Abstract
dsRNA-activated protein kinase (PKR) is activated by viral dsRNAs and phosphorylates eIF2a reducing translation of host and viral mRNA. Although infection with a chimeric West Nile virus (WNV) efficiently induced PKR and eIF2a phosphorylation, infections with natural lineage 1 or 2 strains did not. Investigation of the mechanism of suppression showed that among the cellular PKR inhibitor proteins tested, only Nck, known to interact with inactive PKR, colocalized and co-immunoprecipitated with PKR in WNV-infected cells and PKR phosphorylation did not increase in infected Nck1,2−/− cells. Several WNV stem-loop RNAs efficiently activated PKR in vitro but not in infected cells. WNV infection did not interfere with intracellular PKR activation by poly(I:C) and similar virus yields were produced by control and PKR−/− cells. The results indicate that PKR phosphorylation is not actively suppressed in WNV-infected cells but that PKR is not activated by the viral dsRNA in infected cells.
Keywords: West Nile virus, PKR, dsRNA, eIF2a, Nck
INTRODUCTION
PKR is a serine/threonine kinase composed of an N-terminal regulatory domain that contains two dsRNA binding motifs (DRBMs) and a C-terminal kinase domain (Meurs et al., 1990; Nanduri et al., 1998). These domains are connected by a spacer that provides an interface for dimerization (McKenna et al., 2007). It has been proposed that in the unphosphorylated state, the N-terminal regulatory domain interacts with the C-terminal catalytic domain to inhibit kinase activity (Nanduri et al., 2000). Activation of PKR by dsRNA results in the formation of dimers that are stabilized by autophosphorylation at multiple residues, including Thr446 and Thr451 which are located within the activation loop of the kinase domain and essential for PKR activation (Romano et al., 1998). To date, 18 PKR phosphorylation sites have been identified. Most are serine residues but some are threonine or tyrosine residues (Su et al., 2006; Toth et al., 2006). Active PKR dimers eject the activating dsRNA, presumably, due to phosphorylation of N-terminal residues and then phosphorylate eIF2a (Jammi and Beal, 2001). PKR is constitutively and ubiquitously expressed at low levels due to a kinase conserved sequence (KCS) site in its promoter (Toth et al., 2006). PKR expression is upregulated by Type I IFN which can be produced in response to a viral infection. The majority of PKR is located in the cytoplasm where a portion is associated with ribosomes. Some of the PKR in the nucleus associates with nucleoli (MacQuillan et al., 2009; Tanaka and Samuel, 1994; Toth et al., 2006).
A ternary complex consisting of GTP-eIF2 and a methionyl-tRNA delivers the charged initiator tRNA to the 40S ribosomal subunit of the 43S preinitiation complex but translation initiation requires the hydrolysis of the eIF2-bound GTP to a GDP (Hershey, 1991; Majumdar and Maitra, 2005). Under stress conditions, the alpha-subunit of eIF2 is phosphorylated by one of four eIF2a kinases: general control non-repressed 2 (GCN2), heme-regulated inhibitor (HRI), PKR-like endoplasmic reticulum (ER) kinase (PERK), or PKR (Kaufman, 1999). The eIF2a kinases share a conserved kinase domain that mediates eIF2a phosphorylation, but each responds to a different stress due to its unique regulatory domain (Kaufman, 1999). Phosphorylation of eIF2a on Ser51 leads to the formation of a high-affinity complex with the guanine exchange factor, eIF2B. This inhibits the exchange of GDP with GTP and "stalls" the preinitiation complexes on mRNAs (Sudhakar et al., 2000). Phosphorylation of as little as 20% of eIF2a significantly reduces the synthesis of most cellular proteins (Sudhakar et al., 2000). In virus-infected cells, PKR is activated by viral dsRNA. However, PKR can also be activated by Type I or II IFN by a mechanism mediated by the activated Jaks of the IFN receptor complex (Su et al., 2007), by heparin oligosaccharides, or by IL-3 withdrawal (Toth et al., 2006). PKR activation by peroxide or arsenite treatment is mediated through interaction of the activation domain of PACT with the N-terminal domain of PKR (Ito et al., 1999; Patel et al., 2000).
WNV, a member of the genus Flavivirus within the family Flaviviridae, was first isolated in 1937 from a febrile woman in the West Nile region of Uganda (Brinton, 2002). Until 1999, WNV was mainly confined to Southern Europe, the Middle East, Africa, West and Central Asia, Indonesia and Australia. In 1999, WNV extended into the Western hemisphere where it has since spread rapidly. The majority of WNV infections in humans are asymptomatic. Flu-like symptoms are observed in ~20 % and meningitis, encephalitis and/or paralysis occur in less than 1% of infected individuals (Brinton, 2002; Gubler, 2007). The WNV genome is a positive-sense, single-stranded RNA of ~11 Kb with a 5' cap but no 3' polyA tract. It encodes a single polyprotein that is co- and post-translationally cleaved to generate 3 structural proteins (E, prM and C) and 7 non-structural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b and NS5). The steps of the viral life cycle take place in the cytoplasm. WNV infection does not lead to shut-off of cellular protein synthesis. Viral RNA replication occurs in vesicles formed by invaginations of the ER membranes (Lindenbach, 2007). Nacent virions are assembled though the interaction of viral structural proteins associated with ER membranes with a newly synthesized viral RNA genome followed by budding into the lumen of the ER. Virions are transported through the Golgi system to the cell surface (Brinton, 2002; Gubler, 2007).
PKR has been reported to play a role in NFκB signaling and the control of cell growth through induction of p53 (Garcia et al., 2006) and also to be involved in IFN, PDGF, TNF-α, p38, JNK, STAT1 and IL-1 signaling (Garcia et al., 2006). The involvement of PKR in these multiple cellular processes requires its phosphorylation (Garcia et al., 2006). A number of cellular inhibitors have been identified that can form stable heterocomplexes with PKR and interfere with a step of the PKR activation process: (1) dsRNA recognition (C114 and RPL18), (2) dimerization (p58ipk) or (3) autophosphorylation (Hsp70 and Hsp90)(Garcia et al., 2007). The catalytic subunit of protein phosphatase 1 alpha (PP1a) dephosphorylates PKR resulting in dimer disruption (Tan et al., 2002).
The importance of PKR as a sentinel for the antiviral innate immune response is highlighted by the many reports indicating that most known viruses have evolved mechanisms for inhibiting PKR activity (Garcia et al., 2007). Viral components can either directly inhibit PKR activation or recruit cellular PKR inhibitors. Viral proteins, including Kaposi-sarcoma herpesvirus vIRF2 and LANA2, herpes simplex virus 1 (HSV-1) Us11, Epstein-Barr virus SM, vaccinia virus E3L and hepatitis C virus NS5A and E2 proteins, directly interact with PKR and inhibit either its binding to viral dsRNA or its activation. Overexpression of human papillomavirus E6 protein was reported to induce PKR localization to P-bodies where it is sequestered (Hebner et al., 2006). Viruses, such as adenoviruses and Epstein Barr virus produce small RNA inhibitors of PKR (Langland et al., 2006; Sharp et al., 1993). However, a recent report suggests that adenovirus also overcomes PKR activation by an alternative viral protein mediated mechanism (Spurgeon and Ornelles, 2009). Indirect mechanisms include, recruitment of cellular p58ipk by the influenza NS1 protein into a complex with PKR where it binds to the PKR dimerization interface preventing activation (Lee et al., 1990) and recruitment of PP1a by the HSV protein γ134.5 to dephosphorylate eIF2a (He et al., 1998).
Consistent with our previous data showing that a WNV Eg101 infection does not induce significant eIF2a phosphorylation in BHK cells (Emara and Brinton, 2007), PKR phosphorylation was not significantly induced in rodent cells after infection with either WNV Eg101 or other “natural” lineage 1 or 2 WNV strains. The activation of PKR in cells infected with many other types of viruses resulted in the evolution of viral-mediated processes to suppress PKR activation or activity. Evidence for a WNV-mediated mechanism of PKR suppression was not found. Instead, the results indicate that even though some WNV dsRNAs can activate PKR in vitro, WNV has developed a means to hide its dsRNA from PKR both at early and late times of the infection cycle so that PKR is not activated in infected cells.
RESULTS
PKR phosphorylation is not induced by infection of rodent cells with natural lineage 1 or 2 strains of WNV
PKR can be activated by viral dsRNA and phosphorylates eIF2a leading to attenuation of cell translation (Garcia et al., 2007). We previously reported that WNV-Eg101 infection of BHK cells did not induce significant eIF2a phosphorylation (Emara and Brinton, 2007). To determine whether the low level of eIF2a phosphorylation observed was due to a lack of PKR activation, PKR phosphorylation was initially assessed in mock-infected or WNV Eg101-infected (MOI 5) C3H/He mouse embryo fibroblasts (MEFs) (IFN-responsive). In mock-infected MEFs treated with 100 IU/ml of Type I IFN for 24 h, the levels of PKR and Thr451 phosphoryated (P)-PKR increased significantly (Fig. 1A). Both the PKR and P-PKR levels also increased with time after infection in WNV Eg101-infected MEFs but to lower levels than with IFN treatment.
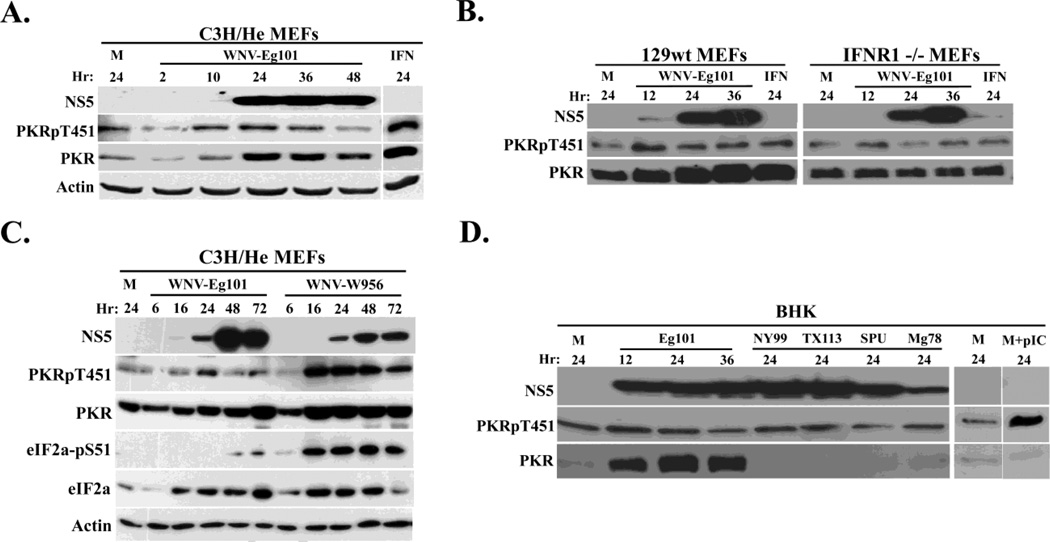
Figure 1. Analysis of PKR phosphorylation in WNV-infected cells.
(A) C3H/He MEFs were mock-infected (M), or infected with WNV Eg101 (MOI of 5) for the indicated times or treated with 100 U/ml universal type I IFN for 24 h (IFN). (B) 129wt or IFNR1−/− MEFs were mock-infected(M) or infected with WNV Eg101 (MOI of 5) for the indicated times or treated with 100 U/ml universal Type I IFN for 24 h.(C) C3H/He MEFs were mock-infected or infected with WNV Eg101 or WNV W956 at a MOI of 1 for the indicated times. (D) BHK cells were mock-infected (M) or infected with WNV Eg101 (MOI of 5) for the indicated times or infected with various strains of WNV (NY99, TX113, SPU and Mg78) for 24 h or transfected with 50 µg/ml poly(IC) for 2 h (pIC). PKR, phopho-Thr451 PKR, eIF2a, phosphor-S51 eIF2a, WNV-NS5 and actin were detected in cell lysates by Western blotting after separation of proteins by 10% SDS-PAGE.
PKR protein expression is known to be upregulated in response to Type I IFN signaling (Tanaka and Samuel, 1994; Toth et al., 2006) and PKR phosphorylation can be induced through direct interactions between PKR and activated JAK1 and/or Tyk2, two components of the Type I IFN receptor complex (Su et al., 2007). We previously reported that IFN-beta expression is upregulated in WNV-infected MEFs by 12 h after infection and that 100 to 600 IU/ml of IFN beta protein are secreted into the infected cell culture fluid (Scherbik et al., 2007). To determine whether the increases in the PKR and P-PKR levels observed in C3H/He MEFs were due to IFN-mediated PKR activation, the upregulation of PKR expression and phosphorylation was compared in IFNR1−/− and control wild type 129 (129wt) MEFs infected with WNV Eg101 at a MOI of 5. Cells treated with 100 IU/ml of Type I universal IFN (PBL Biomedical laboratories, NJ) for 24 h served as a positive control. In C3H/He MEFs, a slight increase in P-PKR and a significant increase in PKR levels compared to mock-infected cells were observed in WNV-infected and IFN-treated 129wt MEFs(Fig. 1B). In contrast, little if any increase in either PKR or P-PKR levels was observed in WNV-infected or IFN-treated IFNR1−/− MEFs (Fig. 1B). These results suggested that the small increase in P-PKR levels observed in WNV-infected MEFs was due to Type I IFN produced and secreted in response to the infection.
In contrast to the low levels of P-PKR induced by a WNV Eg101 infection in MEFs, a lineage 2/1 chimeric infectious clone-derived WNV (W956) induced much higher levels of both P-PKR and PKR (Fig. 1C). The levels of phosphorylated eIF2a were also significantly higher in cells infected with the WNV W956 virus. To determine whether the Eg101 strain was unique in its inability to induce PKR activation, IFN-non-responsive BHK cells were infected with Eg101 or another natural WNV lineage 1 (NY99 or Tx113) or lineage 2 (Mg78 or SPU) strain at a MOI of 5 for 24 h. Mock-infected BHK cells transfected with 50 µg/ml of poly(IC) for 2 h were used as a positive control. Little if any increase in PKR phosphorylation compared to mock levels was observed in BHK cells infected with WNV Eg101 or any of the four additional WNV strains tested (Fig. 1D). Unexpectedly, a dramatic increase in PKR levels was observed in WNV Eg101-infected BHK cells but no increase in PKR levels was observed in cells infected with the other WNV viruses (Fig. 1D). The high degree of upregulation of PKR levels by a WNV Eg101 infection appears to be restricted to BHK cells since Eg101 infection of neither C3H/He nor 129wt MEFs induced greater PKR upregulation than IFN treatment and no increase in total PKR was seen in the WNV Eg101-infected IFNR1−/− MEFs. This effect also did not correlate with the lineage or virulence of the virus strains tested. Due to the Type I IFN insensitivity of BHK cells, the observed PKR upregulation by WNV Eg101 in these cells is expected to be Type I IFN-independent. Although the mechanism of PKR expression upregulation in WNV Eg101-infected BHK cells was not investigated, it was previously reported that the Sp1 and Sp3 transcription factors can upregulate PKR expression in the absence of IFN (Toth et al., 2006). Overall, the data indicate that infection of rodent cells with natural strains of WNV does not result in significant upregulation of PKR phosphorylation.
PKR localization in WNV-infected cells
PKR is typically activated in virus-infected cells by dsRNA viral replication intermediates or hairpin structures within single-stranded viral RNAs (Garcia et al., 2006). WNV RNA replicates in the perinuclear region of infected cells in association with ER membranes (Gillespie et al., 2010; Mackenzie, 2005). To determine whether the cellular distribution of PKR was altered in WNV-infected cells, MEFs and BHK cells were mock-infected or infected with WNV Eg101 at a MOI of 5 for 10 or 24 h. The cells were fixed, permeabilized and incubated with anti-PKR and anti-dsRNA antibody and then visualized by confocal microscopy. PKR was observed to concentrate in the perinuclear region of infected cells and to co-localize with sites of viral RNA replication in both MEFs and BHK cells at 10 h and 24 h after infection (Fig. 2A). This change in PKR cytoplasmic distribution in WNV-infected cells was also observed when only an anti-PKR primary and a specific secondary antibody were used (data not shown). Because PKR serves as a sentinel of the innate immune response, many different types of viruses produce proteins that directly interact with PKR and inhibit its activation (Garcia et al., 2007). To assess PKR colocalization with nonstructural protein components of the WNV replication complex, BHK cells were mock-infected or infected with WNV Eg101 at a MOI of 5 for 24 or 30 h and PKR and the viral NS1, NS3, NS5 proteins were detected by confocal microscopy using viral protein specific antibodies. No co-localization was observed between PKR and NS3, NS5 or NS1 (Fig. 2B). The observed colocalization of PKR with dsRNA but not with individual viral replication complex proteins may be due to the higher signal intensity of the dsRNA antibody and/or to the more restricted distribution of the viral dsRNA replication intermediates compared to those of the nonstructural proteins.
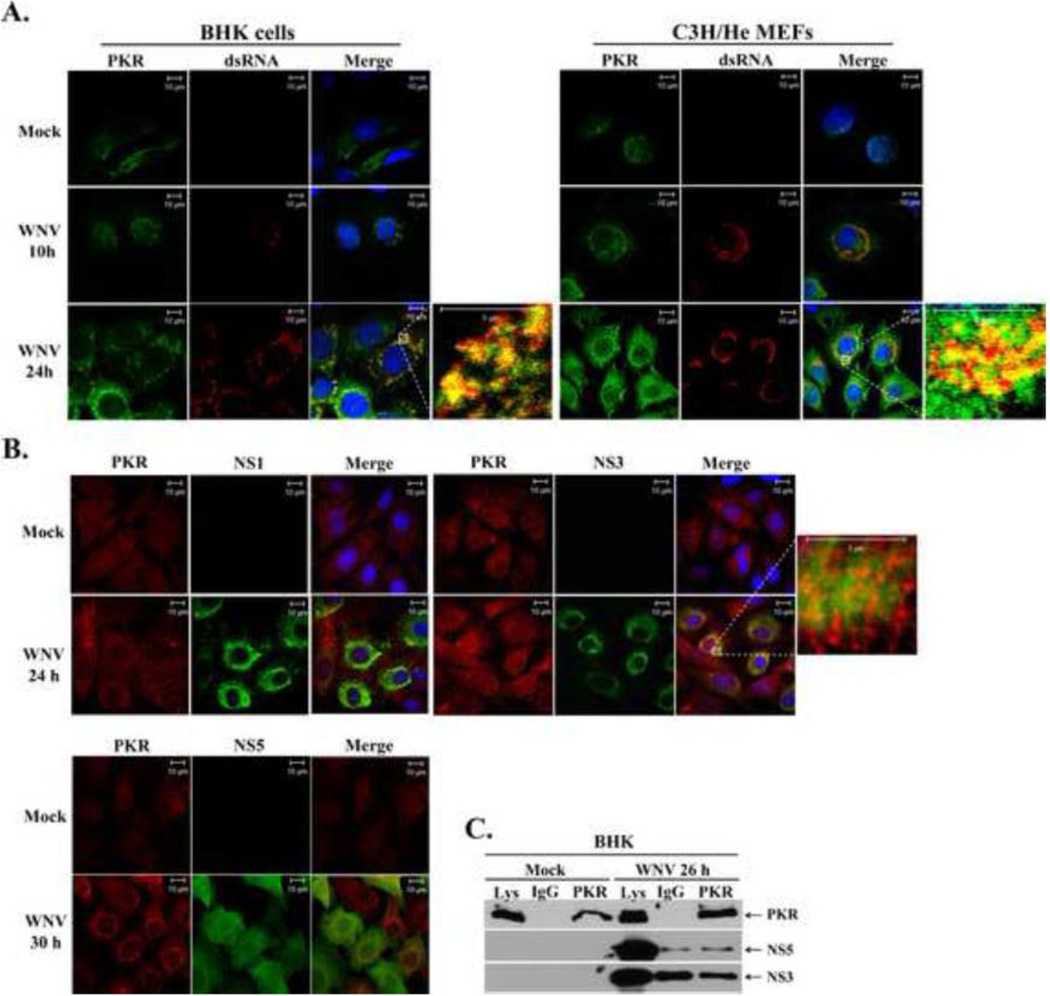
Figure 2. PKR colocalization with sites of WNV replication.
(A) Analysis of PKR colocalization with viral replication complexes. BHK cells and MEFs were mock-infected or infected with WNV Eg101 (MOI of 5). At 10 and 24 h after infection, cells were permeabilized, fixed, and blocked overnight. Cells were stained with anti-PKR and anti-dsRNA antibodies and then AlexaFluor488 (green) and AlexaFluor594 (red) conjugated secondary antibodies, respectively. Cell nuclei were stained with Hoechst. (B) Analysis of PKR colocalization with viral nonstructural proteins. BHK cells and MEFs were mock-infected or infected with WNV Eg101 (MOI of 5). At 24 and 30 h after infection, cells were permeabilized, fixed, and blocked overnight. Cells were stained with anti-PKR and either anti-NS5, anti-NS3 or anti-NS1 antibodies and then AlexaFluor488 (green) and AlexaFluor594 (red) conjugated secondary antibodies, respectively. Cell Nuclei were stained with Hoechst. (C) Analysis of PKR interaction with viral NS3 or NS5 proteins in infected cells. BHK cells were mock-infected (M), or infected with WNV Eg101 (MOI of 5). At 26 h after infection, cells were lysed, S2 fractions were prepared and rabbit anti-PKR antibody or a rabbit IgG was used for immunoprecipitation. Immunoprecipitated proteins were separated by 10% SDS-PAGE and detected by Western blotting using anti-NS5 or anti-NS3 antibodies.
As an additional means of assessing whether PKR associates with viral protein components of the replication complexes, PKR was immunoprecipitated from mock-infected and WNV-infected BHK cells and the precipitates were immunoblotted with anti-NS3 or anti-NS5 antibodies. A species-specific IgG was used as a negative control. Neither NS3 nor NS5 co-immunoprecipitated with PKR (Fig. 2C). These data suggest that although PKR colocalizes with viral replication complexes, it does not interact directly with the NS3 protein, which is a marker for the membrane bound replication complexes, nor with the NS5 (methyl transferase/RNA-dependent RNA polymerase) protein. However, indirect mechanisms of PKR suppression mediated by these viral proteins were not ruled out. Antibodies to additional WNV nonstructural proteins appropriate for confocal microscopy and co-immunoprecipitation were not available; therefore, PKR interaction with these proteins could not be tested.
PKR does not colocalize with known cellular PKR inhibitors in WNV-infected cells
In addition to inhibiting PKR by direct interaction, viral proteins have also been reported to recruit cellular PKR inhibitors or sequester PKR in P bodies (Garcia et al., 2007). To assess whether WNV proteins recruit cellular PKR inhibitors to associate with PKR and inhibit its activation in WNV infected cells, BHK cells were infected with WNV Eg101 at a MOI of 5. At 24 h or 30 h after infection, cells were fixed, permeabilized and incubated with anti-PKR antibody and an antibody to a PKR inhibitor followed by incubation with fluorescent tagged secondary antibodies and visualization by confocal microscopy. Either a mouse monoclonal or a rabbit polyclonal anti-PKR antibody was used depending on the species in which the antibody to the cellular protein being tested was made. The mouse monoclonal antibody detected nuclear PKR more efficiently than the polyclonal antibody. With the exception of PP1a, known cellular PKR inhibitors must remain associated with PKR to mediate their inhibitory effect and therefore would be expected to co-localize with PKR. Neither p58ipk nor PP1a concentrated in the perinuclear regions of WNV-infected cells or colocalized with PKR (Fig. 3). Similarly, PKR did not localize to P-bodies, detected with Dcp1a antibody, in WNV-infected cells (Fig. 3).
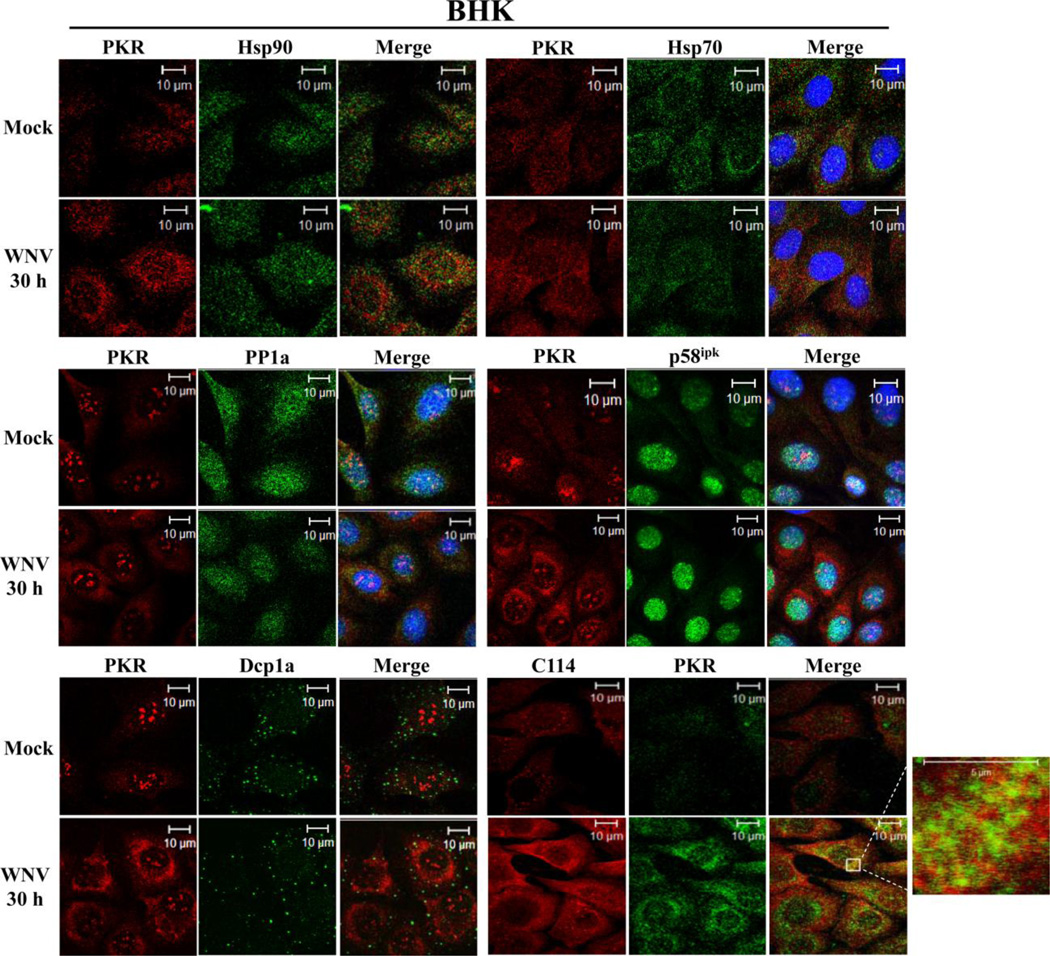
Figure 3. Analysis of PKR colocalization with known cellular PKR inhibitors in WNV-infected cells.
Colocalization of PKR with known cellular PKR inhibitors. BHK cells were mock-infected or infected with WNV Eg101 (MOI of 5). At 30 h after infection, cells were permeabilized, fixed and blocked overnight. Cells were stained with anti-PKR and an antibody to a cellular PKR inhibitor and then with AlexaFluor488 (green) and AlexaFluor594/555 (red) conjugated secondary antibodies. PKR was detected with a mouse monoclonal anti-PKR antibody in the PP1a, p58ipk and Dcp1a experiments and with a rabbit polyclonal antibody in the Hsp70, Hsp90 and C114 experiments.
In uninfected cells, cellular protein inhibitors can bind PKR and modulate its activation. Hsp90 and Hsp70 have both been shown to inhibit PKR by masking its autophosphorylation sites (Donze et al., 2001; Pang et al., 2002; Pratt and Toft, 2003). C114, an IL-11 inducible nuclear dsRNA-binding protein that shuttles between the cytoplasm and nucleus, can inhibit PKR activation through interaction with the PKR DRBMs (Yin et al., 2003). Neither Hsp90, Hsp70, nor C114 were observed to colocalize with PKR in WNV-infected cells (Fig. 3). These data suggest that suppression of PKR phosphorylation in WNV-infected cells is not mediated by any of these cellular PKR inhibitors.
PKR colocalizes with the PKR inhibitor Nck in WNV-infected cells
Nck is an adapter protein that has been reported interact with and directly limit activation of three of the eIF2a kinases PKR, PERK, and HRI, but not GCN2 and specifically modulate eIF2a phosphorylation under various stress conditions (Cardin et al., 2007). Nck binds to only the inactive form of PKR. However, dsRNA can outcompete Nck for PKR binding leading to PKR activation. In a previous in vitro study, 2 µg/ml of poly(I:C) was shown to be sufficient to overcome Nck-mediated inhibition of PKR phosphorylation (Cardin and Larose, 2008). In WNV-infected BHK cells, Nck concentrated in the perinuclear region and colocalized with PKR (Fig. 4A). Nck protein levels were high in uninfected cells and did not increase after WNV-infection in BHK cells (Fig. 4B). Interaction between Nck and PKR was analyzed by co-immunoprecipitation. Lysates made from WNV-infected BHK cells at 24 h after infection were incubated with rabbit anti-PKR antibody or a control nonspecific rabbit IgG and the bound proteins were separated by SDS-PAGE and immunoblotted using a rabbit anti-Nck antibody. Conversely, rabbit anti-Nck antibody or a control nonspecific rabbit IgG was used for immunoprecipitation and mouse-anti-PKR antibody was used for Western blotting. Nck was co-immunoprecipitated by PKR antibody but not by the nonspecific IgG (Fig. 4C, upper panel). Similarly, PKR was co-immunoprecipitated by Nck antibody (Fig. 4C, lower panel). A higher amount of PKR was co-immunoprecipitated by the anti-Nck antibody from WNV-infected cell lysates than from mock-infected cell lysates consistent with the increased level of PKR in these cells. A decrease rather than an increase in the Nck-PKR interaction would be expected if the amplified levels of viral dsRNA present in infected cells by 24 hr had competed with Nck for binding to PKR. The observed colocalization and association of Nck and PKR in WNV-infected cells suggested that most of the PKR that colocalizes with viral dsRNA in infected cells is inactive.
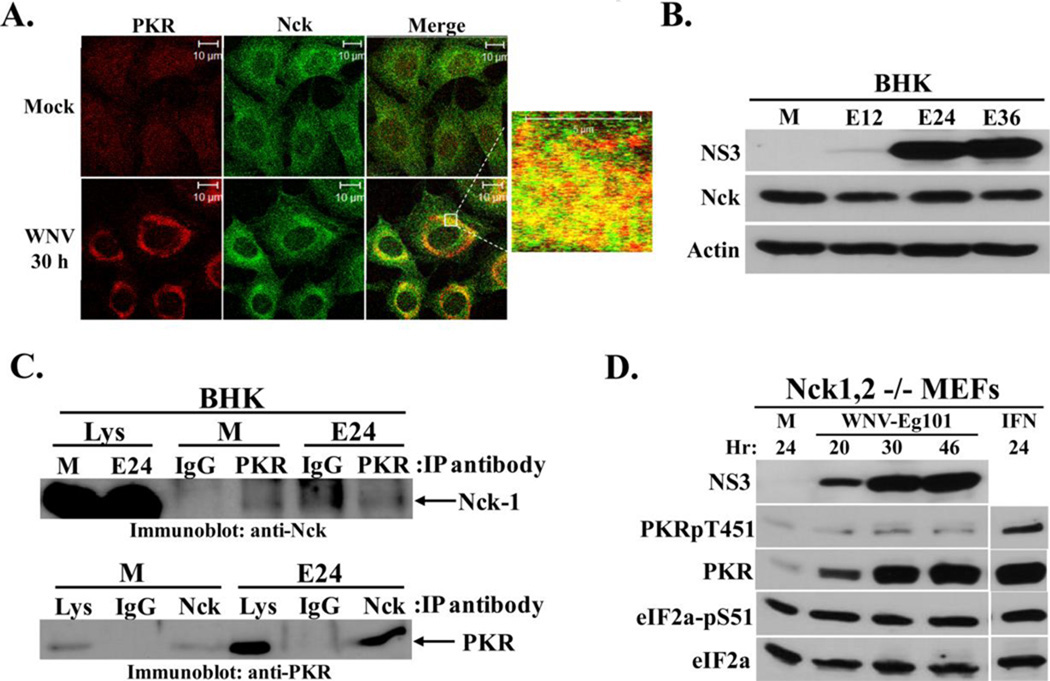
Figure 4. Analysis of Nck-PKR interactions in WNV-infected cells.
(A) Analysis of colocalization of PKR with Nck. BHK cells were mock-infected or infected with WNV Eg101(MOI of 5). At 30 h after infection, cells were permeabilized, fixed and blocked overnight. Cells were stained with mouse monoclonal anti-PKR and rabbit polyclonal anti-Nck antibodies and then with AlexaFluor488 (green) and AlexaFluor594/555 (red) conjugated secondary antibodies. (B) Analysis of Nck protein levels in WNV-infected BHK cells. Cells were mock-infected (M), or infected with WNV Eg101 (MOI of 5) for the indicated times. NS3, Nck-1, and actin were detected by Western blotting after separation of proteins by 10% SDS-PAGE. (C) Co-immunoprecipitation of Nck and PKR. BHK cells were mock-infected (M) or infected with WNV Eg101 (MOI of 5). At 24 h after infection, cells were lysed and S2 fractions were prepared. Rabbit anti-PKR antibody or a rabbit anti-Nck antibody was used for immunoprecipitation; rabbit IgG was used as a control antibody.. Immunoprecipitated proteins were separated by 10% SDS-PAGE and detected by Western blotting using a rabbit anti-Nck antibody (top panel) or a mouse anti-PKR antibody (bottom panel). (D) Analysis of PKR phosphorylation in WNV-infected Nck-knockout cells. Nck-1,2−/− MEFs were mock-infected (M) or infected with WNV Eg101 (MOI of 5) for the indicated times or treated with 100 U/ml universal type I IFN for 24 h (IFN). NS3, total PKR, phopho-Thr451 PKR, total eIF2a and phospho-Ser51 eIF2a were detected by Western blotting after separation of proteins by 10% SDS-PAGE. Blots shown are representative of at least two independent experiments.
Mammalian genomes have two Nck genes. The Nck-1 and Nck-2 proteins encoded by these genes share 68% amino acid homology and have been reported to have redundant functions based on the results of studies with single and double-knock out MEFs (Latreille and Larose, 2006). As an additional means of determining whether Nck plays a direct role in inhibiting PKR activation in WNV infected cells, double knock out Nck1,2−/− MEFs were infected with WNV Eg101 at a MOI of 5. Similar to what was observed with control 129wt and C3H/He MEFs (Fig. 1A and B), PKR levels increased significantly in WNV-infected and IFN-treated Nck1,2−/− MEFs with time after infection (Fig. 4D). However, while only a minimal increase in PKR phosphorylation levels was detected after WNV-infection of Nck1,2−/− MEFs, a significant increase in PKR phosphorylation was observed after IFN-treatment of these cells (Fig. 4D). The results indicate that even when the PKR-associated inhibitor Nck is absent, PKR is still only minimally activated in WNV-infected cells.
The level of phosphorylated eIF2a was high in uninfected Nck1,2−/− MEFs and did not increase after infection or IFN treatment. Nck can form a complex with phosphorylated eIF2a and recruit PP1a to dephosphorylate eIF2a (Latreille and Larose, 2006). The absence of Nck has been reported to increase the sensitivity of PKR, PERK and HRI kinase activation and also to reduce the efficiency of eIF2a phosphorylation by PP1a (Cardin et al., 2007). Consistent with the higher level of eIF2a phosphorylation in Nck1,2−/− MEFs, viral yields from these cells were decreased by about 10 fold (data not shown).
PKR autophosphorylation is induced by viral RNAs in vitro
PKR binds dsRNA in a sequence non-specific manner but the structure of the RNA plays a critical role in PKR activation. The binding of an activating dsRNA to PKR leads to the formation of active PKR dimers followed by the release of the activating dsRNA. Activated PKR dimers have been reported to have reduced affinity for poly(I:C) (Jammi and Beal, 2001; Langland and Jacobs, 1992; Lemaire et al., 2005). Viral PKR inhibitor RNAs, such as adenovirus VAI RNA and Epstein-Bar virus EBER-1 RNA, bind to PKR but their structures inhibit PKR activation (Garcia et al., 2006). Neither of these inhibitor RNAs induced activation of PKR in vitro at any of the concentrations tested (McKenna et al., 2006; McKenna et al., 2007). In competition assays with an activator RNA, optimal inhibition by either VAI RNA or EBER-1 RNA was observed when concentrations of these RNAs were 2–10 times higher than that of an activator RNA.
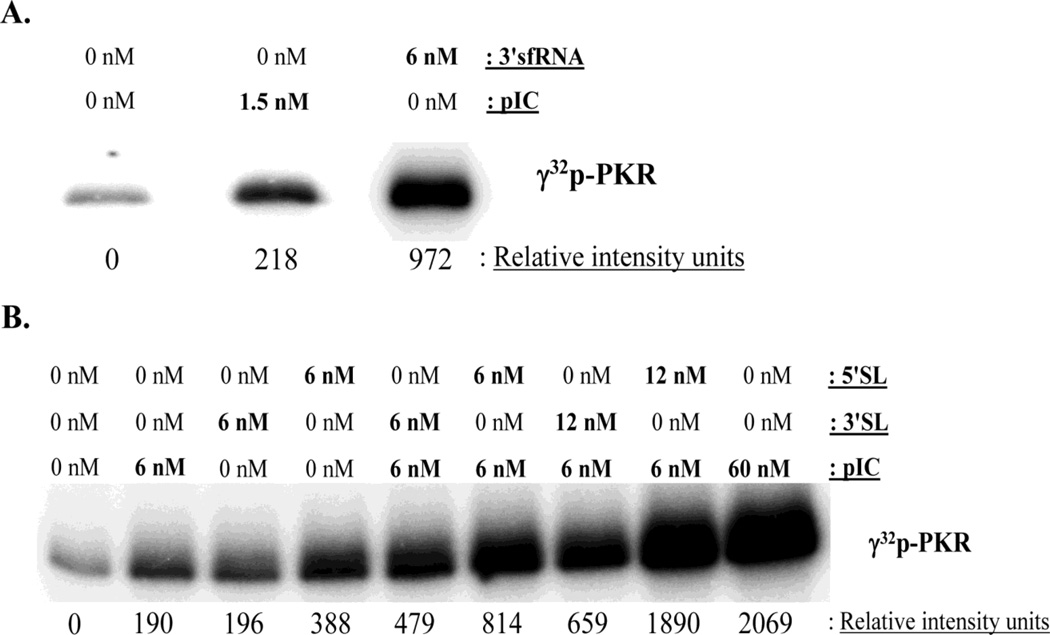
A WNV genomic RNA fragment consisting of the 3' terminal 529 nts (3'sfRNA) accumulates in infected cells as well as in mouse brains (Lin et al., 2004; Scherbik et al., 2006; Urosevic et al., 1997) and is generated by XRN1 5' digestion (Pijlman et al., 2008). The 3'sfRNA is significantly larger (529 nts) than known viral PKR inhibitor RNAs (VAI RNA 136 nts; EBER-1 RNA 157 nts). However, several regions of the mFold predicted structure of the 3'sfRNA (data not shown) have similarity to the VAI and EBER-1 RNA structures (McKenna et al., 2007). The ability of the WNV 3'sfRNA to inhibit PKR activation was tested in an in vitro PKR autophosphorylation assay. Poly(I:C) was used as a control activator RNA. Incubation of either poly(I:C) or 3'sfRNA with purified recombinant PKR induced PKR autophosphorylation (Fig. 5A).
Figure 5. In vitro PKR autophosphorylation assays.
(A) Reaction mixtures containing 150 ng of purified PKR alone or with the indicated concentrations of in vitro transcribed WNV 3'sfRNA or poly(I:C) were incubated as described in Materials and Methods. (B) Reaction mixtures containing 150 ng of purified PKR alone or with the indicated concentrations of in vitro transcribed WNV 3'SL or 5'SL RNA or with poly(I:C) were incubated for 30 min as described in Materials and Methods, or preincubated with the 3'SL or 5'SL for 10 min and then with poly(I:C) for 30 min. γ32P-ATP was included in the reactions as a phosphate donor. Image Gauge 3.2 software was used to measure the relative band intensities of images acquired using a Fuji BAS 2500 analyzer. Results are representative of two independent experiments.
Flavivirus genomic RNAs contain conserved terminal RNA structures (Brinton, 2002; Lindenbach, 2007). Since the structures of the WNV 3' and 5' terminal stem-loop (SL) RNAs are also similar to those of the known viral RNA PKR inhibitors, the ability of these RNAs to inhibit PKR activation was also assessed. Both the 3'SL and 5'SL WNV RNAs induced significant PKR autophosphorylation; the 5' SL RNA induced a 2-fold greater increase in PKR activation than the 3'SL RNA (Fig. 5B). Also, preincubation of either of these RNAs with PKR led to an increase not a decrease in PKR activation by poly(I:C) above that seen with either RNA alone. The data indicate that the WNV 3'sf, 3'SL and 5'SL RNAs can activate PKR in vitro.
Poly(I:C)-mediated PKR activation in WNV-infected BHK cells
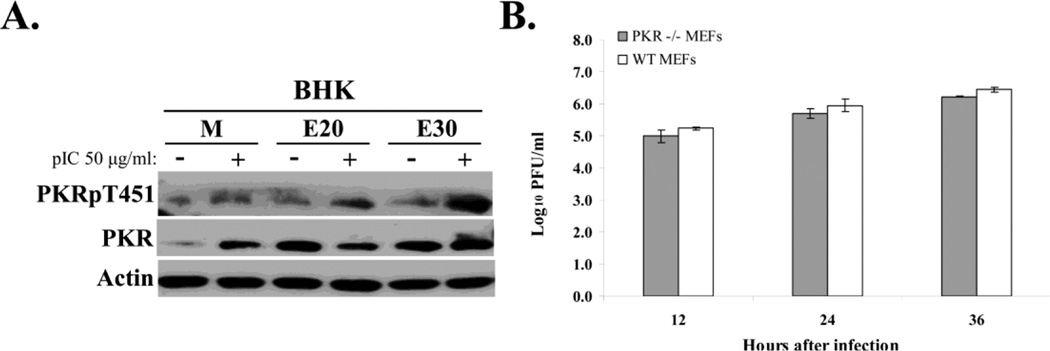
As an additional means of analyzing whether PKR is actively suppressed in WNV-infected BHK cells, the ability of poly(I:C) to activate PKR in infected cells was assessed. In preliminary experiments, the minimum concentration of poly(I:C) required for maximum intracellular PKR activation was determined to be 50 µg/ml (data not shown). BHK cells were mock-infected or infected with WNV Eg101 at a MOI of 5 and at 20 h and 30 h after infection, cells were transfected with 50 µg/ml of poly(I:C) or incubated with transfection reagent alone for 1.5 h. The observed increase in PKR phosphorylation in response to poly(I:C) transfection in infected cells (Fig. 6A) suggested that PKR activation is not actively suppressed in WNV-infected BHK cells. In additional experiments using lower poly(I:C) concentrations (10 or 25 ug/ml), similar levels of PKR activation were observed in infected and uninfected cells (data not shown). The data indicate that WNV infection does not interfere with the ability of PKR to respond to a dsRNA activator.
Figure 6. Analysis of active suppression of PKR activation in infected cells and of PKR anti-flaviviral activity.
(A) Poly(I:C)-mediated PKR autophosphorylation in WNV-infected cells. BHK cells were mock-infected or infected with WNV Eg101 (MOI of 5). At 20 or 30 h after infection, cells were transfected with 50 µg/ml of poly(I:C) in Celfectin II (+) or transfection reagent alone (−) for 1.5 h before cell lysis. PKR, phopho-Thr451 PKR and actin were detected in cell lysates by Western blotting after separation of proteins by 10% SDS-PAGE. (B) Viral yields produced by PKR−/− and wildtype MEFs infected with WNV Eg101 (MOI of 5). Samples of culture fluid were harvested at the indicated times, and infectivity titers were determined by plaque assays done in duplicate on BHK cells. Virus titers are expressed as log10 PFU/ml. Error bars indicate ± standard error of the mean (SEM) (n = 3).
The antiviral effect of PKR in WNV-infected cells was assessed by comparing viral yields from WNV-infected wildtype and PKR−/− MEFs. Virus yields in samples of culture fluid collected at the indicated times after infection with WNV Eg101 (MOI of 5) was determined by plaque assay on BHK cells. Comparable virus titers were observed in both cell types at 12, 24 and 36 h after infection (Fig. 6B) indicating that PKR antiviral activity does not affect WNV yield.
DISCUSSION
PKR is activated by viral dsRNA and exerts its antiviral effect through phosphorylation of eIF2a. Because eIF2a phosphorylation leads to attenuation of protein synthesis, many viruses have evolved mechanisms to avoid, block or suppress PKR activation. Some viruses use alternative translational mechanisms. For example, caliciviruses encode viral proteins that act as cap-analogues while picornaviruses initiate translation from an internal ribosome entry site (IRES) in an eIF2a-independent manner (Lopez-Lastra et al., 2010). Other viruses produce a small RNA or a viral protein that either directly or indirectly inhibits PKR activity (Garcia et al., 2007). WNV genome RNA translation is cap-dependent and so would be susceptible to eIF2a-mediated translation inhibition. However, shut-down of host translation does not occur in flavivirus infected cells (Lindenbach, 2007). We previously reported only low levels of eIF2a phosphorylation in WNV-infected BHK cells (Brinton, 2002; Emara and Brinton, 2007). Consistent with this observation, no increase in PKR phosphorylation in WNV-infected BHK cells and only a slight increase in infected wild type MEFs mediated by IFN was observed in the present study.
The authors of a previous study done with WNV virus-like particles (VLPs) containing WNV replicons concluded that PKR activation provides antiviral protection against WNV (Gilfoy and Mason, 2007). However, a similar number of foci forming units (FFU) were detected in wild type and PKR−/− MEFs 48 h after infection with WNV VLPs and IFN-treatment led to a reduction in FFUs in wild type but not in PKR−/− MEFs suggesting that the PKR activation observed was mediated by IFN and not by WNV dsRNA. The results of this previous study are consistent with those of the present study. An additional previous study, that utilized VLPs containing a WNV replicon with a C-terminal EMCV IRES driving translation of a neomycin gene ORF (Jiang et al., 2010), reported a reduction in virus yield from cells overexpressing PKR compared to control cells. The EMCV IRES was previously shown to activate PKR in vitro and in vivo (Arnaud et al., 2010; Shimoike et al., 2009) and may have contributed to the PKR activation observed.
Suppression of PKR by known cellular inhibitors requires association with PKR (Garcia et al., 2007). Among the cellular inhibitors tested, only Nck colocalized with the PKR that concentrated in the perinuclear region of WNV-infected cells. Since Nck interacts with inactive PKR, the detection of Nck colocalizing with PKR in the perinuclear region of infected cells suggested that the majority of the PKR in this region is in an inactive state. Co-immunoprecipitation of Nck and PKR in both infected and uninfected cells suggested that this interaction was not disrupted by WNV infection. It was previously reported that a minimum of 2 µg/ml of poly(I:C) was sufficient to outcompete Nck binding to PKR (Cardin and Larose, 2008). However, even though the WNV genome RNA and the 3'sfRNA, which were shown to contain several structures that could activate PKR in vitro, as well as viral dsRNA replicative intermediates increase exponentially between 6 and 24 hr after infection, these viral RNAs did not compete with Nck for binding to PKR. The minimal activation of PKR detected in WNV-infected Nck1,2−/− MEFs provided additional evidence that PKR was not activated by viral dsRNA in infected cells and the observation that poly(I:C) induced significant PKR phosphorylation in WNV-infected cells suggested that PKR phosphorylation is not actively suppressed in WNV-infected cells. The similar virus yields produced by wildtype and PKR−/− MEFs infected with WNV provided additional confirmation that PKR does not mediate significant antiviral activity in WNV-infected cells. PKR was reported to associate with ER membranes (Garcia et al., 2007) and the perinuclear concentration of PKR observed in WNV-infected cells could be a by-stander effect of virus-directed ER membrane rearrangement rather than due to recruitment by a viral component.
Although the reasons why viral RNA does not bind to and activate PKR in WNV-infected cells are not known, some of the known characteristics of flavivirus infections provide possible clues. The viral capsid protein forms dimers that associate with ER membranes and viral RNA (Lindenbach, 2007) and these interactions may prevent PKR from binding to the genomic RNA. Also, the WNV genomic 3'SL RNA was previously reported to bind to several cellular proteins that are thought to be required for efficient initiation of viral minus strand RNA synthesis (Brinton, 2002; Davis et al., 2007) and the interactions with these proteins may prevent the interaction of this SL with PKR. The WNV genomic RNA contains a 5' cap, an RNA modification reported to prevent PKR activation by cellular RNAs (Nallagatla et al., 2007). Interactions between the 5'SL and translation factors may mask this structure from detection by PKR. Alternatively, the reported interaction between NS5 and 5' nts of the viral genome (Dong et al., 2008) may prevent the RNA structures in this region from interacting with PKR. Flaviviruses replicate in the cytoplasm and induce extensive ER membrane proliferation and rearrangement. Perinuclear vesicles that are formed by invaginations of the rough ER membrane contain the double-stranded viral RNA replication intermediates and once formed facilitate an exponential amplification of genome RNA (Gillespie et al., 2010; Mackenzie, 2005; Welsch et al., 2009). At later times in the infection cycle, the sequestering of replicating dsRNA in vesicles, the close proximity of viral RNA replication sites to sites of genome RNA translation and packaging and the membrane association of the capsid proteins may all participate in the evasion of WNV dsRNA regions from detection by PKR. However, it is not known how the viral RNAs are hidden from PKR during the early stages of the infection cycle when viral genome and antisense RNA replication occurs symmetrically at low levels (Lindenbach, 2007). Possibly the the viral RNA levels are not high enough to outcompete Nck efficiently.
Viral RNA tertiary interactions may play a role in preventing interaction with PKR. Also, GU wobble pairs in RNAs were previously shown to inhibit PKR activation in vitro (Nallagatla and Bevilacqua, 2008). Not only the number but also the clustering of GU pairs is required to sufficiently alter a dsRNA structure so that it is unable to activate PKR (Nallagatla and Bevilacqua, 2008). More than 500 GU wobble base pairs (bp) were predicted to form in a whole genome fold of the WNV Eg101 RNA (Ann Palmenberg, unpublished data). In the context of the whole genomic RNA, 13 of the 22 GU pairs formed by the small 3'sfRNA sequence are long distance interactions between nts in this region and nts located near the 5' end of the viral RNA and most of these GU pairs are clustered. However, the "free" 3'sfRNA is predicted to form only 12 GU pairs and these are not clustered. The decrease in GU pairs would be expected to increase the ability of the "free" 3'sfRNA to activate PKR. The location of the small 3'sfRNAs that accumulate in infected cells is not known. They may also be “hidden” from PKR though association with multiple cell proteins and/or be sequestered in a cell compartment.
The finding that WNV infections do not actively suppress PKR is a novel mechanism of viral evasion of the antiviral activity of PKR. In contrast to infections with the natural WNV strains tested in this study, a W956 chimeric virus infection efficiently activated PKR. Studies are currently underway utilizing the W956 virus as a tool to gain additional insights about how natural WNV strains avoid activating PKR (Courtney, Scherbik and Brinton, unpublished data).
MATERIALS AND METHODS
Cell lines and viruses
Simian virus 40 (SV40)-transformed C3H/He and C57BL/6 mouse embryo fibroblast (MEF) lines, as well as BHK-21 WI2 cells (Vaheri et al., 1965), were grown as previously described (Scherbik et al., 2006). PKR+/+ and PKR−/− (generated by Charles Weissmann, Scripps Research Institute and provided by Scott Kimball, Pennsylvania State University), SV40-transformed IFNR1−/− and 129 wild-type (129wt) (provided by Herbert Virgin, Washington University, St. Louis, Mo), Nck1,2−/− (provided by Tony Pawson, Samuel Lunenfield Research Institute, Ontario, Canada) MEFs were maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle medium (DMEM) containing high glucose, 10% heat inactivated fetal bovine serum (FBS), 100 µg/ml streptomycin and 100 IU/ml penicillin.
Aliquots of lineage 1 (Eg101, Tx113 and NY99) and lineage 2 (SPU and Mg78) strains of WNV were obtained from Robert Tesh (University of Texas Medical Branch, Galveston, TX). A pool of each virus was prepared by infecting BHK cells at a MOI of 0.1 and harvesting culture fluid 32 h after infection. Clarified culture fluid (1 × 108 PFU/ml, Eg101; 3 × 106 PFU/ml, Tx113; 5 × 107 PFU/ml, NY99; 2 × 107 PFU/ml, SPU; 3 × 106 PFU/ml, Mg78) was aliquoted and stored at −80°C. Plaque assays to measure virus infectivity titers were done in BHK cells as previously described (Scherbik et al., 2006). The construction of the lineage 2/1 chimeric virus genome, referred to as W956, was described previously (Yamshchikov et al., 2001). The W956 genome sequence from the 5' end through the N-terminus of NS5 is from a highly passaged WNV lineage 2 D117B956 strain while the 3' 1496 nts are from the lineage 1 Eg101 strain. A stock of this virus was produced by transfecting BHK cells with 1 µg of in vitro transcribed infectious clone RNA and harvesting culture fluide 72 h after transfection. Clarified culture fluid (~5 × 107 PFU/ml) was aliquoted and stored at −80°C.
Confocal microscopy
BHK cells (2 × 103 cells per well) were seeded on 12.5-mm coverslips, in 24-well plates and 24 h later, the cells were counted and infected with WNV-Eg101 at a MOI of 5. At the indicated times, cells were fixed in 4% paraformaldehyde and then permeabilized using cold 100% methanol. The cells were washed in PBS and incubated overnight at 4°C in blocking buffer (5% heat inactivated horse serum in PBS). The cells were then incubated with a primary antibody diluted in blocking buffer for 1 h at room temperature, washed three times with PBS, and incubated for 1 h at room temperature with AlexaFluor-350, −488, −594 or −555 conjugated secondary antibodies (Invitrogen) diluted in blocking buffer. Cell nuclei were stained with 0.5 µg/ml Hoechst 33258 (Molecular Probes) added during the secondary antibody incubation. Coverslips were mounted with Prolong mounting medium (Invitrogen), and the cells were viewed and photographed with a Zeiss LSM 510 confocal microscope (Zeiss, Germany) using either a 63× or 100× oil immersion objective. The images were merged and analyzed using Zeiss software version 3.2. The same camera settings were used for all images in an experimental series. Primary antibodies used were: mouse monoclonal anti-PKR (B10) (1:100) or rabbit polyclonal anti-PKR (1:100) (Santa Cruz Biotechnology), mouse anti-dsRNA (1:200) (English and Scientific Consulting, Hungary), mouse anti-WNV NS1 (1:100) (Millipore), goat anti-WNV NS3 (1:300) (R&D Systems), mouse anti-WNV NS5 (1:700) (a gift from Pei-Yong Shi, Wadsworth Center, NY State Department of Health, NY), mouse anti-Hsp90 (1:100) (Stressgen), mouse anti-Hsp70 (1:100) (Stressgen), rabbit anti-p58ipk (1:100) (Cell Signaling), rabbit anti- PP1a (1:100) (Cell Signaling), anti-Dcp1a (a gift from J. Lykke-Anderson, University of Colorado, Boulder, CO), rabbit anti-Nck (1:100) (Millipore), mouse anti-C114 (1:100) (Sigma Aldrich) and mouse-anti-RLP18 (1:500) (Sigma Aldrich).
Co-immunoprecipitation assay
BHK cells (2 × 107) were mock-infected or infected with WNV at a MOI of 5. At 26 h after infection, cells were collected in NP-40 lysis buffer [50 mM sodium phosphate (pH 7.2), 150 mM NaCl, 1% Nonidet P-40, and EDTA-free Complete Mini Protease Inhibitor Cocktail (Roche)]. Cell lysates were incubated on ice for 30 min, sonicated, and then centrifuged at 2,000 × g for 5 min at 4°C to make S2 fractions. S2 lysates (500 µl per reaction containing ~500 µg of total protein) were incubated with 1 µg of rabbit anti-PKR-1 or a nonspecific rabbit IgG antibody overnight at 4°C with rotation. Protein G-magnetic beads (New England Biolabs) were added and incubation was continued for 1 h. Beads were collected magnetically, washed seven times with 1 ml of lysis buffer, and proteins were eluted by boiling for 5 min. Proteins were separated by 10% SDS/PAGE, transferred to nitrocellulose membranes, and analyzed by Western blotting using one of the following antibodies: mouse anti-PKR (Santa Cruz), rabbit anti-Nck (Millipore), mouse anti-NS5 (a gift from Pei-Yong Shi, Wadsworth Center, NY State Department of Health, NY) or goat anti-NS3 (R&D systems).
Western blot analysis
Cells were seeded into 12-well plates and grown to ~100% confluency and infected at a MOI of 5. Cells were lysed in RIPA buffer [1X PBS, 1% NP-40, 0.5% sodium deoxycholate, 1% SDS and protease inhibitor cocktail (Roche)] at the indicated times after infection, lysates were collected and 2X Sample Buffer [20% SDS, 25% glycerol, 7% 0.5M Tris-HCl (pH 6.8), 0.5% bromophenol blue, and 5% 2-mercaptoethanol] was added. The samples were boiled for 5 min and proteins were resolved by SDS-PAGE. The proteins were transferred to a nitrocellulose membrane. The membrane was blocked with 5% BSA or 5% non-fat dry milk (NFDM) in 1X TBS + 0.05% Tween-20 (TBST) for 1 h at room temperature. A primary antibody was then incubated with the membrane overnight at 4°C, the membranes were washed 3 times in 1X TBST and then incubated with a secondary antibody diluted in 5% NFDM-TBST for 1 h at room temperature. After washing the membrane twice in 1X TBST and once in 1X TBS, membranes were processed for enhanced chemiluminescence using a Super-Signal West Pico detection kit (Pierce, Rockford, IL) according to the manufacturer's instructions. Membranes were incubated with one of the following primary antibodies: anti-p-eIF2a (Ser51) (Cell Signaling) (1:500 in 5% BSA-TBST) and anti-eIF2a (1:1000 in 5% NFDM-TBST) (Cell Signaling), anti-PKR (1:3000 in 5% NFDM-TBST) (Santa Cruz), anti-PKR-pT451 (1:500 in 5% BSA-TBST) (Millipore), anti-Nck (1:1000 in 5% NFDM-TBST) (Millipore) or anti-actin (1:40,000 in 5% NFDM-TBST) (Abcam). Anti-Rabbit-HRP (1:2000) and anti-Mouse-HRP (1:2000) (Cell Signaling) secondary antibodies were both diluted in 5% NFDM-TBST.
In vitro transcription of viral RNA
WNV3'(+)SL RNA, WNV5'(+)SL RNA, and the WNV 3' small fragment RNA (3'sfRNA) were in vitro transcribed using a MAXIscript in vitro transcription kit (Ambion) in 20-µl reactions containing T7 RNA polymerase (30 U), a PCR purification kit (Qiagen)-purified PCR product (1 µg), 0.8 mM [α-32P]GTP (3,000 Ci/mmol, 10 mCi/ml; Perkin Elmer), and 0.5 mM of CTP, UTP, and ATP. Unlabeled viral RNA was in vitro transcribed as described above, except that 0.5 mM of each of the four nucleoside triphosphates was added to the reaction mixture. The in vitro transcription mixture was incubated at 37°C for 2 h, and transcription was stopped by the addition of DNase I (1 U). The reaction mixture was heated at 95°C for 5 min in 2X Gel Loading Buffer II (Ambion) and the RNA transcripts were purified by electrophoresis on a 6% polyacrylamide gel containing 7 M urea. The wet gel was autoradiographed, the 32P-labeled RNA band as well as unlabeled RNA bands loaded onto adjacent lanes were excised. RNA was eluted from the gel slices by rocking overnight at 4°C in elution buffer [0.5 M NH4OAC, 1 mM EDTA, and 0.2% SDS]. Eluted RNA was filtered through a 0.45-µm cellulose acetate filter unit (Millipore), ethanol-precipitated, resuspended in water, aliquoted, and stored at −80°C. The amount of radioactivity incorporated into each RNA probe was measured in a scintillation counter (model LS6500; Beckman), and the specific activity was estimated using Ambion's specific activity calculator (Ambion). Unlabeled RNA concentrations were calculated based on UV absorbance measured at 260 nm.
RNA secondary structure prediction
The secondary structure of each RNA probe used in this study was predicted using Mfold version 3.1 software (Zuker, 2003).
PKR autophosphorylation assay
Recombinant human PKR expressed and purified from E. coli was a gift from Graem Conn, Emory University (Conn, 2003). Purified PKR (150 µg) was incubated with in vitro transcribed viral RNAs in kinase buffer [5 mM Tris-HCl, (pH 7.6), 1 mM MgCl2, 25 mM KCl, 0.25% Triton-X 100, and RNase Inhibitor (Ambion)] on ice for 10 min. Poly(I:C), 4X Mg/ATP cocktail (Millipore) [20 mM MOPS, pH 7.2, 25 mM β-glycerophosphate, 5 mM EGTA, 1mM Na3VO4, 1 mM dithiothreitol, 75 mM MgCl2, and 0.5 mM ATP] and 10 µCi 32p-γ-ATP (Perkin Elmer) were added to the reactions and incubated at 30°C for 20 minutes. The reactions were stopped by addition of 2X Sample Buffer [20% SDS, 25% glycerol, 7% 0.5M Tris-HCl (pH 6.8), 0.5% bromophenol blue and 5% 2-mercaptoethanol added fresh], and the proteins were resolved by SDS-PAGE. Gels were fixed in 30% methanol and 10% acetic acid, incubated in Autofluor image intensifier solution (National Diagnostics) and then incubated in anti-cracking buffer [7% methanol, 7% acetic acid and 1% glycerol] at room temperature, dried and analyzed using a Fuji BAS 2500 analyzer (Fuji Photo Film Co.) and Image Gauge software (Science Lab, 98, version 3.12; Fuji Photo Film Co.).
Poly(I:C) transfection
To activate intracellular PKR, different concentrations of poly(I:C) (Sigma Aldrich) in the range of 1 to 100 µg were transfected into BHK cell monolayers in 12-well plates. Briefly, 5 µl of poly(I:C) was diluted in 95 µl OptiMEM I Reduced Serum media (Invitrogen) and incubated with 5 µl Celfectin II transfection reagent (Invitrogen) diluted in 95 µl OptiMEM for approximately 20 minutes. The poly(I:C)–Celfectin complexes were added to the cell monolayers (200 µl per well), the volume was increased to 1 ml with OptiMEM and the monolayers were incubated for 1.5 or 2 h at 37°C.
HIGHLIGHTS.
-
>
PKR is upregulated in WNV-infected cells, but PKR phosphorylation is not induced.
-
>
The association of PKR with Nck in WNV-infected cells indicates that PKR is inactive.
-
>
WNV stem-loop RNAs efficiently activate PKR in vitro but not in infected cells.
-
>
WNV infection does not interfere with poly(I:C)-mediated PKR activation.
-
>
WNV yields were similar from wild type and PKR −/− MEFs.
ACKNOWLEDGEMENTS
This work was supported by Public Health Service research grant AI048088 to M.A.B. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank Graeme Conn for providing purified PKR protein and for technical advice and Bronislava Stockman for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D, Wakita T, Meurs EF. Hepatitis C virus controls interferon production through PKR activation. PLoS One. 2010;5:e10575. doi: 10.1371/journal.pone.0010575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton MA. The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annu Rev Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- Cardin E, Larose L. Nck-1 interacts with PKR and modulates its activation by dsRNA. Biochem Biophys Res Commun. 2008;377:231–235. doi: 10.1016/j.bbrc.2008.09.112. [DOI] [PubMed] [Google Scholar]
- Cardin E, Latreille M, Khoury C, Greenwood MT, Larose L. Nck-1 selectively modulates eIF2alphaSer51 phosphorylation by a subset of eIF2alpha-kinases. FEBS J. 2007;274:5865–5875. doi: 10.1111/j.1742-4658.2007.06110.x. [DOI] [PubMed] [Google Scholar]
- Conn GL. Expression of active RNA-activated protein kinase (PKR) in bacteria. Biotechniques. 2003;35:682–684. 686. doi: 10.2144/03354bm03. [DOI] [PubMed] [Google Scholar]
- Davis WG, Blackwell JL, Shi PY, Brinton MA. Interaction between the cellular protein eEF1A and the 3'-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. Journal of virology. 2007;81:10172–10187. doi: 10.1128/JVI.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhang B, Shi PY. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology. 2008;381:123–135. doi: 10.1016/j.virol.2008.07.040. [DOI] [PubMed] [Google Scholar]
- Donze O, Abbas-Terki T, Picard D. The Hsp90 chaperone complex is both a facilitator and a repressor of the dsRNA-dependent kinase PKR. Embo J. 2001;20:3771–3780. doi: 10.1093/emboj/20.14.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci U S A. 2007;104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Gilfoy FD, Mason PW. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. Journal of virology. 2007;81:11148–11158. doi: 10.1128/JVI.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. Journal of virology. 2010;84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ, Kuno G, Markoff J. Flaviviruses. In: David M, Knipe PMH, editors. Fields Virology. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 1153–1252. [Google Scholar]
- He B, Gross M, Roizman B. The gamma134.5 protein of herpes simplex virus 1 has the structural and functional attributes of a protein phosphatase 1 regulatory subunit and is present in a high molecular weight complex with the enzyme in infected cells. J Biol Chem. 1998;273:20737–20743. doi: 10.1074/jbc.273.33.20737. [DOI] [PubMed] [Google Scholar]
- Hebner CM, Wilson R, Rader J, Bidder M, Laimins LA. Human papillomaviruses target the double-stranded RNA protein kinase pathway. J Gen Virol. 2006;87:3183–3193. doi: 10.1099/vir.0.82098-0. [DOI] [PubMed] [Google Scholar]
- Hershey JW. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- Ito T, Yang M, May WS. RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J Biol Chem. 1999;274:15427–15432. doi: 10.1074/jbc.274.22.15427. [DOI] [PubMed] [Google Scholar]
- Jammi NV, Beal PA. Phosphorylation of the RNA-dependent protein kinase regulates its RNA-binding activity. Nucleic Acids Res. 2001;29:3020–3029. doi: 10.1093/nar/29.14.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, Block TM, Guo JT. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. Journal of virology. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–110. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Langland JO, Jacobs BL. Cytosolic double-stranded RNA-dependent protein kinase is likely a dimer of partially phosphorylated Mr = 66,000 subunits. J Biol Chem. 1992;267:10729–10736. [PubMed] [Google Scholar]
- Latreille M, Larose L. Nck in a complex containing the catalytic subunit of protein phosphatase 1 regulates eukaryotic initiation factor 2alpha signaling and cell survival to endoplasmic reticulum stress. J Biol Chem. 2006;281:26633–26644. doi: 10.1074/jbc.M513556200. [DOI] [PubMed] [Google Scholar]
- Lee TG, Tomita J, Hovanessian AG, Katze MG. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of Mr 68,000 from influenza virus-infected cells. Proc Natl Acad Sci U S A. 1990;87:6208–6212. doi: 10.1073/pnas.87.16.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J Mol Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Lin KC, Chang HL, Chang RY. Accumulation of a 3'-terminal genome fragment in Japanese encephalitis virus-infected mammalian and mosquito cells. Journal of virology. 2004;78:5133–5138. doi: 10.1128/JVI.78.10.5133-5138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 1101–1152. [Google Scholar]
- Lopez-Lastra M, Ramdohr P, Letelier A, Vallejos M, Vera-Otarola J, Valiente-Echeverria F. Translation initiation of viral mRNAs. Rev Med Virol. 2010;20:177–195. doi: 10.1002/rmv.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J. Wrapping things up about virus RNA replication. Traffic. 2005;6:967–977. doi: 10.1111/j.1600-0854.2005.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQuillan GC, Caterina P, de Boer B, Allan JE, Platten MA, Reed WD, Jeffrey GP. Ultra-structural localisation of hepatocellular PKR protein using immuno-gold labelling in chronic hepatitis C virus disease. Journal of molecular histology. 2009;40:171–176. doi: 10.1007/s10735-009-9227-0. [DOI] [PubMed] [Google Scholar]
- Majumdar R, Maitra U. Regulation of GTP hydrolysis prior to ribosomal AUG selection during eukaryotic translation initiation. EMBO J. 2005;24:3737–3746. doi: 10.1038/sj.emboj.7600844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna SA, Kim I, Liu CW, Puglisi JD. Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J Mol Biol. 2006;358:1270–1285. doi: 10.1016/j.jmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- McKenna SA, Lindhout DA, Shimoike T, Aitken CE, Puglisi JD. Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J Mol Biol. 2007;372:103–113. doi: 10.1016/j.jmb.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5'-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. Embo J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri S, Rahman F, Williams BR, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q, Christianson TA, Keeble W, Koretsky T, Bagby GC. The anti-apoptotic function of Hsp70 in the interferon-inducible double-stranded RNA-dependent protein kinase-mediated death signaling pathway requires the Fanconi anemia protein, FANCC. J Biol Chem. 2002;277:49638–49643. doi: 10.1074/jbc.M209386200. [DOI] [PubMed] [Google Scholar]
- Patel CV, Handy I, Goldsmith T, Patel RC. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J Biol Chem. 2000;275:37993–37998. doi: 10.1074/jbc.M004762200. [DOI] [PubMed] [Google Scholar]
- Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Experimental biology and medicine (Maywood, N.J. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Romano PR, Garcia-Barrio MT, Zhang X, Wang Q, Taylor DR, Zhang F, Herring C, Mathews MB, Qin J, Hinnebusch AG. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol Cell Biol. 1998;18:2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. RNase L plays a role in the antiviral response to West Nile virus. Journal of virology. 2006;80:2987–2999. doi: 10.1128/JVI.80.6.2987-2999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherbik SV, Stockman BM, Brinton MA. Differential expression of interferon (IFN) regulatory factors and IFN-stimulated genes at early times after West Nile virus infection of mouse embryo fibroblasts. Journal of virology. 2007;81:12005–12018. doi: 10.1128/JVI.01359-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp TV, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud CG, Hilse K, Clemens MJ. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoike T, McKenna SA, Lindhout DA, Puglisi JD. Translational insensitivity to potent activation of PKR by HCV IRES RNA. Antiviral Res. 2009;83:228–237. doi: 10.1016/j.antiviral.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Spurgeon ME, Ornelles DA. The adenovirus E1B 55-kilodalton and E4 open reading frame 6 proteins limit phosphorylation of eIF2alpha during the late phase of infection. Journal of virology. 2009;83:9970–9982. doi: 10.1128/JVI.01113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Wang S, Baltzis D, Qu LK, Raven JF, Li S, Wong AH, Koromilas AE. Interferons induce tyrosine phosphorylation of the eIF2alpha kinase PKR through activation of Jak1 and Tyk2. EMBO Rep. 2007;8:265–270. doi: 10.1038/sj.embor.7400891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Wang S, Baltzis D, Qu LK, Wong AH, Koromilas AE. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2alpha RNA-dependent protein kinase. Proc Natl Acad Sci U S A. 2006;103:63–68. doi: 10.1073/pnas.0508207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhakar A, Ramachandran A, Ghosh S, Hasnain SE, Kaufman RJ, Ramaiah KV. Phosphorylation of serine 51 in initiation factor 2 alpha (eIF2 alpha) promotes complex formation between eIF2 alpha(P) and eIF2B and causes inhibition in the guanine nucleotide exchange activity of eIF2B. Biochemistry. 2000;39:12929–12938. doi: 10.1021/bi0008682. [DOI] [PubMed] [Google Scholar]
- Tan SL, Tareen SU, Melville MW, Blakely CM, Katze MG. The direct binding of the catalytic subunit of protein phosphatase 1 to the PKR protein kinase is necessary but not sufficient for inactivation and disruption of enzyme dimer formation. J Biol Chem. 2002;277:36109–36117. doi: 10.1074/jbc.M205109200. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Samuel CE. Mechanism of interferon action: structure of the mouse PKR gene encoding the interferon-inducible RNA-dependent protein kinase. Proc Natl Acad Sci U S A. 1994;91:7995–7999. doi: 10.1073/pnas.91.17.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- Urosevic N, van Maanen M, Mansfield JP, Mackenzie JS, Shellam GR. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. J Gen Virol. 1997;78(Pt 1):23–29. doi: 10.1099/0022-1317-78-1-23. [DOI] [PubMed] [Google Scholar]
- Vaheri A, Sedwick WD, Plotkin SA, Maes R. Cytopathic effect of rubella virus in RHK21 cells and growth to high titers in suspension culture. Virology. 1965;27:239–241. doi: 10.1016/0042-6822(65)90170-4. [DOI] [PubMed] [Google Scholar]
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamshchikov VF, Wengler G, Perelygin AA, Brinton MA, Compans RW. An infectious clone of the West Nile flavivirus. Virology. 2001;281:294–304. doi: 10.1006/viro.2000.0795. [DOI] [PubMed] [Google Scholar]
- Yin Z, Haynie J, Williams BR, Yang YC. C114 is a novel IL-11-inducible nuclear double-stranded RNA-binding protein that inhibits protein kinase R. J Biol Chem. 2003;278:22838–22845. doi: 10.1074/jbc.M212969200. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]