Abstract
Mammalian orthoreoviruses (reoviruses) are highly tractable models for studies of viral replication and pathogenesis. The versatility of reovirus as an experimental model has been enhanced by development of a plasmid-based reverse genetics system. Infectious reovirus can be recovered from cells transfected with plasmids encoding cDNAs of each reovirus gene segment using a strategy that does not require helper virus and is independent of selection. In this system, transcription of each gene segment is driven by bacteriophage T7 RNA polymerase, which can be supplied transiently by recombinant vaccinia virus (rDIs-T7pol) or by cells that constitutively express the enzyme. Reverse genetics systems have been developed for two prototype reovirus strains, type 1 Lang (T1L) and type 3 Dearing (T3D). Each reovirus cDNA was encoded on an independent plasmid for the first-generation rescue system. The efficiency of virus recovery was enhanced in a second-generation system by combining the cDNAs for multiple reovirus gene segments onto single plasmids to reduce the number of plasmids from 10 to 4. The reduction in plasmid number and the use of baby hamster kidney cells that express T7 RNA polymerase increased the efficiency of viral rescue, reduced the incubation time required to recover infectious virus, and eliminated potential biosafety concerns associated with the use of recombinant vaccinia virus. Reovirus reverse genetics has been used to introduce mutations into viral capsid and nonstructural components to study viral protein-structure activity relationships and can be exploited to engineer recombinant reoviruses for vaccine and oncolytic applications.
Keywords: reovirus, reverse genetics, dsRNA, T7 RNA polymerase, reassortment
1. Introduction
The ability to engineer viruses that contain specific sequence modifications is the foundation for developing powerful reagents that serve contemporary virology. Consequently, a strategy to recover infectious virus from plasmids encoding cloned cDNAs corresponding to the viral genome is among the most important technological advances for a virological experimental system. Plasmid-based reverse genetics systems facilitate structure-f unction analysis of viral proteins, studies to define replication and packaging signals, incorporation of markers to track viruses in cell culture and in vivo, and development of antiviral vaccines and therapeutics. Recovering virus from cDNA limits accumulation of mutations that occur during maintenance of stocks by serial passage, increases the reproducibility of experiments, and permits standardization of assays between laboratories. Although a plasmid-based system exists for nearly every major virus group, development of an entirely plasmid-based system for recovering viable virus had eluded members of the Reoviridae family until 2007.
Viruses of the Reoviridae contain double-stranded (ds) RNA genomes of 9 to 12 gene segments (1). These viruses infect a wide range of eukaryotic hosts, including mammals, birds, insects, and plants (2). The Reoviridae includes rotaviruses, the most common diarrheal pathogen of children (3), bluetongue virus, an economically important pathogen of sheep and cattle (4), and mammalian orthoreoviruses (simply called reoviruses here), which are important experimental models for studies of dsRNA virus replication and pathogenesis (2). Reovirus was the first member of the Reoviridae for which a plasmid-based reverse genetics system was developed (5). Bluetongue virus can now be recovered following transfection of cells with in vitro-transcribed RNAs (6). Single-gene replacement systems that require helper virus, selection, or both have been developed for rotavirus (7-9). Here, we describe the development and use of a plasmid-based reverse genetics system for reovirus.
Reoviruses are nonenveloped, icosahedral viruses that contain 10 segments of dsRNA (1). Viral genomic positive-sense RNAs are capped at the 5′ end and not polyadenylated (1). Virions are composed of two concentric protein shells, the outer capsid and core. Reovirus entry into host cells is initiated by the attachment of the σ1 protein to cell-surface receptors (10, 11). The σ1 protein targets at least two different receptors, sialic acid-containing glycans (12-15) and junctional adhesion molecule-A (16-18). After receptor binding, virions are internalized via a β1 integrin-dependent process into endosomes (19), where virion disassembly occurs (19, 20). During disassembly, attachment protein σ1 and outer-capsid protein σ3 are removed, exposing the μ1 protein, which is subsequently cleaved. Proteolysis of μ1 results in conformational rearrangements that facilitate endosomal membrane rupture and delivery of transcriptionally active core particles into the cytoplasm (21, 22). Primary transcription occurs within the viral core, and nascent RNAs are translated or encapsidated into new viral cores, where they serve as templates for negative-strand synthesis. Within new viral cores, secondary rounds of transcription can occur. Outer-capsid proteins coalesce onto nascent particles to generate progeny virions, which are released by a mechanism that is not clear.
Through forward genetics and reassortant analysis, studies of reovirus have yielded a wealth of information about mechanisms that underlie viral replication and pathogenesis. However, the ability to introduce specific changes into the genome of Reoviridae viruses has been limited. Antibody-dependent selection (23) and helper virus-based systems (24, 25) have been used to replace single gene segments, but these strategies are limited by the availability of antibodies and potential contamination from the helper virus. RNAi-mediated inhibition of viral gene expression can be complemented by transfection of a plasmid encoding a copy of the gene with the siRNA target sequence altered to prevent siRNA-mediated degradation (24, 26, 27). This strategy is limited by the efficiency of viral gene knockdown and the transfection efficiency of the complementation plasmid. Moreover, none of these strategies leads to selection-free incorporation of the gene segment into progeny virions.
2. Reovirus plasmid-based reverse genetics system
2.1 Theory
Development of a plasmid-based reverse genetics system for members of the Birnaviridae family, which contain two genomic dsRNA segments, suggested that delivery of viral positive-strand RNA alone could launch successful viral progeny production for a dsRNA virus (28). Furthermore, introduction of message-sense RNA isolated from virions of Reoviridae family members leads to the production of infectious progeny virions in some cases (6, 29, 30). Together, these data suggested that a plasmid-based reverse genetics system for reovirus would be feasible if the transcripts produced from the plasmid physically mimicked viral message-sense RNAs.
To engineer plasmids that would yield replicas of positive-sense reovirus RNAs, cDNAs for each gene segment from reovirus strains T1L and T3D were introduced into plasmids at sites flanked by the promoter sequence for T7 RNA polymerase and the hepatitis delta virus (HDV) ribozyme (Fig. 1A). Reovirus can be recovered from cells in which T7 RNA polymerase is delivered transiently by infection with a recombinant vaccinia (31) or from cells that constitutively express the enzyme (32) . T7 RNA polymerase initiates transcription at a defined guanosine residue (33). All reovirus transcripts contain the nucleotide sequence GCUA at the 5′ terminus. Use of T7 RNA polymerase to catalyze transcription of reovirus RNAs thus produces transcripts with native reovirus 5′ termini. Self-cleavage of the HDV ribozyme will generate RNAs with native reovirus 3′ termini (25). RNAs will be translated by host cell ribosomes to produce reovirus proteins. In cells transfected with plasmids encoding all ten reovirus gene segments, the viral gene products will join with reovirus RNAs to form viral replication complexes, mediate negative-strand synthesis, and ultimately assemble infectious virions. Reverse genetics systems for negative-sense RNA viruses require co-transfection of plasmids expressing nucleocapsid and polymerase proteins to facilitate negative-strand RNA synthesis from plasmid-derived positive-sense RNA molecules (34). However, inclusion of expression plasmids encoding reovirus proteins are not required for, nor do they enhance, recovery of recombinant reoviruses (T. Kobayshi and T. Dermody, unpublished results).
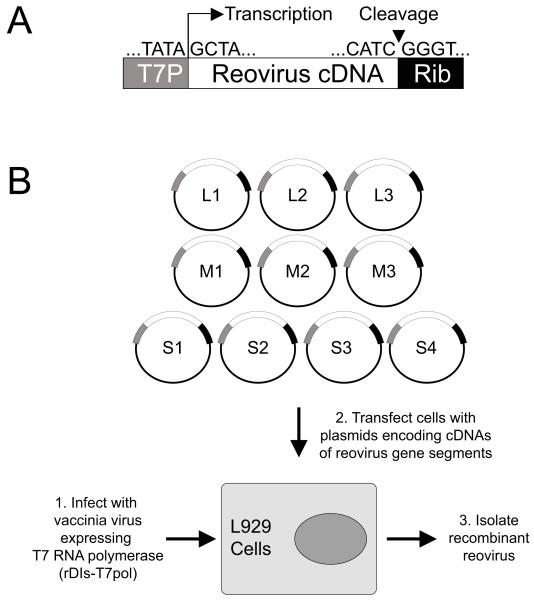
Fig. 1.
(A) Schematic of a plasmid encoding a prototype reovirus gene segment cDNA. Cloned cDNAs representing each of the ten reovirus dsRNA gene segments are flanked by the promoter sequence for bacteriophage T7 RNA polymerase and the antigenomic HDV ribozyme. (B) Schematic of approach. (1) L cells are infected with rDIs-T7pol. (2) The cells are transfected with cDNA constructs. Nascent transcripts correspond to viral mRNAs containing the native 5′ end. Self cleavage by the HDV ribozyme generates the native 3′ end. (3.) Following 2-5 days of incubation, transfected cells are lysed by freeze-thaw, and viable virus is isolated by plaque assay using murine L cells.
Very few cells produce virus immediately following transfection, as only 2-3 infectious centers per 106 cells are detected 24 hours post-transfection (5). We think there are two main reasons for this effect. First, the percentage of cells that take up all ten plasmids is likely small. Second, RNAs transcribed directly from plasmids will not contain a 5′ cap, and the translation efficiency of these RNAs will be lower than that of RNAs transcribed by the virus. Virus produced from infectious centers will eventually infect adjacent cells in the culture, resulting in amplification of the virus generated from plasmids. Recovery of recombinant reoviruses requires multiple rounds of replication following plasmid transfection.
Recombinant viruses are designated by the prefix “rs” to denote recombinant strain (eg., rsT1L and rsT3D). To confirm that viruses isolated following plasmid-based rescue are recombinant and not the result of contaminating virus, silent mutations have been introduced into the cDNAs encoding the L3 and L1 genes of T1L and T3D, respectively, which can be detected by sequence analysis (5, 35).
2.2 Preparation of plasmids
Each gene segment from T1L and T3D was converted to cDNA by reverse transcription and cloned into plasmids that encode an ampicillin-resistance gene (5, 35, 36). Plasmids are transformed into the DH5α strain of Escherichia coli and maintained and amplified under ampicillin selection. All plasmids are grown at 37°C, except those encoding the T3D L1 gene segment, which are grown at 30°C. Plasmids are purified from bacterial cultures using endotoxin-free maxiprep and midiprep-plasmid purification systems (Qiagen).
2.3 Preparation of rDIs-T7pol
The first-generation reovirus reverse genetics system relies on a recombinant strain of vaccinia virus (rDIs-T7pol) to deliver T7 RNA polymerase into cells (31). rDIs-T7pol replicates efficiently in chicken embryo fibroblasts (CEFs) but is defective for growth in mammalian cells (31).
Seed tissue culture-treated 100 mm dishes (Costar) with 1 × 106 CEF cells per dish one day prior to infection. CEF cells are maintained in MED199 (Caisson Laboratories) supplemented to contain 10% 1X tryptose phosphate broth (EMD Chemicals), 5% fetal bovine serum, 1% chicken serum , 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 25 ng/ml of amphotericin B (all supplements from Gibco). The cells should be ~ 90% confluent at the time of use.
The growth medium is removed, and cells are infected with rDIs-T7pol suspended in 3 ml serum-free MED199 at an MOI of 1 TCID50 per cell. Incubate the cells at 37°C for 1 hour, rocking every 15 minutes.
The inoculum is removed and replaced with 10 ml of complete MED199. The cells are incubated at 37°C until the entire cell sheet exhibits cytopathic effects. This ranges from 3 to 7 days.
The cells are harvested by scraping into the medium and pelleted by centrifugation at 200 × g at 4°C for 10 minutes.
The cells are re-suspended in 1 ml of MED199 and subjected to three freeze/thaw cycles. Virus is dispersed using a cup sonicator. The virus is sonicated on ice for 2 minutes followed by rest on ice for an additional 2 minutes. The sonication step is repeated until the pellet is no longer visible. Virus is divided into 0.5 ml aliquots and stored at −80°C.
The titer of a freshly-thawed aliquot of rDIs-T7 pol is quantified by TCID50 assay on CEF cells (37).
2.4 First-generation reverse genetics system
L929 (L) cells are infected with rDIs-T7pol and transfected with 10 plasmids, each corresponding to a single reovirus gene segment (Fig. 1B). A 10-plasmid system has been developed for both T1L and T3D (5, 35). rsT1L can be recovered as early as 24 hours post-transfection; however, rsT3D progeny do not appear until 48 hours (5, 35). The basis for this difference is not known. The titer of recovered virus increases over time, peaking between 3 and 5 days.
Seed tissue culture-treated 60 mm dishes (Costar) with 3 × 106 L cells per dish one day prior to rescue. L cells are maintained in Joklik’s modified Eagle’s minimal essential medium (MEM) (Irvine Scientific) supplemented to contain 10% fetal bovine serum, 2 mM L-glutamine, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 25 ng/ml of amphotericin B. The cells should be ~ 90% confluent at the time of use.
Disperse freshly thawed rDIs-T7pol using a cup sonicator. The virus is sonicated on ice for 2 minutes followed by rest on ice for an additional 2 minutes. The sonication step is repeated until cell debris is no longer visible.
The growth medium is removed, and cells are infected with rDIs-T7pol suspended in 1 ml OPTI-MEM I (Invitrogen) at an MOI of 0.5 TCID50 per cell.
Incubate the cells at 37°C for 1 hour, rocking every 15 minutes.
For each rescue reaction, add 750 μl OPTI-MEM I to a 1.5 ml microcentrifuge tube. Add 53.25 μl TransIT-LT1 (Mirus) directly into the OPTI-MEM I (ratio of 3 μl TransIT-LT1 per 1 μg of DNA). Do not touch the walls of the tube with the pipet tip. Mix completely by gentle pipetting or vortex for 2 seconds. Incubate at room temperature (RT) for 20 minutes.
- Combine the following amounts of plasmid DNA in a microcentrifuge tube. A total of 17.75 μg of plasmid DNA is used. These plasmid concentrations were empirically determined to yield the highest titers of recombinant progeny.
- 2 μg each of pT7-L1, pT7-L2, and pT7-L3
- 1.75 μg each of pT7-M1, pT7-M2, and pT7-M3
- 2 μg of pBacT7-S1
- 1.5 μg each of pT7-S2, pT7-S3, and pT7-S4
Add the plasmid DNA directly to the diluted OPTI-MEM I/TransIT-LT1 and mix by gentle pipetting or vortex for 2 seconds. Incubate at room temperature for 30 minutes.
Aspirate the rDIs-T7pol inoculum and wash the cells with 2 ml of complete growth medium.
Add 5 ml of complete growth medium to the culture dish.
Add the transfection mix to the cells in a drop-wise manner.
Incubate the cells at 37°C for 5 days.
Subject cells to two freeze/thaw cycles to release intracellular virus. Recombinant viruses are isolated by plaque assay using L cells (38). Individual plaques are picked and amplified in L cells to create a passage 1 (P1) stock.
To confirm the sequence of the virus, viral RNA is extracted from virions and subjected to Onestep RT-PCR (Qiagen) using primers specific for the gene segment of interest. PCR products are analyzed following electrophoresis in Tris-borate-EDTA agarose gels or purified and subjected directly to sequence analysis. The presence of a noncoding signature mutation in the L3 or L1 gene of viruses generated by plasmid-based rescue also is confirmed using RT-PCR and gene-specific primers for T1L and T3D, respectively (5, 35).
2.5 Second-generation reverse genetics system
To reduce the number of plasmids required to recover reovirus by plasmid-based rescue, multiple cDNAs were combined into single plasmids. For each virus strain, different combinations of gene segments were cloned into four plasmids (Table 1). Reducing the number of plasmids increased the efficiency of rescue for both rsT1L and rsT3D, and virus can be recovered at earlier times compared to the 10-plasmid system (35). Both rsT1L and rsT3D can be isolated by 24 hours post-transfection, and peak titers are usually achieved by 48 hours. This increase in virus recovery most likely results from a higher transfection efficiency leading to an increased number of cells receiving all ten reovirus gene segments. The procedure for rescuing virus using the 4-plasmid system is nearly identical to that for the 10-plasmid system. The only modification is the use of an equivalent amount of each plasmid. A total of 17.75 μg of plasmid DNA is used per rescue reaction (4.44 μ g of each plasmid).
Table 1.
cDNAs for the indicated gene segments were combined for the 4-plasmid reverse genetics systems for rsT1L and rsT3D.
| Virus strain | Gene segments on plasmid |
|---|---|
| T1L | L1/M2 |
| L2/M3 | |
| L3/S3 | |
| M1/S1/S2/S4 | |
| T3D | L1/S1 |
| L2/M3 | |
| L3/M1 | |
| M2/S2/S3/S4 |
3. Reovirus rescue in BHK-T7 cells
To further streamline the rescue protocol, we use baby hamster kidney cells engineered to stably express T7 RNA polymerase (BHK-T7) (32). Virus recovery from transfected BHK-T7 cells is comparable to the levels obtained using rDIs-T7pol for both rsT1L and rsT3D (35). Moreover, recovering virus in BHK-T7 cells simplifies the protocol and eliminates potential biohazards associated with rDIs-T7pol.
Seed tissue culture-treated 60 mm dishes with 3 × 106 BHK-T7 cells 1 day prior to rescue. BHK-T7 cells are maintained in Dulbecco’s modified Eagle’s MEM (Invitrogen) supplemented to contain 10% fetal bovine serum, 2 mM L-glutamine, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 25 ng/ml of amphotericin B. The BHK-T7 cells were generated using a plasmid expressing T7 RNA polymerase and a neomycin resistance gene (32). To maintain selective pressure, the growth medium is supplemented with geneticin (1 mg/ml) during alternating passages in culture. The cells are plated without geneticin for virus rescue. The cells should be ~ 90% confluent at the time of use.
For each rescue reaction, add 750 μl OPTI-MEM I to a 1.5 ml microcentrifuge tube. Add 53.25 μl TransIT-LT1 (Mirus) directly into the OPTI-MEM I (ratio of 3 μl TransIT-LT1 per 1 μg of DNA). Do not touch the walls of the tube with the pipet tip. Mix completely by gentle pipetting or vortex for 2 seconds. Incubate at room temperature (RT) for 20 minutes.
- Combine the plasmid DNA in a microcentrifuge tube. A total of 17.75 μg of plasmid DNA is used for all transfections. For the 10-plasmid system, use the following mounts of plasmid.
- 2 μg each of pT7-L1, pT7-L2, and pT7-L3
- 1.75 μg each of pT7-M1, pT7-M2, and pT7-M3
- 2 μg of pBacT7-S1
- 1.5 μg each of pT7-S2, pT7-S3, and pT7-S4
Add the plasmid DNA directly to the diluted OPTI-MEM I/TransIT-LT1 and mix by gentle pipetting or vortexing for 2 seconds. Incubate at room temperature for 30 minutes.
Aspirate the medium and add 5 ml of complete growth medium to the culture dish.
Add the transfection mix to the cells in a drop-wise manner.
Incubate the cells at 37°C for 2 days.
Subject cells to two freeze/thaw cycles to release intracellular virus. Recombinant viruses are isolated by plaque assay using L cells (38). Individual plaques are picked and amplified in L cells to create a passage 1 (P1) stock.
To confirm the sequence of the virus, viral RNA is extracted from virions and subjected to Onestep RT-PCR (Qiagen). PCR products are analyzed following electrophoresis in Tris-borate-EDTA agarose gels or purified and subjected directly to sequence analysis. The presence of a noncoding signature mutation in the L3 or L1 gene of viruses generated by plasmid-based rescue also is confirmed using RT-PCR and gene-specific primers for T1L and T3D, respectively (5, 35).
4. Generation of reovirus mutants and reassortants
To generate viral mutants and reassortants, we modify the 4-plasmid system to isolate the gene segment of interest. Primer-based mutagenesis is used to alter a gene segment encoded on a single plasmid. This approach requires using single gene-encoding plasmids for each of the other gene segments that were originally combined with the targeted gene segment. Depending on the gene segment to be altered, rescue is performed with as many as seven plasmids or as few as five. Similarly, generation of reassortant viruses containing a combination of gene segments from strains T1L and T3D usually requires the use of plasmids encoding single gene segments. For example, a single-gene reassortant containing the S1 gene from T3D and nine gene segments from T1L would require the T3D S1 gene to be supplied on a single plasmid. Because the plasmid encoding T1L S1 also contains T1L M1, S2, and S4, these gene segments also must to be supplied in single gene form. Thus, seven plasmids would be required for rescue: T1L L1/M2, T1L L2/M3, T1L L3/S3, T1L M1, T3D S1, T1L S2, and T1L S4. For mutant and reassortant viruses, the rescue protocol is identical to that described for rsT1L and rsT3D. The only modification is that the total quantity of DNA remains 17.75 μg divided evenly among the number of plasmids used. This strategy has been used to successfully recover viruses with mutations in reovirus structural (5, 39, 40) and nonstructural proteins (41-43) and numerous reassortant viruses (5, 41, 43).
5. Troubleshooting
Although the reovirus plasmid-based reverse genetics system is efficient for rescue of viable recombinant reoviruses, instances arise when infections reovirus cannot be recovered. Failure to recover virus is attributable to two possibilities, one biological and the other technical. First, the introduced mutation may be lethal for the virus. Viruses encoding lethal mutations, i.e., changes that render the virus incapable of replicating due to nonfunctional proteins or nucleic acids, cannot be recovered using this technique. In this situation, our laboratory has adopted the convention that a mutant virus that cannot be rescued following three attempts is considered to be non-viable. However, for those mutant viruses that cannot be recovered, it may be possible to generate infectious particles by supplying a wild-type version of the mutant protein during the rescue procedure (5). Such particles would be fit to complete steps in the viral infectious cycle until the activity of the encoded mutant protein or RNA is required. Such a strategy may be useful in defining functions of some viral proteins and RNAs.
The second possibility is that at least one aspect of the rescue process has failed. In these instances, plasmid purity and concentration, transfection efficiency, and T7 activity are possible impediments to rescue. Each of these individual steps can be assessed to determine the cause of the problem. To distinguish between the two possibilities, it is imperative to include positive and negative controls in each rescue attempt. Rescuing wild-type virus is our standard positive control. For the negative control, we recommend omitting one of the plasmids.
6. Concluding remarks
Since its discovery more than 50 years ago, reovirus has been used to make numerous important contributions to the field of virology. A combination of forward genetic approaches, reassortant genetics, and sequence analysis has provided an abundance of information about viral determinants that govern reovirus tropism, cell entry, replication, and disease (1). The development of a plasmid-based reverse genetics system has enabled a more precise exploration of reovirus biology. Coupled with a robust mouse model of disease, the reovirus reverse genetics technology will foster enhanced understanding of pathogen-host interactions. Moreover, reovirus reverse genetics also will allow for more effective design of reovirus for clinical applications. Reovirus infects mucosal surfaces, induces potent humoral and cell-mediated immune responses, is naturally attenuated, and now can be manipulated to express vaccine epitop es (1). These key features make reovirus an ideal vaccine vector. Reovirus shows promise as an oncolytic therapeutic (44, 45) and currently is being employed in Phase III clinical trials to treat head and neck cancer (46). The ability to engineer reoviruses with specific alterations will allow the design and development of safer and more effective reovirus-based vaccines and cancer therapeutics.
Research Highlights.
-
>
We describe a plasmid-based reverse genetics system for mammalian reovirus.
-
>
Transcription of reovirus gene segments is catalyzed by T7 RNA polymerase.
-
>
Reducing plasmid number by combining viral gene segments enhances virus recovery.
-
>
Using cells that express T7 RNA polymerase increases the efficiency of virus rescue.
-
>
This approach can be exploited for vaccine and oncolytic applications.
Acknowledgements
This work was supported by Public Health Service awards T32 CA09385 (K.W.B.), F32 AI075776 (K.W.B.), R01 AI32539, R01 AI50080, R01 AI76983, R37 AI38296, and the Elizabeth B. Lamb Center for Pediatric Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schiff LA, Nibert ML, Tyler KL. In: Fields Virology. Knipe DM, Howley PM, editors. Vol. 2. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1853–915. [Google Scholar]
- 2.Nibert ML, Schiff LA. In: Fields Virology. Howley PM, editor. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 1679–728. [Google Scholar]
- 3.Parashar UD, Hummelman EG, Bresee JS, Miller MA, Glass RI. Emerg. Infect. Dis. 2003;9:565–72. doi: 10.3201/eid0905.020562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy P. In: Field’s Virology. Howley PM, editor. Lippincott, Williams, and Wilkins; Philadelphia: 2001. [Google Scholar]
- 5.Kobayashi T, Antar AAR, Boehme KW, Danthi P, Eby EA, Guglielmi KM, Holm GH, Johnson EM, Maginnis MS, Naik S, Skelton WB, Wetzel JD, Wilson GJ, Chappell JD, Dermody TS. Cell Host Microbe. 2007;1:147–57. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyce M, Celma CC, Roy P. J. Virol. 2008;82:8339–48. doi: 10.1128/JVI.00808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komoto S, Sasaki J, Taniguchi K. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4646–51. doi: 10.1073/pnas.0509385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komoto S, Kugita M, Sasaki J, Taniguchi K. J. Virol. 2008;82:6753–7. doi: 10.1128/JVI.00601-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Troupin C, Dehee A, Schnuriger A, Vende P, Poncet D, Garbarg-Chenon A. J. Virol. :846711–9. doi: 10.1128/JVI.00547-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiner HL, Ault KA, Fields BN. J. Immunol. 1980;124:2143–48. [PubMed] [Google Scholar]
- 11.Lee PW, Hayes EC, Joklik WK. Virology. 1981;108:156–63. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong GD, Paul RW, Lee PW. Virology. 1984;138:37–48. doi: 10.1016/0042-6822(84)90145-4. [DOI] [PubMed] [Google Scholar]
- 13.Dermody TS, Nibert ML, Bassel-Duby R, Fields BN. J. Virol. 1990;64:5173–76. doi: 10.1128/jvi.64.10.5173-5176.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul RW, Lee PW. Virology. 1987;159:94–101. doi: 10.1016/0042-6822(87)90351-5. [DOI] [PubMed] [Google Scholar]
- 15.Pacitti A, Gentsch JR. J. Virol. 1987;61:1407–15. doi: 10.1128/jvi.61.5.1407-1415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barton ES, Forrest JC, Connolly JL, Chappell JD, Liu Y, Schnell FJ, Nusrat A, Parkos CA, Dermody TS. Cell. 2001;104:441–51. doi: 10.1016/s0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- 17.Campbell JA, Shelling P, Wetzel JD, Johnson EM, Wilson GAR, Forrest JC, Aurrand-Lions M, Imhof B, Stehle T, Dermody TS. J. Virol. 2005;79:7967–78. doi: 10.1128/JVI.79.13.7967-7978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prota AE, Campbell JA, Schelling P, Forrest JC, Peters TR, Watson MJ, Aurrand-Lions M, Imhof B, Dermody TS, Stehle T. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5366–71. doi: 10.1073/pnas.0937718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maginnis MS, Forrest JC, Kopecky-Bromberg SA, Dickeson SK, Santoro SA, Zutter MM, Nemerow GR, Bergelson JM, Dermody TS. J. Virol. 2006;80:2760–70. doi: 10.1128/JVI.80.6.2760-2770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebert DH, Deussing J, Peters C, Dermody TS. J Biol Chem. 2002;277:24609–17. doi: 10.1074/jbc.M201107200. [DOI] [PubMed] [Google Scholar]
- 21.Nibert ML, Odegard AL, Agosto MA, Chandran K, Schiff LA. J. Mol. Biol. 2005;345:461–74. doi: 10.1016/j.jmb.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Odegard AL, Chandran K, Zhang X, Parker JS, Baker TS, Nibert ML. J Virol. 2004;78:8732–45. doi: 10.1128/JVI.78.16.8732-8745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Den Wollenberg DJ, Van Den Hengel SK, Dautzenberg IJ, Kranenburg O, Hoeben RC. Expert Opin. Biol. Ther. 2009;9:1509–20. doi: 10.1517/14712590903307370. [DOI] [PubMed] [Google Scholar]
- 24.Trask SD, Taraporewala ZF, Boehme KW, Dermody TS, Patton JT. Proc. Natl. Acad. Sci. U. S. A. :10718652–7. doi: 10.1073/pnas.1011948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roner MR, Joklik WK. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8036–41. doi: 10.1073/pnas.131203198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi T, Chappell JD, Danthi P, Dermody TS. J. Virol. 2006;80:9053–63. doi: 10.1128/JVI.00276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi T, Ooms LS, Chappell JD, Dermody TS. J. Virol. 2009;83:2892–906. doi: 10.1128/JVI.01495-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao K, Vakharia VN. J. Virol. 1998;72:8913–20. doi: 10.1128/jvi.72.11.8913-8920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roner MR, Sutphin LA, Joklik WK. Virology. 1990;179:845–52. doi: 10.1016/0042-6822(90)90153-i. [DOI] [PubMed] [Google Scholar]
- 30.Boyce M, Roy P. J. Virol. 2007;81:2179–86. doi: 10.1128/JVI.01819-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii K, Ueda Y, Matsuo K, Matsuura Y, Kitamura T, Kato K, Izumi Y, Someya K, Ohsu T, Honda M, Miyamura T. Virology. 2002;302:433–44. doi: 10.1006/viro.2002.1622. [DOI] [PubMed] [Google Scholar]
- 32.Buchholz UJ, Finke S, Conzelmann KK. J. Virol. 1999;73:251–9. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Nucleic Acids Res. 1987;15:8783–98. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9345–50. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi T, Ooms LS, Ikizler M, Chappell JD, Dermody TS. Virology. 2010:194–200. doi: 10.1016/j.virol.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chappell JD, Gunn VL, Wetzel JD, Baer GS, Dermody TS. J. Virol. 1997;71:1834–41. doi: 10.1128/jvi.71.3.1834-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed LJ, Muench H. American Journal of Hygiene. 1938;27:493–97. [Google Scholar]
- 38.Virgin HW, IV, Bassel-Duby R, Fields BN, Tyler KL. J. Virol. 1988;62:4594–604. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danthi P, Coffey CM, Parker JS, Abel TW, Dermody TS. PLoS Path. 2008;4:e1000248. doi: 10.1371/journal.ppat.1000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danthi P, Kobayashi T, Holm GH, Hansberger MW, Abel TW, Dermody TS. J. Virol. 2008;82:161–72. doi: 10.1128/JVI.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boehme KW, Guglielmi KM, Dermody TS. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19986–91. doi: 10.1073/pnas.0907412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zurney J, Kobayashi T, Holm GH, Dermody TS, Sherry B. J. Virol. 2009;83:2178–87. doi: 10.1128/JVI.01787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ooms LS, Kobayashi T, Dermody TS, Chappell JD. J. Biol. Chem. 2010;285:41604–13. doi: 10.1074/jbc.M110.176255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoeckel J, Hay JG. Curr. Opin. Mol. Ther. 2006;8:249–60. [PubMed] [Google Scholar]
- 45.Twigger K, Vidal L, White CL, De Bono JS, Bhide S, Coffey M, Thompson B, Vile RG, Heinemann L, Pandha HS, Errington F, Melcher AA, Harrington KJ. Clin. Cancer Res. 2008;14:912–23. doi: 10.1158/1078-0432.CCR-07-1400. [DOI] [PubMed] [Google Scholar]
- 46.Lal R, Harris D, Postel-Vinay S, de Bono J. Curr. Opin. Mol. Ther. 2009;11:532–9. [PubMed] [Google Scholar]