blot analysis, its expression level exhibited a prominent variability among glaucomatous donors. This cytoplasmic ubiquitin-editing zinc finger protein plays a key role in the negative regulation of TNF-α signaling by functioning as a dual inhibitor of NF-κB activation and TNF-α–mediated apoptosis.50 By antagonizing interactions with ubiquitin-conjugating enzymes, TNFAIP3 may inactivate various molecules downstream of TNFR1,51 block JNK activation, and inhibit proteolytic cleavage of caspase 8.52,53 TNFAIP3-mediated inhibition of the caspase cascade effectively protects neurons from postischemic apoptosis in the CNS.54 In addition to antiapoptotic activities, TNFAIP3 is involved in the negative feedback regulation of NF-κB signaling by its interaction with various upstream signaling molecules, modulating their ubiquitination and proteasome-mediated degradation.55,56 TNFAIP3, acting through NF-κB signaling, restricts innate and adaptive immune responses and ensures the transient nature of inflammatory signaling. Consequently, reduced TNFAIP3 expression is suggested to predispose to autoimmunity as well as increasing the susceptibility to neuronal injury.50,57 The essential role of TNFAIP3 in the regulation of apoptosis and NF-κB signaling has been clearly demonstrated with the generation of TNFAIP3 knockout mice, which develop severe inflammation in multiple organs and die prematurely at 3 to 6 weeks of age. TNFAIP3-deficient cells fail to terminate TNF-α–induced NF-κB activation and become more susceptible to TNF-α–mediated apoptosis.58 In addition, the RNA interference-mediated downregulation of TNFAIP3 in human dendritic cells results in enhanced stimulation of cytotoxic T cells and inhibition of regulatory T cells.59,60 Given the key functions of TNFAIP3 in the regulation of cell death and the prevention of autoimmunity, it would be interesting to determine whether aberrations in its expression may increase RGC susceptibility to TNF-α–mediated apoptosis or may alter the intensity or duration of immune responses in glaucoma.
Consistent with previous experimental findings, recent genetic studies have demonstrated several mutations in the human TNFAIP3 locus as risk alleles for multiple autoimmune diseases in humans.50 Findings of these studies motivated us to determine whether the variability in TNFAIP3 expression among glaucomatous donors reflects a similar association. We therefore initiated analyses of genetic and epigenetic differences across these samples. Despite the lack of any detectable genomic variation correlated to individual differences in protein expression, our data obtained from bisulfate sequencing demonstrated that the methylation of cytosine nucleotides in the TNFAIP3 promoter is correlated with the variability in retinal protein expression among glaucomatous donors. Although bisulfate sequencing is inherently challenging, as the electropherograms demonstrated, potential problems such as bisulfate treatment-related DNA degradation, incomplete conversion, or differential PCR amplification rates of converted and unconverted sequences did not occur in our hands. Cytosine nucleotide methylation is one of the most critical epigenetic mechanisms for gene silencing described for TNFAIP3. This gene has been shown to be inactivated because of partial methylation of several CpG sites upstream of exon 1.61 In the DNA extracted from glaucomatous retinas, we observed methylation of only one of these sites but detected the methylation of several cytosine residues not followed by guanine residues. Although cytosine methylation of the CpG dinucleotide is well documented,62 non-CpG methylation has more recently been described,63,64 and emerging data indicate that this type of methylation may result from de novo methylation mediated by the methyltransferases DNMT3a and DNMT3b.65,66 It is of interest to note that differential methylation may also be related to differential demethylation caused by demethylase,67 glycosylase,68 or other related enzymatic activities.69 It appeals further studies to determine whether detected methylation differences in the TNFAIP3 promoter may represent an epigenetic modification that could modify a person's response to TNF-α–mediated processes in glaucoma.
Diverse Consequences of TNF-α Signaling in Glaucoma
By highlighting various proteins linked to TNF-α/TNFR1 signaling in the glaucomatous human retina, findings of this study support that a complex cross-talk relationship between multiple signaling pathways determines diverse consequences of TNF-α signaling.3 Factors determining opposing effects of TNF-α signaling also include the type of receptor preferentially used. Two cell surface receptors, p55 (TNFR1) and p75 (TNFR2), mediate biological activities of TNF-α. These two receptors are co-expressed on most cell types and feed into diverse signaling pathways according to differences in their intracellular domains. A death domain in TNFR1, not present in TNFR2, leads to apoptotic cell death, whereas signaling through TNFR2 leads primarily to cell proliferation. Similarly, TNFR1 has been found to augment neuronal death and TNFR2 has been found to promote neuroprotection in a retinal ischemia model in knockout mice.70 No increase was detectable in the expression of TNFR2 in the glaucomatous human retina. A recent study71 of an experimental rat glaucoma model has supported that signaling through TNFR2 may be neurotoxic through a paracrine mechanism by increasing the glial production of neurotoxic proteins, including TNF-α. Another study72 has similarly shown that activation of this receptor may trigger RGC death through a non-cell–autonomous signaling pathway by inducing TNF-α production in Müller cells. These findings collectively suggest that our proteomic data supportive of TNF-α–mediated cell death signaling in human glaucoma may predominantly reflect TNFR1 signaling. Regarding inflammation signaling, studies using receptor-specific antibodies,73 ligands,74 and knockout mice75–77 have indicated that TNFR1 is the primary signaling receptor on most cell types through which the majority of inflammatory responses classically attributed to TNF-α occur. Furthermore, soluble TNF-α, more than its membrane-bound form, is required to generate neuroinflammation,78 which is the principal ligand for TNFR1.79 Thus, TNFR1 appears to be the primary receptor for both neurodegenerative and inflammatory consequences of TNF-α signaling in glaucoma. However, any contribution of TNFR2 signaling to retinal proteomic components in glaucoma must be further determined by loss-of-function studies.
Diverse functional characteristics of TNF-α may also be attributed to time-dependent factors and the duration of TNF-α signaling. Because activated glial cells are the major source of increased TNF-α production in glaucoma, the activation status of different glial cell types and the time course of specific glial responses are particularly important. Our parallel studies using a cell-specific proteomic approach in animal models should facilitate improved understanding of the receptor type-related, cell type-specific, and time-dependent components of TNF-α signaling and should help identify new treatment targets for glaucoma.
Footnotes
Supported in part by National Eye Institute Grants R01 EY013813, R01 EY017131 (GT), and EY019485 (MHK), The Robert W. Rounsavall, Jr. Family Foundation, Inc. (GT), and by an unrestricted grant from Research to Prevent Blindness Inc. (Department of Ophthalmology and Visual Sciences).
Disclosure: X. Yang, None; C. Luo, None; J. Cai, None; D.W. Powell, None; D. Yu, None; M.H. Kuehn, None; G. Tezel, None
References
- 1. Tezel G. The role of glia, mitochondria, and the immune system in glaucoma. Invest Ophthalmol Vis Sci. 2009;50:1001–1012 [DOI] [PubMed] [Google Scholar]
- 2. Tezel G. The immune response in glaucoma: a perspective on the roles of oxidative stress. Exp Eye Res. 2011;93:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tezel G. TNF-alpha signaling in glaucomatous neurodegeneration. Prog Brain Res. 2008;173:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tezel G, Yang X, Luo C, et al. Oxidative stress and the regulation of complement activation in human glaucoma. Invest Ophthalmol Vis Sci. 2010;51:5071–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luo C, Yang X, Kain AD, Powell DW, Kuehn MH, Tezel G. Glaucomatous tissue stress and the regulation of immune response through glial toll-like receptor signaling. Invest Ophthalmol Vis Sci. 2010;51:5697–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tezel G, Luo C, Yang X. Accelerated aging in glaucoma: immunohistochemical assessment of advanced glycation end products in the human retina and optic nerve head. Invest Ophthalmol Vis Sci. 2007;48:1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cummins TD, Barati MT, Coventry SC, Salyer SA, Klein JB, Powell DW. Quantitative mass spectrometry of diabetic kidney tubules identifies GRAP as a novel regulator of TGF-beta signaling. Biochim Biophys Acta. 2010;1804:653–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392 [DOI] [PubMed] [Google Scholar]
- 9. Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658 [DOI] [PubMed] [Google Scholar]
- 10. Powell DW, Weaver CM, Jennings JL, et al. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol. 2004;24:7249–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Merchant ML, Cummins TD, Wilkey DW, et al. Proteomic analysis of renal calculi indicates an important role for inflammatory processes in calcium stone formation. Am J Physiol Renal Physiol. 2008;295:F1254–F1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431 [DOI] [PubMed] [Google Scholar]
- 13. Bouwmeester T, Bauch A, Ruffner H, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105 [DOI] [PubMed] [Google Scholar]
- 14. Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308 [DOI] [PubMed] [Google Scholar]
- 15. Libby RT, Li Y, Savinova OV, et al. Susceptibility to neurodegeneration in a glaucoma is modified by bax gene dosage. PLoS Genet. 2005;1:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490 [DOI] [PubMed] [Google Scholar]
- 17. Huang W, Dobberfuhl A, Filippopoulos T, et al. Transcriptional upregulation and activation of initiating caspases in experimental glaucoma. Am J Pathol. 2005;167:673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Napankangas U, Lindqvist N, Lindholm D, Hallbook F. Rat retinal ganglion cells upregulate the pro-apoptotic BH3-only protein Bim after optic nerve transection. Brain Res Mol Brain Res. 2003;120:30–37 [DOI] [PubMed] [Google Scholar]
- 19. McKernan DP, Cotter TG. A critical role for Bim in retinal ganglion cell death. J Neurochem. 2007;102:922–930 [DOI] [PubMed] [Google Scholar]
- 20. Nickells RW. Variations in the rheostat model of apoptosis: what studies of retinal ganglion cell death tell us about the functions of the Bcl2 family proteins. Exp Eye Res. 2010;91:2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529 [DOI] [PubMed] [Google Scholar]
- 22. Doh SH, Kim JH, Lee KM, Park HY, Park CK. Retinal ganglion cell death induced by endoplasmic reticulum stress in a chronic glaucoma model. Brain Res. 2010;1308:158–166 [DOI] [PubMed] [Google Scholar]
- 23. Malhotra JD, Miao H, Zhang K, et al. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc Natl Acad Sci U S A. 2008;105:18525–18530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harding HP, Zhang Y, Zeng H, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633 [DOI] [PubMed] [Google Scholar]
- 25. Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang W, Fileta J, Rawe I, Qu J, Grosskreutz CL. Calpain activation in experimental glaucoma. Invest Ophthalmol Vis Sci. 2010;51:3049–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakagawa T, Yuan J. Cross-talk between two cysteine protease families: activation of caspase-12 by calpain in apoptosis. J Cell Biol. 2000;150:887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death [see comments]. Science. 1996;274:782–784 [DOI] [PubMed] [Google Scholar]
- 29. Nakano M, Knowlton AA, Yokoyama T, Lesslauer W, Mann DL. Tumor necrosis factor-alpha-induced expression of heat shock protein 72 in adult feline cardiac myocytes. Am J Physiol. 1996;270:H1231–H1239 [DOI] [PubMed] [Google Scholar]
- 30. Tang G, Minemoto Y, Dibling B, et al. Inhibition of JNK activation through NF-kappaB target genes. Nature. 2001;414:313–317 [DOI] [PubMed] [Google Scholar]
- 31. Harari OA, Liao JK. NF-kappaB and innate immunity in ischemic stroke. Ann N Y Acad Sci. 2010;1207:32–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sarnico I, Lanzillotta A, Benarese M, et al. NF-kappaB dimers in the regulation of neuronal survival. Int Rev Neurobiol. 2009;85:351–362 [DOI] [PubMed] [Google Scholar]
- 33. Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362 [DOI] [PubMed] [Google Scholar]
- 34. Guo D, Dunbar JD, Yang CH, Pfeffer LM, Donner DB. Induction of Jak/STAT signaling by activation of the type 1 TNF receptor. J Immunol. 1998;160:2742–2750 [PubMed] [Google Scholar]
- 35. Wang Y, Wu TR, Cai S, Welte T, Chin YE. Stat1 as a component of tumor necrosis factor alpha receptor 1-TRADD signaling complex to inhibit NF-kappaB activation. Mol Cell Biol. 2000;20:4505–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang DY, Ray A, Rodgers K, et al. Global gene expression changes in rat retinal ganglion cells in experimental glaucoma. Invest Ophthalmol Vis Sci. 2010;51:4084–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson EC, Doser TA, Cepurna WO, et al. Cell proliferation and interleukin-6-type cytokine signaling are implicated by gene expression responses in early optic nerve head injury in rat glaucoma. Invest Ophthalmol Vis Sci. 2011;52:504–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426 [DOI] [PubMed] [Google Scholar]
- 39. Ogura Y, Sutterwala FS, Flavell RA. The inflammasome: first line of the immune response to cell stress. Cell. 2006;126:659–662 [DOI] [PubMed] [Google Scholar]
- 40. Jain N, Sudhakar C, Swarup G. Tumor necrosis factor-alpha-induced caspase-1 gene expression = role of p73. FEBS Lett J. 2007;274:4396–4407 [DOI] [PubMed] [Google Scholar]
- 41. Franchi L, Eigenbrod T, Nunez G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol. 2009;183:792–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gris D, Ye Z, Iocca HA, et al. NLRP3 plays a critical role in the development of experimental autoimmune encephalomyelitis by mediating Th1 and Th17 responses. J Immunol. 2010;185:974–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jha S, Srivastava SY, Brickey WJ, et al. The inflammasome sensor, NLRP3, regulates CNS inflammation and demyelination via caspase-1 and interleukin-18. J Neurosci. 2010;30:15811–15820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Irmler M, Thome M, Hahne M, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195 [DOI] [PubMed] [Google Scholar]
- 45. Chang DW, Xing Z, Pan Y, et al. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21:3704–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rezaie T, Child A, Hitchings R, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079 [DOI] [PubMed] [Google Scholar]
- 47. Wang JT, Kunzevitzky NJ, Dugas JC, Cameron M, Barres BA, Goldberg JL. Disease gene candidates revealed by expression profiling of retinal ganglion cell development. J Neurosci. 2007;27:8593–8603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. De Marco N, Buono M, Troise F, Diez-Roux G. Optineurin increases cell survival and translocates to the nucleus in a Rab8-dependent manner upon an apoptotic stimulus. J Biol Chem. 2006;281:16147–16156 [DOI] [PubMed] [Google Scholar]
- 49. Zhu G, Wu CJ, Zhao Y, Ashwell JD. Optineurin negatively regulates TNFalpha- induced NF-kappaB activation by competing with NEMO for ubiquitinated RIP. Curr Biol. 2007;17:1438–1443 [DOI] [PubMed] [Google Scholar]
- 50. Vereecke L, Beyaert R, van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 2009;30:383–391 [DOI] [PubMed] [Google Scholar]
- 51. Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lademann U, Kallunki T, Jaattela M. A20 zinc finger protein inhibits TNF-induced apoptosis and stress response early in the signaling cascades and independently of binding to TRAF2 or 14–3-3 proteins. Cell Death Differ. 2001;8:265–272 [DOI] [PubMed] [Google Scholar]
- 53. He KL, Ting AT. A20 inhibits tumor necrosis factor (TNF) alpha-induced apoptosis by disrupting recruitment of TRADD and RIP to the TNF receptor 1 complex in Jurkat T cells. Mol Cell Biol. 2002;22:6034–6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu L, Miao H, Hou Y, Zhang B, Guo L. Neuroprotective effect of A20 on TNF-induced postischemic apoptosis. Neurochem Res. 2006;31:21–32 [DOI] [PubMed] [Google Scholar]
- 55. Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappaB activation. Proc Natl Acad Sci U S A. 1996;93:6721–6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Heyninck K, De Valck D, Vanden Berghe W, et al. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol. 1999;145:1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tavares RM, Turer EE, Liu CL, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee EG, Boone DL, Chai S, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Song XT, Evel-Kabler K, Shen L, Rollins L, Huang XF, Chen SY. A20 is an antigen presentation attenuator, and its inhibition overcomes regulatory T cell-mediated suppression. Nat Med. 2008;14:258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Breckpot K, Aerts-Toegaert C, Heirman C, et al. Attenuated expression of A20 markedly increases the efficacy of double-stranded RNA-activated dendritic cells as an anti-cancer vaccine. J Immunol. 2009;182:860–870 [DOI] [PubMed] [Google Scholar]
- 61. Chanudet E, Huang Y, Ichimura K, et al. A20 is targeted by promoter methylation, deletion and inactivating mutation in MALT lymphoma. Leukemia. 2010;24:483–487 [DOI] [PubMed] [Google Scholar]
- 62. Gruenbaum Y, Stein R, Cedar H, Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981;124:67–71 [DOI] [PubMed] [Google Scholar]
- 63. Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Han H, Cortez CC, Yang X, Nichols PW, Jones PA, Liang G. DNA methylation directly silences genes with non-CpG island promoters and establishes a nucleosome occupied promoter. Hum Mol Genet. 2011;20:4299–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002;289:41–48 [DOI] [PubMed] [Google Scholar]
- 66. Barres R, Osler ME, Yan J, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198 [DOI] [PubMed] [Google Scholar]
- 67. Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583 [DOI] [PubMed] [Google Scholar]
- 68. Cortellino S, Xu J, Sannai M, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fuso A, Ferraguti G, Grandoni F, et al. Early demethylation of non-CpG, CpC-rich, elements in the myogenin 5′-flanking region = a priming effect on the spreading of active demethylation. Cell Cycle. 2010;9:3965–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fontaine V, Mohand-Said S, Hanoteau N, Fuchs C, Pfizenmaier K, Eisel U. Neurodegenerative and neuroprotective effects of tumor necrosis factor (TNF) in retinal ischemia: opposite roles of TNF receptor 1 and TNF receptor 2. J Neurosci. 2002;22:RC216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bai Y, Dergham P, Nedev H, et al. Chronic and acute models of retinal neurodegeneration TrkA activity are neuroprotective whereas p75NTR activity is neurotoxic through a paracrine mechanism. J Biol Chem. 2010;285:39392–39400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lebrun-Julien F, Bertrand MJ, De Backer O, et al. ProNGF induces TNFalpha-dependent death of retinal ganglion cells through a p75NTR non-cell-autonomous signaling pathway. Proc Natl Acad Sci U S A. 2010;107:3817–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sheehan KC, Pinckard JK, Arthur CD, Dehner LP, Goeddel DV, Schreiber RD. Monoclonal antibodies specific for murine p55 and p75 tumor necrosis factor receptors: identification of a novel in vivo role for p75. J Exp Med. 1995;181:607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Van Ostade X, Vandenabeele P, Everaerdt B, et al. Human TNF mutants with selective activity on the p55 receptor. Nature. 1993;361:266–269 [DOI] [PubMed] [Google Scholar]
- 75. Rothe J, Lesslauer W, Lotscher H, et al. Mice lacking the tumour necrosis factor receptor 1 are resistant to TNF-mediated toxicity but highly susceptible to infection by Listeria monocytogenes. Nature. 1993;364:798–802 [DOI] [PubMed] [Google Scholar]
- 76. Barbara JA, Smith WB, Gamble JR, et al. Dissociation of TNF-alpha cytotoxic and proinflammatory activities by p55 receptor- and p75 receptor-selective TNF-alpha mutants. EMBO J. 1994;13:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Peschon JJ, Torrance DS, Stocking KL, et al. TNF receptor-deficient mice reveal divergent roles for p55 and p75 in several models of inflammation. J Immunol. 1998;160:943–952 [PubMed] [Google Scholar]
- 78. Ruuls SR, Hoek RM, Ngo VN, et al. Membrane-bound TNF supports secondary lymphoid organ structure but is subservient to secreted TNF in driving autoimmune inflammation. Immunity. 2001;15:533–543 [DOI] [PubMed] [Google Scholar]
- 79. Grell M, Douni E, Wajant H, et al. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell. 1995;83:793–802 [DOI] [PubMed] [Google Scholar]