Abstract
With rapid advances in our knowledge of the human genome and increasing availability of high-throughput investigative technology, genome-wide association (GWA) studies have recently gained marked popularity. As an unbiased approach to identifying genomic regions of importance in complex human disease, the results of such studies have the potential to illuminate novel causal pathways, guide mechanistic research, and aid in prediction of disease risk. The use of a genome-wide approach presents considerable methodological and statistical challenges, and properly conducted studies are essential to avoid false-positive results. A total of 22 GWA studies have been published in pulmonary medicine thus far, implicating several intriguing genomic regions in the determination of pulmonary function measures, onset of asthma, and susceptibility to chronic obstructive pulmonary disease. Many questions remain, however, as most identified genetic variants contribute only nominally to overall disease risk, genetic disease mechanisms remain uncertain, and disease-associated variants are not consistent across studies. Perhaps most fundamentally, the association signals identified have not yet been traced to causal variants. This perspective will review the current state of GWA studies in pulmonary disease. We begin with an introduction to the hypothesis, principles, and limitations of this type of genome-wide approach, highlight key points from available studies, and conclude by addressing future approaches to better understand the genetics of complex pulmonary disease.
Keywords: genetics, chronic obstructive pulmonary disease, asthma
Since publication of the first genome-wide association (GWA) study in 2005 (1), the GWA technique has rapidly gained popularity as a strategy to investigate the genetic basis of complex disease. In excess of 1,200 GWA studies are currently published, reporting on over 200 human traits and disease states (2). As a result of such studies, novel insights into disease pathogenesis have been achieved for a diverse range of conditions, from macular degeneration to inflammatory bowel disease. For the purpose of this article, we will consider the definition of a GWA study as one in which a minimum of 100,000 individual variants were genotyped in the initial patient sample. As of February 4, 2011, a total of 22 GWA studies have been published in the realm of pulmonary medicine, including 3 studies that have evaluated genes associated with pulmonary function measures (3–5), 13 that have focused on the genetic basis of asthma (6–18), and 3 that have reported susceptibility loci in chronic obstructive pulmonary disease (COPD) (19–21). In addition, interstitial pulmonary fibrosis (IPF) (22), cystic fibrosis (CF) (23), and sarcoidosis (24) have each been evaluated in one GWA study. The patient population, initial sample size, and key findings for each of the pulmonary GWA studies are illustrated in Table 1.
TABLE 1.
CHARACTERISTICS OF GENOME-WIDE ASSOCIATION STUDIES PUBLISHED IN PULMONARY MEDICINE
| Study | Phenotype | Initial Sample No,/Ancestry | Associated Genes* | OR or Beta Estimate (95% CI) |
| Wilk, 2009 | Pulmonary function | 7,691 individuals Northern European | HHIP (FEV1/FVC) | 1% increase (NR) |
| Repapi, 2009 | Pulmonary function | 20,288 individuals | AGER (FEV1/FVC) | 0.09% variance explained (NR) |
| Northern European | DAAM2 (FEV1/FVC) | 0.07% variance explained (NR) | ||
| GSTCD (FEV1) | 0.14% variance explained (NR) | |||
| HHIP (FEV1/FVC) | 0.27% variance explained (NR) | |||
| HTR4 (FEV1) | 0.07% variance explained (NR) | |||
| THSD4 (FEV1/FVC) | 0.09% variance explained (NR) | |||
| TNS1 (FEV1) | 0.07% variance explained (NR) | |||
| Hancock, 2009 | Pulmonary function | 20,890 individuals | ADAM19 (FEV1/FVC) | 0.38% increase (NR) |
| Northern European | AGER, PPT2 (FEV1/FVC) | 1% increase (NR) | ||
| FAM13A (FEV1/FVC) | 0.3% increase (NR) | |||
| GPR126 (FEV1/FVC) | 0.42% increase (NR) | |||
| HHIP (FEV1/FVC) | 0.52% increase (NR) | |||
| HTR4 (FEV1/FVC) | 0.4% increase (NR) | |||
| INTS12, NPNT, FLJ20184, GSTCD (FEV1) | 64.7mL increase (NR) | |||
| PTCH1 (FEV1/FVC) | 0.5% increase (NR) | |||
| Pillai, 2009 | COPD | 823 cases | CHRNA3, CHRNA5, IREB2 | 1.4 (1.18–1.67) |
| 810 smoking controls | PSMA4, NP_001013641.2, Q9UD29 | 1.38 (1.17–1.63) | ||
| Northern European | HHIP | |||
| Cho, 2010 | COPD | 2,940 cases | CHRNA3, CHRNA5, IREB2 | 1.30 (1.18–1.43) |
| 1,380 smoking controls | FAM13A | 1.32 (1.19–1.47) | ||
| Northern European | HHIP | 1.25 (1.14–1.37) | ||
| Kong, 2010 | COPD | 2,542 individuals | AC006320.2 | 1.47 (NR) |
| Northern European | ADARB2 | 1.47 (NR) | ||
| AL139 | 2.05 Hounsfield unit increase (NR) | |||
| BICD1 | 1.46 (NR) | |||
| CDH6 | 1.83 Hounsfield unit increase (NR) | |||
| CSMD1 | 2.19 (NR) | |||
| PARD3 | 1.85 Hounsfield unit increase (NR) | |||
| Moffatt, 2007 | Asthma, childhood onset | 994 cases | ORMDL3 | 1.45 (1.17–1.81) |
| 1,243 controls | ||||
| Northern European | ||||
| Hancock, 2009 | Asthma, childhood onset | 492 case trios | TLE4, CHCHD9 | 1.64 (1.32–2.04) |
| Mexican | ||||
| Himes, 2009 | Asthma, childhood onset | 422 cases | PDE4D | 1.18 (1.08–1.30) |
| 1,533 controls | ||||
| Northern European | ||||
| Kim, 2009 | Asthma, toluene induced | 84 cases | CTTNA3 | 5.0 (2.36–10.6) |
| 263 controls | Intergenic | 5.29 (2.41–11.61) | ||
| Korean | Intergenic | 5.20 (2.47–10.92) | ||
| Mathias, 2009 | Asthma | 464 cases | NA† | NR |
| 471 controls | ||||
| African | ||||
| Sleiman, 2010 | Asthma, childhood onset | 793 cases | DENND1B | 1.43 (NR) |
| 1988 controls | ||||
| Northern European | ||||
| Li, 2010 | Asthma, severe | 607 cases | MAVS | NR |
| 3,294 controls | SCG3 | NR | ||
| Northern European | RAD50 | 1.64 (1.36–1.97) | ||
| Himes, 2010 | Asthma, childhood onset | 359 cases | NA | NR |
| 846 controls | ||||
| Northern European | ||||
| Moffatt, 2010 | Asthma | 10,365 cases | GSDMA | 1.17 (1.11–1.23) |
| 16,110 controls | GSDMB | 1.18 (1.11–1.23) | ||
| Northern European | HLA-DQ | 1.18 (1.13–1.24) | ||
| IL2RB | 1.12 (1.08–1.16) | |||
| IL13 | 1.15 (1.09–1.20) | |||
| IL18R1 | 1.15 (1.10–1.20) | |||
| IL33 | 1.20 (1.13–1.28) | |||
| RORA | 1.18 (1.11–1.25) | |||
| SLC22A5 | 1.11 (1.06–1.15) | |||
| SMAD3 | 1.12 (1.09–1.16) | |||
| Dewan, 2010 | Asthma, childhood onset | 66 cases | NA | NR |
| 42 controls | ||||
| Various | ||||
| Ferreira, 2010 | Asthma | 986 cases | ORMDL3 | 1.33 (NR) |
| 1846 controls | ||||
| Northern European | ||||
| Kim, 2010 | Asthma, aspirin intolerant | 80 cases | NA | NR |
| 100 controls | ||||
| Korean | ||||
| Ege, 2011 | Asthma and atopy | 850 atopic asthma | NA | NR |
| 348 asthma only | ||||
| 510 controls | ||||
| Central European | ||||
| Gu, 2009 | Cystic fibrosis severity | 160 severe cases | NA | NR |
| 160 mild cases | ||||
| Northern European | ||||
| Hoffman, 2008 | Sarcoidosis | 499 cases | NA | NR |
| 490 controls | ||||
| Northern European | ||||
| Mushiroda, 2008 | Pulmonary fibrosis | 159 cases | TERT | 2.11 (1.61–2.78) |
| 934 controls | ||||
| Japanese |
Definition of abbreviations: CI = confidence interval; NA = not applicable; NR = not reported; OR = odds ratio.
Listed associated genes are limited to those with a P value less than 1 × 10−5 in the overall study population (initial + replication).
No genomic region surpassed a P value less than 1 × 10−5 in the overall study population, although a gene may have been reported, and may have been demonstrated to have biologic plausibility.
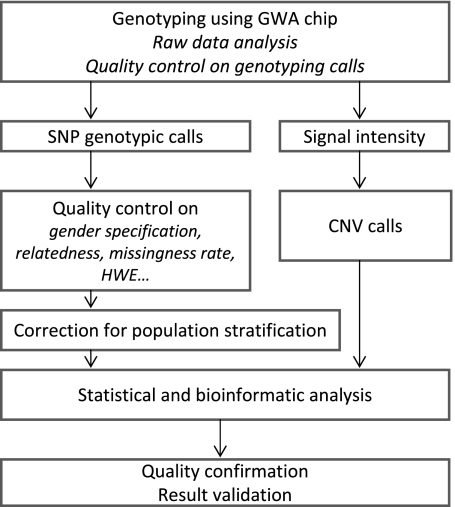
The fundamental principle underlying the GWA approach is the “common disease, common variant” hypothesis—the idea that common diseases are attributable, at least in part, to allelic variants present in more than 5% of the population. Although several GWA study designs have been employed, due to cost and time constraints the majority of available studies are of the case–control design. The work flow of a typical case–control GWA study is delineated in Figure 1. Ultimately, disease susceptibility is mapped to a particular genomic region when the frequency of a variant differs significantly between affected and unaffected individuals. A positive association may either reflect a direct effect of the genotyped variant or, more likely, can relate to an ungenotyped variant that is in high linkage disequilibrium (LD) or closely inherited with that observed.
Figure 1.
Major steps necessary for completion of a genome-wide association study. CNV = copy number variation; GWA = genome-wide association; HWE = Hardy-Weinberg equilibrium; SNP = single-nucleotide polymorphism.
As GWA studies increased in popularity, several statistical and methodological concerns became apparent that, in some cases, challenged the validity of the study results (25). These concerns include introduction of bias due to disease misclassification, population stratification, and genotyping errors, in addition to issues surrounding adjustment for the enormous number of multiple comparisons performed in such analyses (26).
Meticulous phenotypic classification of diseased cases and selection of an appropriate disease-free control group are critical initial steps in complex disease mapping. Ancestral markers should be used to further correct for inherent differences in population structure resulting from ethnic admixture (27). To ensure that genotyping data are of the highest quality, a platform with sufficient density to provide adequate coverage of the human genome is imperative, as are rigorous quality control standards, including checks on single-nucleotide polymorphism (SNP) call rate and maintenance of Hardy-Weinberg equilibrium. In the context of GWA studies, it is now more typical to take the perspective of “total evidence,” and to trust associations that are well beyond a broadly accepted significance threshold. By consensus, the statistical threshold for declaring genome-wide significance is P < 1 × 10−8 to P < 1 × 10−7 after application of the Bonferroni correction or another method by which to account for multiple comparisons (25, 28). Finally, demonstration of functional plausibility using in vivo or in vitro experimental models remains critical to determine if a specific gene or gene variant is actually involved in the generation of the observed phenotype and to further elucidate its specific biological mechanism of action.
Although some initial GWA studies failed to fully account for these design issues, most modern studies have been well executed and meet demanding methodological and statistical standards. With this general understanding of the basic hypothesis, principles, and limitations underlying the genome-wide approach, we now offer a perspective on the current state of GWA studies in pulmonary medicine, highlight the knowledge gained to date, and consider opportunities to further expand our understanding of the genetics of complex pulmonary disease.
Determinants of Pulmonary Function
Three GWA studies evaluating genetic relationships with measures of pulmonary function have been performed (3–5). The first involved an analysis of the ratio of FEV1 to FVC in patients enrolled in the Framingham Heart Study, whereas the latter two involved meta-analyses of multiple cohorts to identify genetic predictors of both FEV1 and FEV1/FVC ratio. All three included very large sample sizes, appropriate validation cohorts, adjustment for smoking status, and, subsequently, all identified highly significant variants associated with objective pulmonary function measures.
The first study by Wilk and colleagues (3) identified several SNPs on chromosome 4 near the gene for hedgehog interacting protein (HHIP) that met criteria for genome-wide significance, and were validated upon replication in an independent cohort. As these SNPs lie in an intergenic region, the HHIP gene is not directly implicated, and its overall significance remains unclear. However, given its known importance in signaling during fetal lung development (29) and previously documented association with height determination (30), HHIP remains an intriguing candidate gene. In addition, the functional importance of noncoding regulatory elements and intergenic region transcription in the control of downstream gene regulation is increasingly recognized (31–33), and this may provide a mechanism by which the identified intergenic SNPs alter gene, and thus trait, expression.
The two more recent studies both represent large-scale, collaborative meta-analyses that pool phenotypic and genotypic data on tens of thousands of subjects and model genetic predictors of FEV1 and FEV1/FVC (4, 5). Because of the very large sample size, these studies have increased power to detect genetic variants with lower allele frequencies and modest effect sizes. Importantly, each of these two follow-up studies confirmed a significant association between the intergenic region upstream from HHIP and FEV1/FVC ratio. In addition, several other gene variants were also identified that predicted FEV1 or FEV1/FVC ratio (Table 1). Advanced glycosylation end product–specific receptor (AGER) was found to be significantly associated with FEV1/FVC, and C-terminal domain containing glutathione S-transferase (GSTCD) and 5-hydroxytryptamine receptor 4 (HTR4) were found significantly associated with FEV1 across both of these large-scale studies. Many of the identified genes associated with lung function are biologically plausible and are involved in cell attachment, migration, and epithelial–extracellular matrix interaction, and thus could contribute to lung morphogenesis and remodeling (5).
In summary, these studies, relying upon objective and reproducible measures of lung function, demonstrate that FEV1 and FEV1/FVC ratio are heritable traits regulated by genetic variation at several key loci. Additional innovative work employing ancestry informative genetic markers has established that lung function differs according to genetic ancestry and, furthermore, suggests that incorporating measures of genetic ancestry into normative equations of lung function may provide more accurate predictions than those based on self-reported ethnicity (34).
Interestingly, despite the large sample sizes employed in the GWA studies of pulmonary function to date, all of the identified gene variants exhibit extremely small effect sizes, and thus contribute only minimally to variation in lung function among the population. This finding would suggest that additional undiscovered genetic factors, unmeasured gene–gene interactions, or as-of-yet poorly understood environmental influences may account for the majority of the observed variation.
COPD Susceptibility
As the third leading cause of death in the United States, COPD represents a major threat to public health (35). The observation that only a subset of smokers develops COPD strongly suggests that disease susceptibility is under genetic influence. Thus far, three GWA studies for COPD have been published (19–21) and, together, this line of research has generated exciting hypotheses related to the genetic susceptibility of this highly prevalent disease.
Two studies phenotyped COPD in several hundreds to thousands of smokers using accepted criteria for airflow obstruction, and compared identified patients with COPD to smoking control subjects with normal lung function (19, 20). Each employed a multistage replication design to follow-up findings of interest in similar populations, using a similar phenotype, the same genetic model, and demonstrating the same direction of association. Notably, both studies identified and replicated SNPs within a genomic region on chromosome 15 spanning several genes of interest to include α-nicotinic acetylcholine receptors 3–5 (CHRNA3–5), iron-responsive element-binding protein 2 (IREB2), and surfactant protein B–binding protein (Q9 UD29). Deep sequencing of Q9 UD29 in a small number of patients with COPD homozygous for the risk allele eliminated this as the source of polymorphic variance (19), bringing the focus to CHRNA3–5 and IREB2 as potential candidate genes in COPD susceptibility.
As the CHRNA3–5 locus was previously reported to be associated with smoking behavior (36), results were adjusted for smoking exposure, and remained highly significant. In one cohort, carriers of the risk allele at a single SNP locus within this genomic region had a population-attributable risk for COPD of 12.2% overall. This attributable risk increased to 14.3% in current smokers, and fell to 3.1% in former smokers. CHRNA3–5 encodes a family of nicotinic acetylcholine receptor proteins (19). Although it is well known that cholinergic activity within the airway initiates bronchial smooth muscle contraction and mucous secretion, additional evidence suggests that these receptors are also active in bronchial epithelial cells, lymphocytes, and neutrophils, and may inhibit neutrophil apoptosis, thus contributing to the proinflammatory milieu important in COPD pathogenesis (37).
In addition to the CHRNA3–5 locus, the second and larger of the two studies (20) identified and replicated family with sequence similarity 13 member A (FAM13A) as a potential candidate gene for COPD susceptibility. Very little is known about the function or regulation of this gene and its protein product. An association between COPD and HHIP was also noted in multiple independent populations in each of these studies, falling at or just below the threshold for genome-wide significance. This observation raises the possibility that genes that influence variation in lung function across the population also influence susceptibility to selected pulmonary diseases. Although an intriguing idea, most studies of lung disease to date would suggest that this is actually not the case, but, clearly, further investigation into the overlap between the genetic regulation of lung function in health and disease are needed (38).
The third study to evaluate genetic associations with COPD (21) applied a phenotyping strategy intended to specifically identify those patients with emphysematous lung disease. More than likely, this alternate phenotype accounted for, or at least contributed to, the strikingly different results generated when compared with the aforementioned studies (Table 1). In this study, radiologist interpretations of thoracic computed tomography scans were used to quantitatively and qualitatively characterize the presence and degree of emphysema in over 2,000 individuals with COPD. Although the study did not detect an association with quantitative emphysema, pooled analyses suggested a near-significant association between the genomic region encompassing bicaudal D homolog 1 (BICD1) and qualitative emphysema score. The strength of this association is supported, given that the frequency of the risk allele increased with increasing amounts of qualitative emphysema. Although the study is not without limitations, it is the only study to specifically address an emphysematous endophenotype, and can be viewed as hypothesis generating for follow-up research.
Asthma
The preponderance of the GWA studies in pulmonary medicine thus far has been aimed at exploring genetic associations with asthma (6–18). Identification of genetic susceptibility to asthma is complicated by heterogeneity of the clinical phenotype, variable approaches to diagnose and define asthma, and high probability that multiple genetic and environmental factors contribute to disease development. In addition, there are discrepancies in the burden of asthma among different ethnicities, and it is possible that the gene variants influencing susceptibility in these populations are divergent from those in populations of Northern European descent (39).
The majority of studies have considered the impact of genetics on childhood-onset asthma (6–8, 10, 11, 13, 15). The first included a family-based panel of children of Northern European descent with physician-diagnosed asthma (6). Several SNPs on the long arm of chromosome 17 were identified and validated in 2 independent populations. As the region of interest spanned several genes, the authors subsequently evaluated transcript levels for 14 of the 19 genes lying within the identified region using B cell–derived Epstein-Barr virus transformed lymphoblastoid cell lines from probands and their siblings. Transcripts in one gene, ORM1-like protein 3 (ORMDL3), were strongly and consistently positively associated with the dominant SNPs identified in the GWA study (6).
ORMDL3 encodes an endoplasmic reticulum–resident transmembrane protein, and demonstrates a wide tissue distribution, but is particularly high in cells participating in the inflammatory response. After the GWA-generated hypothesis, highly innovative research has revealed potential mechanisms by which ORMDL3 may influence asthma susceptibility. Polymorphisms in genes at the 17q21 locus were recently demonstrated to correlate with cord blood ORMDL3 expression and, furthermore, were found to influence IL-17 secretion from stimulated cord blood mononuclear cells, suggesting a potential role for ORMDL3 in early T cell regulation (40). In addition, others have shown that the ORMDL3 gene product alters endoplasmic reticulum–mediated calcium signaling, and thus can potentially disrupt protein folding and promote activation of endogenous inflammatory pathways (41).
Because the initial GWA study implicating its importance in asthma susceptibility, ORMDL3 and variants in neighboring genes, gasdermin A and B (GSDMA-B), have been highly replicated in several case–control series, and appear to be fairly specific for asthma onset in childhood, particularly in those of Northern European descent. Estimates indicate that the disease-associated markers identified in ORMDL3/GSDMA-B may account for between 29.5 and 38% of the variance in childhood asthma expression, and, as such, are likely to greatly influence the burden of disease in the population (6, 14). These markers, however, perform poorly as predictors of individual disease risk (14).
An additional large study evaluating physician-diagnosed, persistent childhood asthma in North American children of European ancestry not only replicated the above genomic region, but also led to novel findings (11). Two independent replication populations were employed to validate initial findings. The first included subjects of European descent with childhood onset asthma—very similar to the discovery sample—whereas the second encompassed North American children of African ancestry. Eight SNPs tagging to a genomic region on chromosome 1 attained genome-wide significance in the initial sample. A total of 12 additional SNPs in strong LD were also associated with asthma, and all 20 polymorphisms mapped to a single LD block spanning the gene for DENN/MADD domain containing 1b (DENND1B). Interestingly, although several of the implicated SNPs were validated to be statistically significant in both independent replication populations, the risk allele associated with asthma at each SNP in the cohort of African descent was the alternative allele to that associated with asthma in the discovery set of European descent.
The challenge of studying genetic associations in populations of African descent was further demonstrated in a study of both subjects with asthma from the Baltimore–Washington, D.C. area self-identified to be of African descent and African Caribbean probands and their families (10). Although 3 of the greater than 600,000 evaluated SNPs met genome-wide significance for asthma association in the North American cohort, the minor allele frequency for each of these was less than 1% in both cases and controls, and, ultimately, none were substantiated in the African Caribbean population or in four other replication populations, including North Americans of African descent.
Although these negative findings may be due to inadequate power given modest sample sizes, incomplete or dissimilar phenotypes, or subtle population admixture, other factors may have contributed. The genomic coverage offered by commercially available genotyping platforms may be limited in African populations due to less extensive LD between variants. Although imputation can be, and was, employed in an attempt to enrich genomic coverage, even this may be of limited use in populations of African origin, as the HapMap panel may be poorly representative of the haplotype structure in the population of interest.
Beyond childhood-onset asthma, a large, consortium-based GWA study involving greater than 10,000 subjects with physician-diagnosed asthma aimed to identify associations with additional asthma subtypes, including later-onset asthma (age at onset ≥ 16 yr), severe asthma, and occupational asthma (14). Common alleles in several genomic regions were associated with disease risk at all ages, including IL-1 receptor–like 1/IL-18 receptor 1 (IL1RL1/IL18R1), the HLA-DQ region of the major histocompatibility complex gene, IL-33 (IL33), SMAD family member 3 (SMAD3), and IL-2 receptor β (IL2RB). Although none of these associations were specific to severe or occupational asthma, the HLA-DQ region was observed to have a slightly stronger association with later-onset asthma.
It is likely that different subsets of genes influence disease susceptibility versus disease severity. Only one study of asthma phenotypes was designed to specifically assess genetic associations with severe asthma and asthma-related quantitative trait (12). This study included cases with physician-characterized severe asthma who also met additional criteria based on use of medication and urgent or emergent care services. Controls for the evaluation of asthma-related quantitative traits were fully phenotyped with no family or personal history of asthma and normal pulmonary function without bronchial hyperresponsiveness or bronchodilator response. Although no SNP surpassed the threshold for genome-wide significance, the RAD50 homolog–IL-13 (RAD50-IL13) regions demonstrated the strongest evidence for association with the severe asthma phenotype and asthma-related quantitative traits.
The study of asthma severity is inherently difficult given profound disease heterogeneity. Although classically grouped as mild, moderate, or severe according to society-based guidelines, a recent hierarchical cluster analysis of multiple clinical variables in subjects participating in the Severe Asthma Research Program identified five distinct subtypes of asthma severity. For example, cluster 1 represented those with early-onset atopic asthma, normal lung function, and requirement for two or fewer controller medications, whereas cluster 3 was a unique group of mostly older, more obese women with late-onset nonatopic asthma, only moderate reduction in FEV1, and frequent oral corticosteroid requirement (42). The identification of these novel disease clusters suggests that future GWA studies aimed at identifying genes important in asthma severity are likely to require more detailed phenotyping efforts, including longitudinal data on lung function, exacerbations, and medication requirements. In addition, the use of gene expression patterns in combination with this thorough clinical phenotyping approach may even further enhance GWA success (38).
Other Pulmonary Diseases
Beyond COPD and asthma, GWA studies for many important and prevalent pulmonary diseases are either limited (as is the case for IPF, sarcoidosis, and CF) or lacking entirely. A small GWA study evaluating Japanese subjects with IPF implicated telomerase reverse transcriptase (TERT) in the development of sporadic IPF (22). TERT contributes to telomere replication and stabilization, and is especially important in cells undergoing high turnover or relying on progenitor reserves, as may be the case for bronchial epithelium. This finding is of particular interest, as telomerase mutations had previously been identified to account for some familial cases of IPF in White subjects (43).
The strongest genetic association linked to sarcoidosis in a single GWA study of German subjects was annexin A11 (ANXA11) (24), and this association was recently confirmed in a similar independent series of German subjects with both radiologic and biopsy features consistent with pulmonary sarcoidosis (44). Similarly, a GWA study of lung disease severity in CF evaluating a small sample of patients implicated IFN-related developmental regulator 1 (IFRD1) as a potential disease-modifying gene (23). IFRD1 is biologically plausible, as it has been shown to regulate neutrophil effector function, and deficiency is associated with delayed airway bacterial clearance. Although initial results from these three small studies are intriguing, large-scale collaborative follow-up studies are essential to further characterize the genetic impact on these important pulmonary diseases.
Conclusions and Future Directions
The GWA approach has facilitated several novel discoveries in pulmonary medicine. With the exception of the innovative research revealing specific polymorphisms and functional mechanisms that may mediate the influence of ORMDL3 on asthma susceptibility, the biological insights of these discoveries have been limited in part by an inability to identify the precise genetic variants responsible for reported associations. In most cases, it is not possible to be certain of what gene or genes underlie the observed association, nor ascertain the nature of their involvement. In this regard it is also interesting to highlight the peculiar example of variants for childhood asthma in which the direction of effect of risk alleles is reversed in populations of European and African ancestry. This observation is difficult to reconcile with a model of common variants with modest effects on risk, and could reflect “synthetic associations” in which different rare variants in different populations have distinct patterns of associations with more common variants (45). This possibility emphasizes both the challenge in interpreting GWA study results and the importance of continued investigation to determine the variants that underlie detected association signals.
Another important point is that the genetic variants identified to be associated with common pulmonary diseases or pulmonary function appear to contribute only minimally to overall disease risk or trait heritability. Potential explanations for this so called “missing heritability” are many, including likely contributions from as-of-yet unevaluated rare variants, epigenetic modifications resulting in altered gene expression, or poorly understood gene–gene and gene–environment interactions. In particular, interest in rare variants is growing, because most GWA studies, not just those in pulmonary medicine, have failed to explain the preponderance of the susceptibility to many common human diseases (46).
By some estimates, approximately 60% of SNPs in the human genome have a mean allele frequency of less than 5%, and these rarer variants or combinations thereof could explain development of many common diseases. To evaluate variants with a frequency of less than about 1%, whole-genome sequencing will be necessary. Next-generation sequencing machines have remarkably greater capacity and speed when compared with older technologies, thus making whole-genome or whole-exome sequencing in large numbers of cases and controls an approaching reality (47). As a proof of concept, such approaches have already been used to identify the genetic basis of rare Mendelian diseases, such as Miller syndrome (48) and X-linked leukoencephalopathy (49).
Although the relevance of these results to common multigenic diseases remains uncertain, several strategies have been proposed by which whole-genome sequencing approaches could be performed in smaller, highly selected samples. For example, study designs that leverage carefully selected cases with extreme phenotypes of disease or employ family-based studies, including affected and unaffected individuals, could provide a powerful approach to identifying rare disease-causing variants (47). Still others propose that the examination of rare variants by next-generation sequencing will be insufficient to further our understanding of disease heritability. Rather, they suggest moving beyond single-gene analyses to enhanced analytical modeling of complex gene–gene interactions (38).
Certainly a comprehensive approach will be necessary to further leverage the GWA method to study complex pulmonary diseases in the future. Multicenter collaborative efforts to generate large longitudinal cohorts or the addition of gene expression profiles to create more in-depth clinical phenotyping will be critical. Examples of such studies currently underway include COPDGene and the EVE asthma genetics consortium; their results should continue to increase our understanding of the genetic basis of these common diseases and help reconcile differences observed in earlier studies of asthma and COPD. In addition, multicenter work is ongoing to evaluate the genetic basis for pulmonary conditions that have not previously been subject to GWA, such as acute lung injury (50, 51). Furthermore, ongoing efforts will use information from the 1,000 Genomes Project to construct genotyping platforms that offer more substantial coverage of variants in populations of African descent, therefore allowing more effective use of GWA studies in non-European samples, particularly important given the increasing burden of asthma in diverse ethnic populations (46, 52). In the future, next-generation sequencing will offer the potential for even greater depth of knowledge with regard to the underlying basis of many common lung conditions. Finally, an enhanced understanding of gene–gene interactions and gene–environment interactions will be gained through improved analytic techniques and ambitious endeavors, such as the Environmental Genome Project led by the National Institute of Environmental Health Sciences (53). These techniques will be essential given that gene–environment effects may, by design, be obscured in large consortia data sets.
Despite this limitation, a clear strength of the GWA studies conducted to date in pulmonary medicine has been the assembly of collaborative and multidisciplinary teams of investigators across multiple institutions. Although the path to fully uncovering the genetic basis of pulmonary disease has only begun, these teams, with rapidly advancing technology and cutting-edge bioinformatic approaches hold the potential to substantially impact our understanding of the genetic basis of many complex diseases in the future. Eventually, continued pursuit of this research will translate into clinically useful information through identifying novel causal pathways and new therapeutic targets, and by generating knowledge of individual patterns of disease predisposition, thus facilitating personalized risk assessment and disease management.
Footnotes
Supported by National Institutes of Health grants 1P50-HL084917-01 (project 3 [S.M.P.]; training grant [J.L.T.]), R01 HL081619 (J.C.), and 1K24 HL91140–01A2 (S.M.P.).
Originally Published in Press as DOI: 10.1164/rccm.201106-0971PP on July 28, 2011
Author Disclosure: J.L.T., D.B.G. and D.G. do not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.C. was a consultant for and received institutional grant support from GlaxoSmithKline. He worked as an expert witness in Brake Worker litigation for various law firms. S.M.P. receives research support through the Biomarker Factory LLC.
References
- 1.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005;308:385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hindorff LAJH, Hall PN, Mehta JP, Manolio TA. A catalog of published genome-wide association studies [Internet; accessed 2011 Mar 21]. Available from: http://www.genome/gov/gwastudies
- 3.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, Myers RH, Borecki IB, Silverman EK, Weiss ST, et al. A genome-wide association study of pulmonary function measures in the Framingham heart study. PLoS Genet 2009;5:e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, Zhao JH, Ramasamy A, Zhai G, Vitart V, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet 2010;42:36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YM, Chen TH, Barr RG, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet 2010;42:45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007;448:470–473 [DOI] [PubMed] [Google Scholar]
- 7.Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY, Li H, del Rio-Navarro BE, Willis-Owen SA, Weiss ST, et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in Mexican children. PLoS Genet 2009;5:e1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, Wilk JB, Willis-Owen SA, Klanderman B, Lasky-Su J, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet 2009;84:581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SH, Cho BY, Park CS, Shin ES, Cho EY, Yang EM, Kim CW, Hong CS, Lee JE, Park HS. Alpha-t-catenin (CTNNA3) gene was identified as a risk variant for toluene diisocyanate-induced asthma by genome-wide association analysis. Clin Exp Allergy 2009;39:203–212 [DOI] [PubMed] [Google Scholar]
- 10.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, Tsai YJ, Yang M, Campbell M, Foster C, et al. A genome-wide association study on african-ancestry populations for asthma. J Allergy Clin Immunol 2010;125:336–346 e334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, Wang K, Rafaels NM, Michel S, Bonnelykke K, et al. Variants of dennd1b associated with asthma in children. N Engl J Med 2010;362:36–44 [DOI] [PubMed] [Google Scholar]
- 12.Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, Bleecker ER. Genome-wide association study of asthma identifies rad50-il13 and hla-dr/dq regions. J Allergy Clin Immunol 2010;125:328–335 e311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himes BE, Lasky-Su J, Wu AC, Wilk JB, Hunninghake GM, Klanderman B, Murphy AJ, Lazarus R, Soto-Quiros ME, Avila L, et al. Asthma-susceptibility variants identified using probands in case–control and family-based analyses. BMC Med Genet 2010;11:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 2010;363:1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeWan AT, Triche EW, Xu X, Hsu LI, Zhao C, Belanger K, Hellenbrand K, Willis-Owen SA, Moffatt M, Cookson WO, et al. PDE11A associations with asthma: results of a genome-wide association scan. J Allergy Clin Immunol 2010;126:871–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira MA, McRae AF, Medland SE, Nyholt DR, Gordon SD, Wright MJ, Henders AK, Madden PA, Visscher PM, Wray NR, et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur J Hum Genet 2011;19:458–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ege MJ, Strachan DP, Cookson WO, Moffatt MF, Gut I, Lathrop M, Kabesch M, Genuneit J, Buchele G, Sozanska B, et al. Gene–environment interaction for childhood asthma and exposure to farming in Central Europe. J Allergy Clin Immunol 2011;127:138–144 [DOI] [PubMed] [Google Scholar]
- 18.Kim JH, Park BL, Cheong HS, Bae JS, Park JS, Jang AS, Uh ST, Choi JS, Kim YH, Kim MK, et al. Genome-wide and follow-up studies identify CEP68 gene variants associated with risk of aspirin-intolerant asthma. PLoS ONE 2010;5:e13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 2009;5:e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet 2010;42:200–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong X, Cho MH, Anderson W, Coxson HO, Muller N, Washko G, Hoffman EA, Bakke P, Gulsvik A, Lomas DA, et al. Genome-wide association study identifies BICD1 as a susceptibility gene for emphysema. Am J Respir Crit Care Med 2011;183:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mushiroda T, Wattanapokayakit S, Takahashi A, Nukiwa T, Kudoh S, Ogura T, Taniguchi H, Kubo M, Kamatani N, Nakamura Y. A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. J Med Genet 2008;45:654–656 [DOI] [PubMed] [Google Scholar]
- 23.Gu Y, Harley IT, Henderson LB, Aronow BJ, Vietor I, Huber LA, Harley JB, Kilpatrick JR, Langefeld CD, Williams AH, et al. Identification of IFRD1 as a modifier gene for cystic fibrosis lung disease. Nature 2009;458:1039–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann S, Franke A, Fischer A, Jacobs G, Nothnagel M, Gaede KI, Schurmann M, Muller-Quernheim J, Krawczak M, Rosenstiel P, et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat Genet 2008;40:1103–1106 [DOI] [PubMed] [Google Scholar]
- 25.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008;9:356–369 [DOI] [PubMed] [Google Scholar]
- 26.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA 2008;299:1335–1344 [DOI] [PubMed] [Google Scholar]
- 27.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 2005;6:95–108 [DOI] [PubMed] [Google Scholar]
- 28.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 2008;32:381–385 [DOI] [PubMed] [Google Scholar]
- 29.Cardoso WV, Lü J. Regulation of early lung morphogenesis: questions, facts and controversies. Development 2006;133:1611–1624 [DOI] [PubMed] [Google Scholar]
- 30.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet 2008;40:575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 2010;28:1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature 2007;447:799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 2010;28:817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar R, Seibold MA, Aldrich MC, Williams LK, Reiner AP, Colangelo L, Galanter J, Gignoux C, Hu D, Sen S, et al. Genetic ancestry in lung-function predictions. N Engl J Med 2010;363:321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minino A. Death in the United States, 2009 [Internet; accessed 2011 Jul 13]. Available from: http://www.cdc.gov/nchs/data/databriefs/db64.pdf [PubMed]
- 36.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008;452:638–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wessler I, Kirkpatrick CJ, Racké K. Non-neuronal acetylcholine, a locally acting molecule, widely distributed in biological systems: expression and function in humans. Pharmacol Ther 1998;77:59–79 [DOI] [PubMed] [Google Scholar]
- 38.Weiss ST. What genes tell us about the pathogenesis of asthma and chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2010;181:1170–1173 [DOI] [PubMed] [Google Scholar]
- 39.Barnes KC. Genetic epidemiology of health disparities in allergy and clinical immunology. J Allergy Clin Immunol 2006;117:243–254 [DOI] [PubMed] [Google Scholar]
- 40.Lluis A, Schedel M, Liu J, Illi S, Depner M, von Mutius E, Kabesch M, Schaub B. Asthma-associated polymorphisms in 17q21 influence cord blood ORMDL3 and GSDMA gene expression and IL-17 secretion. J Allergy Clin Immunol 2011;127:1587–1594 [DOI] [PubMed] [Google Scholar]
- 41.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum–mediated calcium signaling and cellular stress. Hum Mol Genet 2010;19:111–121 [DOI] [PubMed] [Google Scholar]
- 42.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, D'Agostino R, Jr, Castro M, Curran-Everett D, Fitzpatrick AM, et al. National Heart Lung, Blood Institute's Severe Asthma Research Program. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med 2010;181:315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA, III, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med 2007;356:1317–1326 [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Pabst S, Kubisch C, Grohé C, Wollnik B. First independent replication study confirms the strong genetic association of ANXA11 with sarcoidosis. Thorax 2010;65:939–940 [DOI] [PubMed] [Google Scholar]
- 45.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol 2010;8:e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature 2009;461:747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 2010;11:415–425 [DOI] [PubMed] [Google Scholar]
- 48.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat Genet 2010;42:30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsurusaki Y, Osaka H, Hamanoue H, Shimbo H, Tsuji M, Doi H, Saitsu H, Matsumoto N, Miyake N. Rapid detection of a mutation causing X-linked leucoencephalopathy by exome sequencing. J Med Genet 2011;48:606-609 [DOI] [PubMed] [Google Scholar]
- 50.Christie JD, Wurfel MM, O'Keefe G, Christiani DC, Ware LB, Calfee CS, Meyer N, Bradfield J, Kim C, Li M, et al. Genome wide association (GWA) identifies functional susceptibility loci for trauma-associated acute lung injury [abstract]. Am J Respir Crit Care Med 2010;181:A1025 [Google Scholar]
- 51.Wurfel MM, Christie JD, Holden TD, O'Keefe GE, Calfee CS, Matthay MA, Hakonarson H, Jarvik GP, Crosslin DR, Lin X, et al. ARDSNet Investigators. Genome-wide association study of acute lung injury risk loci in the ISPAAR consortium [abstract]. Am J Respir Crit Care Med 2011;183:A5536 [Google Scholar]
- 52.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collman G. Environmental genome project [Internet; accessed 2011 Mar 7]. Available from: http://www.niehs.nih.gov/research/supported/programs/egp/