Abstract
Immuno-magnetic separation has become an essential tool for high throughout and low cost isolation of biomolecules and cells from heterogeneous samples. However, as magnetic selection is essentially a “black-and-white” assay, its application has been largely restricted to single-target and single-parameter studies. To address this issue, we have developed an immuno-magnetic separation technology that can quickly sort multiple targets at high yield and purity using selectively displaceable DNA linkers. We envision that this technology will be readily adopted for experiments requiring high throughput selection of multiple targets, or further adapted for selection of a single target based on multiple surface epitopes.
Magnetic separation technologies have played a critical role in a variety of biomedical applications ranging from molecular diagnostics to cell-based therapy.1 In contrast to other separation technologies, such as spatial separation via microarrays2 or optical separation using fluorescence-activated cell sorting,3 magnetic separation offers major advantages in terms of throughput and cost.4,5 However, as selection is based on a single parameter (magnetization), only one target can be isolated at a time. Thus, intricate protocols are necessary to separate multiple targets from a sample (multi-target), or to isolate a single target based on multiple surface epitopes (multi-parameter).6 Given that emerging research demands interrogation of increasingly complex and heterogeneous systems, in particular within the fields of immunology and oncology, there is a clear need for innovative magnetic separation technologies that enable multiplexed target sorting with high throughput, purity, and yield.
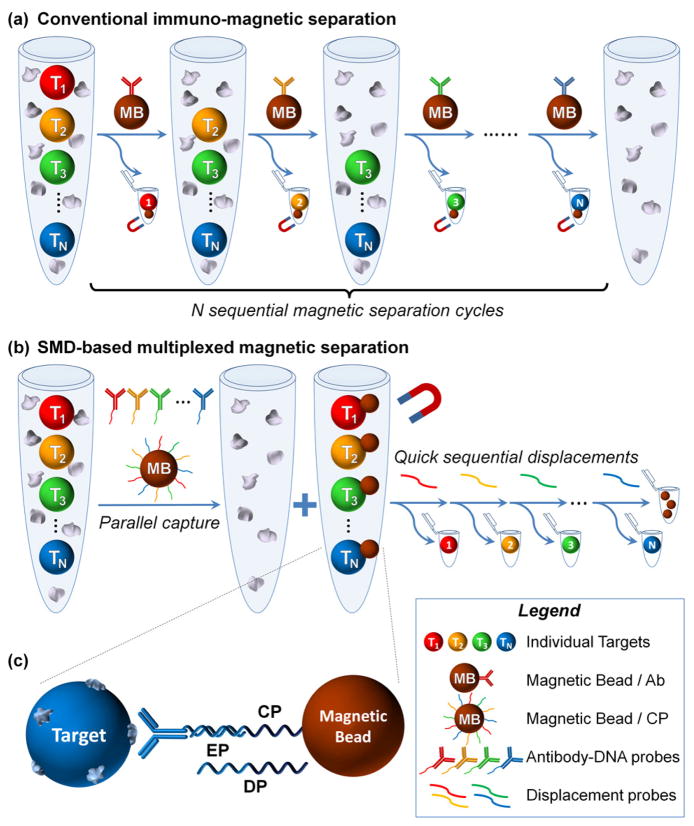
Several strategies have been proposed to incorporate multiplexing potential into magnetic separation. One promising approach is to use the size tunable properties of magnetic nanoparticles for simultaneous isolation of several targets.7 For example, Adams et al. described a multitarget MACS, which applied microfluidics and high-gradient magnetic fields to separate 2 bacterial targets using 2 distinct magnetic tags at >90% purity and >500 fold enrichment.8 However, multi-target sorting through ‘physical’ encoding of magnetic particles requires sophisticated instrumentation and remains highly limited by the number of discrete magnetic tags that can be reliably separated. In a more straightforward approach, multiplexed separation can be achieved through multiple sequential rounds of single-target magnetic selection (Figure 1a). As an example, Semple et al. used this method to sort CD4+ and CD19+ lymphocytes in a 4-hour procedure.9 Yet, despite its simplicity, not only is sequential sorting time-consuming, lengthy separation protocols often result in an alteration of the biological state of the target (e.g. gene expression and/or viability of cells),10 rendering such an approach unsuitable for many applications.
Figure 1.
Schematic of multi-target immuno-magnetic sorting. (a) Conventional sorting of multiple targets involves lengthy sequential magnetic isolation steps. (b) In contrast, SMD-based sorting technology captures all targets of interest simultaneously, followed by a rapid sorting through release of MB-Target link. (c) Target is captured through immuno-recognition by DNA-encoded antibody and partial hybridization with CP on MB. Selective target release is achieved through sequence-specific EP displacement due to a more favorable hybridization between CP and DP.
Complementary to the challenge of spatial or temporal segregation of target-carrying magnetic particles is the issue of incorporating multiplexing capability within the target capture method itself. Magnetic selection can be applied in one of two formats: (1) direct selection, where the affinity ligand is directly coupled to the magnetic nanoparticle, and (2) indirect selection, where targets are first incubated with an excess of primary affinity ligand and then captured by magnetic particles via secondary affinity ligand. As the indirect method allows for optimal affinity ligand orientation on target, a ‘signal amplification’ effect is observed, improving yield and purity.5 Furthermore, indirect method enables utilization of a wide range of commercial affinity ligands without the need for further modification. At the same time, this approach is particularly challenging to multiplex, given the limitations in selectivity of primary-secondary affinity ligands (e.g. biotin-streptavidin and primary-secondary antibody links). In this regard, DNA-antibody conjugates represent a powerful tool for multiplexed indirect selection, first demonstrated by Heath et al. on DNA microarray platform,2 and recently applied for characterization of secreted proteins from single cells, opening exciting opportunities in study of human immune cell responses.11 However, the small surface area of microarray chips hampers large-scale sorting applications. In this context, incorporation of molecular encoding into the conventionally single-parameter magnetic selection platform holds the key to achieving truly multiplexed, high-throughput target sorting.
Here, we report a rapid multi-target immuno-magnetic separation technology that combines extensive multiplexing capacity of DNA-antibody conjugates and high selectivity, throughput, and simplicity of magnetic isolation by employing a unique approach through strand-mediated displacement (SMD) of DNA linkers. Our key insight is that the combination of spatial and temporal segregation could offer simultaneous selection of multiple target populations from a heterogeneous sample, followed by quick isolation of individual targets through SMD, inspired by the fast kinetics and selectivity of SMD in DNA motors and walkers.12 The major steps of SMD for multi-target sorting are illustrated in Figure 1b. In the first step, antibodies encoded with distinct DNA sequences (encoding probe, EP) bind their identifying antigen on target populations. Next, magnetic beads (MBs) coated with capture probes (CPs), which partially hybridize with their corresponding EPs, enable simultaneous magnetic selection of all targets of interest. In the second step, the magnetically enriched targets are released one population at a time through SMD upon addition of a displacement probe (DP). Probe assembly is schematically illustrated in Figure 1c. The DP binds selectively to an exposed ‘toehold’ region on the CP, and then ‘unzips’ the EP from its original binding site due to the longer complementarity between CP and DP, thus breaking the MB-target link. In this way, multiple targets can be quickly isolated through a serial addition of DPs. In direct contrast to the conventional multi-cycle magnetic separation (Figure 1a), SMD technology employs only a single round of slow immuno-recognition, and the following SMDs are a remarkably rapid process (with displacement half-time of only ~0.2 seconds13). Therefore, with sorting of N targets (N>1), our technology should offer significantly shortened assay time (1 × N hours vs. 1 hour + rapid SMDs), which is highly desired for preserving the native state and bio-functionality of isolated targets.
Although SMD-based magnetic sorting represents a platform technology applicable to a wide range of analytes, proof-of-concept work reported here most closely resembles conditions necessary for sorting of live cells, which is a significant application of this technology. In our setup, four-color fluorescent beads of size similar to mammalian cells are used as a model system for development of the SMD technology. Fluorescent beads are easy to identify and count, and are thus ideal for technology characterization and validation. Three of the colored beads are surface-modified with a species-specific IgG, which serves as an identifying antigen; the fourth bead is unmodified and serves as an impurity population to remove (Figure S1). To demonstrate multi-target enrichment and isolation, 3 sets of DNA sequences are designed with 16 base-pair overlap between CP and EP and a 6 base-pair toehold region, allowing for a total of 22 base-pair DP binding (Table S1).
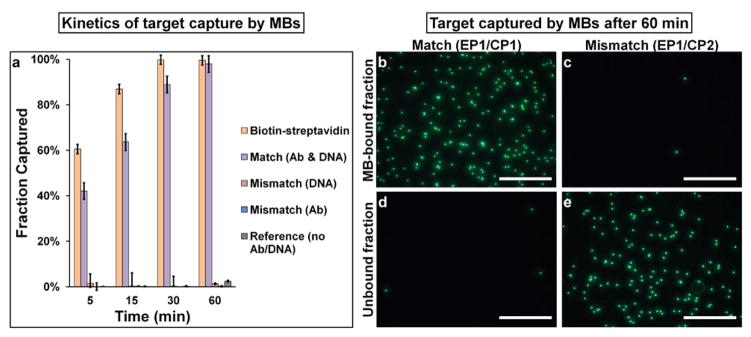
Considering that the length of target separation methods can dramatically impact biological characteristics of the target, we first investigated the kinetics and specificity of the magnetic capture and SMD-based release using single-color (green) microspheres. As shown in Figure 2a, target capture through hybridization between EPs and CPs slightly lags behind that mediated by the gold standard streptavidin-biotin interaction, but both reach nearly complete target capture at 1 hour time point (>98%). Representative fluorescence micrographs taken for the unbound (supernatant) and MB-bound fractions for matched and mismatched DNA sequences after a 60-min incubation period are shown in Figure 2 b–e. Clearly, the matched EP1/CP1 pairs lead to nearly complete capture of target beads in the MB-bound fraction, whereas the mismatched sequences (CP2) produce only negligible non-specific binding. High specificity of target capture through DNA hybridization was observed with a set of systematically designed negative controls (Figure 2a, Table S2, Figure S2).
Figure 2.
Kinetics and specificity of target capture. (a) Quantitative analysis of target capture through DNA hybridization. Average of 3 separate experiments is shown. Error bars indicate standard deviations. (b–e) Qualitative evaluation of target capture with fluorescence microscopy. Targets (green) are retained in MB-bound fraction for complementary EP1/CP1 link (b,d), but not for non-complementary EP1/CP2 link (c,e). Scale bar 250 μm.
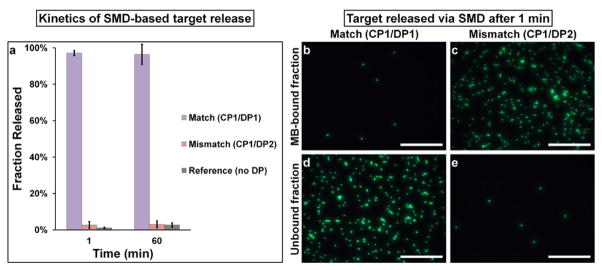
Following magnetic capture via DNA linkers, we probed the specificity and kinetics of target release via SMD (Table S3). Target release was measured after 1 min or 60 min incubation periods (Figure 3a). Remarkably, even after 1 min of SMD, release of target beads into the supernatant is nearly complete (97 %) for complementary DP, while only minimal nonspecific release (< 3 %) is observed for the mismatched DP and reference, demonstrating high selectivity and speed of SMD despite potential issues, such as steric hindrance. Fluorescence microscopy (Figure 3b–e) and further control studies (Figure S3) corroborate this conclusion. The outstanding kinetics and selectivity of SMD results from three fundamental features of this technology. First, as mentioned earlier, DNA displacement with longer complementary strands is an extremely selective and fast process. Second, diffusion of small DNA strands to microbeads is much faster than diffusion between two microparticles. Third, a profound impact originates from the differential concentrations in the separation reaction. In the case of conventional immuno-magnetic separation, the two ‘reactants’ are MBs and targets, which are often used in fM to pM concentration range.10,14 In our SMD approach, on the other hand, the two ‘reactants’ are MB-target complexes and single-stranded DNA with concentration typically in the μM range, which is six to nine orders of magnitude higher than that of MBs, thus promoting the reaction rate. Noteworthy, there is virtually no non-specific target release for 1 min and 60 min incubation periods, indicating that the 16 base pair EP-CP overlap offers sufficient long-term stability (under gentle rotating agitation). Yet, vigorous washing results in more noticeable non-specific target release with DNA links compared to biotin-streptavidin bond (data not shown). Such behavior is not surprising, as DNA-DNA binding strength (20–50 pN for 10–30 base pairs)15 is considerably lower than that of biotin-streptavidin bond (300 pN).16 Poor bond stability can be partially addressed by using longer DNA probes, while maintaining binding specificity through careful sequence design. Furthermore, this effect may be negligible for separation of smaller analytes (proteins, bacteria), as shear forces decrease along with particle size.
Figure 3.
Kinetics and specificity of target release. (a) Quantitative analysis of SMD-based target release kinetics. Average of 3 separate experiments is shown. Error bars indicate one standard deviation. (be) Fluorescence microscopy evaluation of target release yield and specificity. Nearly complete release of targets (green beads) into supernatant is obtained with complementary (b,d)but not non-complementary (c,e) DPs. Scale bar 250 μm.
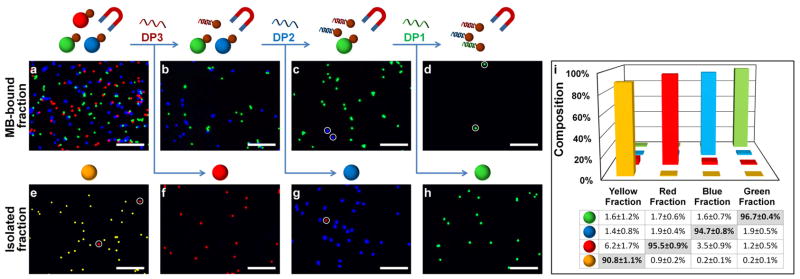
Next, we proceeded to demonstrate the utility of SMD technology for quick sorting of multiple targets from a mixed sample, transforming conventional single-parameter magnetic separation into a multiplexed format. To begin, four populations of fluorescent beads are pooled in a single sample at even proportions. Beads of the three primary colors are tagged with distinct antigens on the surface (green with rabbit IgG, blue with mouse IgG, and red with human IgG). Unmodified yellow beads serve as an impurity for removal. Three antibodies specifically recognizing those surface antigens are tagged with unique encoding oligonucleotides (EP1, EP2, and EP3) and incubated with the mixture sample. In parallel, MBs are modified with CP1, CP2, and CP3, complementary to each EP. Following the procedure schematically illustrated in Figure 1b, the red, blue and green beads are simultaneously enriched magnetically and subsequently isolated one at a time by addition of their cognate DPs. Qualitative evaluation with fluorescence microscopy (Figure 4a–h) and quantitative analysis with flow cytometry (Figure 4i) reveal excellent purity (Red: 95.5%; Blue: 94.7%, and Green: 96.7%) and reproducibility (standard deviation < 1%) for each of the isolated fractions. We also calculated the overall yield of each target (Red: 68.2±13.8%, Blue: 74.5±8.5%, and Green: 61.4±8.2%) by dividing the number of beads collected by that in the reference sample, which did not undergo the magnetic separation procedure. The sources of the loss may include dead volume in pipette tips, retention of beads on centrifuge tube walls/caps, incomplete magnetic capture, nonspecific release, and incomplete release of targets. These parameters deserve future optimization especially for separation and characterization of rare targets, such as stem cells or circulating tumor cells. Nevertheless, the yield and throughput reported herein marks a considerable improvement over previous multi-target magnetic selection methods,8 while requiring no sophisticated instrumentation.
Figure 4.
Rapid multi-target SMD-based sorting. (a–h) Fluorescence microscopy of MB-bound (a–d) and isolated (e–h) fluorescent bead fractions at different stages of SMD-based sorting. Immuno-magnetic isolation of 3 targets (red, green, and blue beads) retains targets within MB-bound fraction (a), leaving “impurity” (yellow beads) in supernatant (e). Sequential introduction of target-specific DPs leads to selective release of a target into supernatant (f,g,h), leaving non-displaced targets in MB-bound fraction (b,c,d). Impurities are indicated by white circles. Images are processed according to procedure outlined in Figure S4. Scale bar 100 μm (i) Quantitative analysis of purity of isolated fractions with flow-cytometry. Average and standard deviation from 3 separate experiments are shown.
To further highlight the benefits of SMD technology, we contrasted our approach to conventional sequential immuno-magnetic sorting (Figure S5). Here, using biotinylated antibodies and streptavidin-coated MBs, we again isolated 3 targets from an initial mixture of 4 bead populations. Interestingly, not only is this protocol time consuming (taking 5 hours rather than 1.5 hours with SMD-based sorting), using a single secondary affinity label (streptavidin) results in a marked decrease in target purity, as any antibody-biotin labeled target not captured during its corresponding magnetic selection step becomes an impurity for subsequent targets. In contrast, SMD sorting protocol demonstrates higher purities because complete target release via SMD is more favorable than complete target capture by MBs due to improved reactant kinetics, and unique DPs prohibit cross-reactivity between sequential isolation steps. At the same time, both procedures share major sources of target loss, thus offering similar overall yields for each target.
Finally, we tested the dynamic range for multi-target cell sorting, as large variation in target concentration can be encountered with certain practical applications. Here, we found that changing the ratio of two targets from 1:1 to 1:100 resulted in a considerable drop in purity of rare target during both capture (98% to 43%) and release (98% to 20%) steps (Figure S6). Inferring from the control studies (Figure S2 and S3) and similar performance of biotin-streptavidin mediated capture (Figure S6), we believe that major causes for this effect are antibody cross-reactivity, incomplete washing, and rupture of DNA links. Therefore, further protocol optimization is required for applications where targets are present at high dynamic range.
In summary, we have developed a simple yet robust multi-target immuno-magnetic separation technology based on the clever concept of DNA strand-mediated displacement. Magnetic separation serves as a high-throughput platform for sorting of a wide array of targets, while DNA-antibody conjugates enable highly multiplexed indirect selection, which confers important benefits of high target yield and purity. Finally, rapid target sorting is enabled by SMD technology via fast DNA binding and displacement, fast diffusion of relatively small DPs, and high concentration of DNA reactants. Overall, combination of these critical components provides a unique solution to a long-standing problem in magnetic separation: multi-target sorting at high yield, purity, and throughput.
We believe that versatility of SMD-based separation platform will enable a number of powerful applications, such as live cell sorting, as both target capture through DNA hybridization and SMD can be carried out in a range of cell- and bio-compatible buffers and at ambient or chilled conditions.2 Further, SMD technology might streamline implementation of conventional immuno-magnetic selection where MBs must be removed to avoid interference with further analysis or adverse effects on target biological state,17 as well as to allow further isolation via a different surface epitope of the same target, thus enabling multi-parameter selection. Finally, availability of DNA-antibody conjugates on target surface following MB release should enable isolation of rare targets by applying several selective rounds of magnetic capture and SMD release.
Supplementary Material
Acknowledgments
This work was supported in part by NIH (R01CA131797), NSF (0645080), and the UW Department of Bioengineering. X.H.G. thanks the NSF for a Faculty Early Career Development award (CAREER). P.Z. thanks the NSF for Graduate Research Fellowship (DGE-0718124).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Abbreviations; Experimental materials and methods; Validation of antigen coverage on target beads; Study of target capture and release specificity; Multi-target sorting via standard sequential magnetic capture; Evaluation of SMD technology for separation of targets at varying abundance. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.(a) Almand B, Resser JR, Lindman B, Nadaf S, Clark JI, Kwon ED, Carbone DP, Gabrilovich DI. Clin Cancer Res. 2000;6:1755–1766. [PubMed] [Google Scholar]; (b) Olsvik O, Popovic T, Skjerve E, Cudjoe KS, Hornes E, Ugelstad J, Uhlen M. Clin Microbiol Rev. 1994;7:43–54. doi: 10.1128/cmr.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mackensen A, Meidenbauer N, Vogl S, Laumer M, Berger J, Andreesen R. J Clin Oncol. 2006;24:5060–5069. doi: 10.1200/JCO.2006.07.1100. [DOI] [PubMed] [Google Scholar]; (d) Stamm C, Westphal B, Kleine HD, Petzsch M, Kittner C, Klinge H, Schümichen C, Nienaber CA, Freund M, Steinhoff G. The Lancet. 2003;361:45–46. doi: 10.1016/S0140-6736(03)12110-1. [DOI] [PubMed] [Google Scholar]
- 2.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. J Am Chem Soc. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Bonner WA, Hulett HR, Sweet RG, Herzenberg LA. Rev Sci Instrum. 1972;43:404–409. doi: 10.1063/1.1685647. [DOI] [PubMed] [Google Scholar]; (b) Fu AY, Spence C, Scherer A, Arnold FH, Quake SR. Nat Biotechnol. 1999;17:1109–1111. doi: 10.1038/15095. [DOI] [PubMed] [Google Scholar]
- 4.Miltenyi S, Müller W, Weichel W, Radbruch A. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 5.Šafařík I, Šafaříková M. J Chromatogr B. 1999;722:33–53. [PubMed] [Google Scholar]
- 6.Pruszak J, Sonntag KC, Aung MH, Sanchez-Pernaute R, Isacson O. Stem Cells. 2007;25:2257–68. doi: 10.1634/stemcells.2006-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Chalmers JJ, Zborowski M, Sun L, Moore L. Biotechnol Prog. 1998;14:141–148. doi: 10.1021/bp970140l. [DOI] [PubMed] [Google Scholar]; (b) Inglis DW, Riehn R, Austin RH, Sturm JC. Appl Phys Lett. 2004;85:5093–5095. [Google Scholar]; (c) Liu C, Lagae L, Wirix-Speetjens R, Borghs G. J Appl Phys. 2007;101:024913. [Google Scholar]; (d) Yellen BB, Erb RM, Son HS, Hewlin JR, Shang H, Lee GU. Lab Chip. 2007;7:1681–1688. doi: 10.1039/b713547e. [DOI] [PubMed] [Google Scholar]; (e) Yavuz C, Mayo JT, Yu W, Prakash A, Falkner J, Yean S, Cong L, Shipley H, Kan A, Tomson M, Natelson D, Colvin V. Science. 2006;314:964–967. doi: 10.1126/science.1131475. [DOI] [PubMed] [Google Scholar]
- 8.Adams JD, Kim U, Soh HT. Proc Natl Acad Sci U S A. 2008;105:18165–18170. doi: 10.1073/pnas.0809795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semple JW, Allen D, Chang W, Castaldi P, Freedman J. Cytometry. 1993;14:955–60. doi: 10.1002/cyto.990140816. [DOI] [PubMed] [Google Scholar]
- 10.Tiwari A, Punshon G, Kidane A, Hamilton G, Seifalian AM. Cell Biol Toxicol. 2003;19:265–272. doi: 10.1023/b:cbto.0000004929.37511.ed. [DOI] [PubMed] [Google Scholar]
- 11.Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, Ribas A, Heath JR. Nat Med. 2011;17:738–43. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Bath J, Turberfield AJ. Nat Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]; (b) Chen Y, Mao C. J Am Chem Soc. 2004;126:8626–8627. doi: 10.1021/ja047991r. [DOI] [PubMed] [Google Scholar]; (c) Shin JS, Pierce NA. J Am Chem Soc. 2004;126:10834–10835. doi: 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]; (d) Zhang DY, Seelig G. Nat Chem. 2011;3:103–13. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 13.Yurke B, Mills A. Genet Program Evolvable Mach. 2003;4:111–122. [Google Scholar]
- 14.Pilling D, Kitas GD, Salmon M, Bacon PA. J Immunol Methods. 1989;122:235–41. doi: 10.1016/0022-1759(89)90269-x. [DOI] [PubMed] [Google Scholar]
- 15.Strunz T, Oroszlan K, Schafer R, Guntherodt HJ. Proc Natl Acad Sci U S A. 1999;96:11277–11282. doi: 10.1073/pnas.96.20.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zlatanova J, Lindsay SM, Leuba SH. Prog Biophys Mol Biol. 2000;74:37–61. doi: 10.1016/s0079-6107(00)00014-6. [DOI] [PubMed] [Google Scholar]
- 17.(a) Nie CQ, Bernard NJ, Schofield L, Hansen DS. Infect Immun. 2007;75:2275–2282. doi: 10.1128/IAI.01783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Roberts SJ, Ng BY, Filler RB, Lewis J, Glusac EJ, Hayday AC, Tigelaar RE, Girardi M. Proc Natl Acad Sci U S A. 2007;104:6770–6775. doi: 10.1073/pnas.0604982104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Russell MS, Iskandar M, Mykytczuk OL, Nash JHE, Krishnan L, Sad S. J Immunol. 2007;179:211–220. doi: 10.4049/jimmunol.179.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.