Abstract
Construction of DNA fragment libraries for next-generation sequencing can prove challenging, especially for samples with low DNA yield. Protocols devised to circumvent the problems associated with low starting quantities of DNA can result in amplification biases that skew the distribution of genomes in metagenomic data. Moreover, sample throughput can be slow, as current library construction techniques are time-consuming. This study evaluated Nextera, a new transposon-based method that is designed for quick production of DNA fragment libraries from a small quantity of DNA. The sequence read distribution across nine phage genomes in a mock viral assemblage met predictions for six of the least-abundant phages; however, the rank order of the most abundant phages differed slightly from predictions. De novo genome assemblies from Nextera libraries provided long contigs spanning over half of the phage genome; in four cases where full-length genome sequences were available for comparison, consensus sequences were found to match over 99% of the genome with near-perfect identity. Analysis of areas of low and high sequence coverage within phage genomes indicated that GC content may influence coverage of sequences from Nextera libraries. Comparisons of phage genomes prepared using both Nextera and a standard 454 FLX Titanium library preparation protocol suggested that the coverage biases according to GC content observed within the Nextera libraries were largely attributable to bias in the Nextera protocol rather than to the 454 sequencing technology. Nevertheless, given suitable sequence coverage, the Nextera protocol produced high-quality data for genomic studies. For metagenomics analyses, effects of GC amplification bias would need to be considered; however, the library preparation standardization that Nextera provides should benefit comparative metagenomic analyses.
INTRODUCTION
The extensive availability and low cost of high-throughput DNA sequencing (HTS) has revolutionized the life sciences, enabling the sequencing of large genomes and opening entirely new approaches to gene expression analysis, mutation mapping, and analysis of noncoding RNAs (15–17, 21). In microbiology, HTS has given rise to the new subdiscipline of metagenomics, which utilizes sequence information to explore the ecology and functional capabilities of entire microbial communities. By avoiding cultivation or enrichment, metagenomics allows relatively unbiased assessment of the taxonomic composition and genetic content of an autochthonous microbial community (27).
Methodological requirements for any metagenomic analysis include (i) confirmation that the original sample is free from contamination with allochthonous or nontarget microorganisms; (ii) unbiased isolation of nucleic acids from all populations of microorganisms within a sample; and (iii) isolation of enough nucleic acid to meet the requirements of a given HTS platform (e.g., Illumina or 454). In particular, obtaining sufficient amounts of nucleic acid has been a challenge for those environments where microbial density is low or for microorganisms with small genome sizes such as viruses. While 454 sequencing of genomes from cultivated viruses can be achieved using as little as 1 ng of DNA (11), sequencing of larger genomes or viral metagenomes from environmental samples requires a larger amount of starting template.
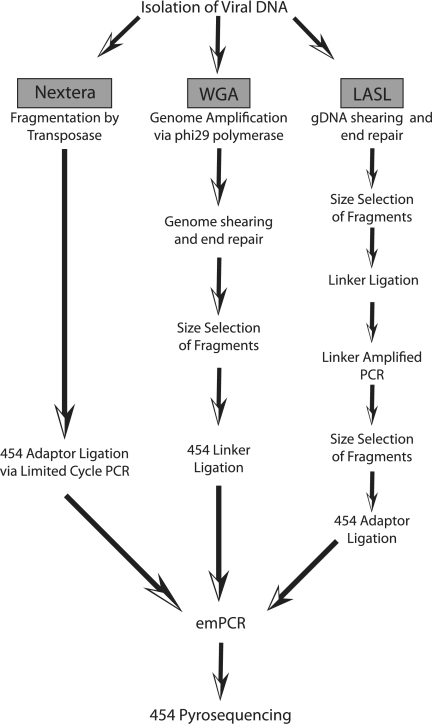
To date, issues of yield have been addressed by strategies designed to nonselectively amplify environmental DNA. For example, the linker adapter shotgun library (LASL) protocol involves shearing of environmental DNA followed by ligation of adapter sequences and subsequent PCR amplification of DNA fragments by the use of primers targeted to the adapter sequences (5, 25) (Fig. 1). Despite the success of this approach, the protocol is time-consuming and contains several independent steps, some of which (e.g., shearing and gel purification of fragments) can pose risks for cross-sample contamination (5). An alternative and simpler approach to amplification of environmental DNA has been to use commercially available whole-genome amplification kits, e.g., REPLI-g (Qiagen) or Genomiphi (GE Healthcare) (9, 23). These kits utilize the highly processive, strand-displacing DNA polymerase of bacteriophage phi29 to produce microgram quantities of DNA from as little as 10 ng of initial DNA template (8) (Fig. 1). Despite the efficiency and ease of use of multiple-displacement amplification (MDA) procedures in metagenomics, this approach can introduce significant bias in downstream sequence libraries. For example, circular genomes are preferentially amplified over linear ones (24); moreover, as a consequence of strand displacement and the formation of multiple replication forks, chimeric sequences can occur within metagenome libraries created from MDA-treated environmental DNA, making comparisons between metagenomes prepared using this protocol essentially nonquantitative (14, 20, 28). Thus, an easy and quick protocol that produces sufficient high-quality fragmented DNA for HTS from small starting quantities of initial sample DNA is highly desirable for those researchers interested in applying metagenomic approaches to the study of microorganisms within environmental samples.
Fig. 1.
Overview of sequencing strategies used to prepare viral genomic DNA samples for 454 sequencing.
Nextera is a new product developed by Epicentre Biotechnologies (Madison, WI) that performs fragmentation and bar-coding of DNA libraries for the 454 and Illumina sequencing platforms. The Nextera kit prepares DNA for sequencing in two main steps (Fig. 1). First, 50 ng of starting DNA is fragmented by a transposome (transposase and transposon end complex), with simultaneous addition of a 19-bp inverted repeat to each end of the fragment during the reaction. In the second step, the fragmented DNA is amplified by limited-cycle PCR, using primers targeted to the transposon sequence. These primers add the specific adapters and bar codes (if desired) to the fragments. After DNA purification, about 500 ng of sheared and adapter-ligated DNA is isolated and ready for sequencing. The Nextera technology provides advantages compared to the LASL and MDA approaches, as it is less time-consuming, requiring around 2 h to prepare fragmented and amplified DNA; also, because the kit does not utilize the phi29 polymerase for amplification (as in the MDA approach), it avoids the amplification biases inherent in the use of that enzyme.
A series of in vitro experiments were designed to evaluate the ability of the Nextera protocol to provide sequence data suitable for both de novo genome assembly and metagenomic analyses of microbial community composition. In these experiments, double-stranded DNA (dsDNA) viruses were utilized as a model system, as the small genome size allowed for high-coverage sampling with relatively small amounts of raw sequence data. Although these experiments were restricted to 454 pyrosequencing and bacteriophage DNA, these evaluations of Nextera performance should be broadly applicable to other next-generation DNA sequencing platforms (e.g., Illumina and Ion Torrent) and other scientific applications (e.g., environmental metagenomics) that are challenged by low DNA yields.
MATERIALS AND METHODS
Creation of mock metagenome.
A mock metagenome sample composed of nine genome-sequenced phages, enterobacteriophage T4 (GenBank accession no. NC_000866), mycobacteriophages Catera (NC_008207), Fruitloop (NC_011288), Gumball (NC_011290), Omega (NC_004688), Porky (NC_011055), Solon (NC_011267), and cyanophages P-SS2 (NC_013021) and S-SM1 (GU071094.1), was constructed to assess the ability of the Nextera kit to provide data on the frequency distribution of viral genotypes within environmental samples. These phages are considered “known,” as reference genome sequences were available for comparative analyses. Genome equivalents of phage DNA were calculated from the molecular weight of each complete genome. Phage genomic DNA (gDNA) was mixed in differing proportions to simulate the unequal distribution of viruses within a natural sample (see Table 3). Phage gDNA was diluted in TE buffer (10 mM TRIS-HCl, 1 mM EDTA, pH 8) and quantified using a Qubit Quant-iT dsDNA high-sensitivity (HS) assay kit (Invitrogen; Carlsbad, CA) according to manufacturer's protocol. These concentrations were used to calculate the volume of each phage gDNA dilution needed for addition to the mock metagenome sample. The sample mixture was brought to a final volume of 200 μl and a final concentration of 26 ng/μl. Predicted coverage for each phage in the mock community was calculated as the average read length multiplied by the target number of reads for each genome divided by the genome size.
Table 3.
Predicted versus experimental coverage and percent abundance of reads for each phage in the mock metagenome
| Phage | Virus typea | Genome size (bp) | GC content (%) | Predicted result |
Experimental result |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Recruitment to reference sequence |
Mock assembly |
|||||||||||
| No. of reads | Coverage | Abundance of reads (%) | No. of reads | Coverageb (± SD) | Abundance of reads (%) | No. of contigs | Longest contig (bp) | N50 contig (bp) | ||||
| T4 | C | 166,000 | 35 | 20,000 | 47 | 28.1 | 31,049 | 61.6 (14.6) | 48.2 | 3 | 169,170 | 169,170 |
| Catera | M | 153,766 | 65 | 14,000 | 36 | 19.7 | 7,592 | 18.0 (8.6) | 11.8 | 4 | 85,298 | 85,298 |
| Fruitloop | M | 58,471 | 62 | 12,000 | 82 | 16.8 | 8,745 | 53.3 (18.9) | 13.6 | 1 | 58,308 | 58,308 |
| Gumball | M | 64,807 | 60 | 10,000 | 62 | 14.0 | 9,999 | 56.4 (18.8) | 15.5 | 1 | 64,807 | 64,807 |
| Omega | M | 110,865 | 61 | 8,000 | 29 | 11.2 | 3,362 | 10.9 (5.2) | 5.2 | 6 | 41,626 | 20,153 |
| Porky | M | 76,312 | 63 | 4,000 | 21 | 5.6 | 2,038 | 9.7 (5.8) | 3.2 | 7 | 34,434 | 17,944 |
| Solon | M | 49,487 | 64 | 2,000 | 16 | 2.8 | 715 | 5.2 (3.4) | 1.1 | 12 | 10,570 | 7,363 |
| P-SS2 | Cy | 107,530 | 52 | 1,000 | 3.7 | 1.4 | 757 | 2.5 (2.2) | 1.2 | 81 | 2,924 | 1,313 |
| S-SM1 | Cy | 174,079 | 41 | 200 | 0.5 | 0.3 | 205 | 0.4 (0.9) | 0.3 | 50 | 915 | 463 |
| Total | 71,200 | 64,718 (257 unassigned, 64,462 assigned) | 172 (6 unassigned, 166 assigned) | |||||||||
C, coliphage; M, mycobacteriophage; Cy, cyanophage.
Coverage is defined as number of bases per position in the consensus sequence.
Preparation of Nextera libraries for 454 sequencing.
Bar-coded DNA fragment libraries were prepared using a (Roche 454-compatible) Nextera DNA sample preparation kit (Epicentre Biotechnologies, Madison, WI). Fifty nanograms of sample DNA was fragmented utilizing 1 μl of transposome with 4 μl of high-molecular-weight buffer. Tagmentation reactions (i.e., DNA fragmentation and addition of the 19-bp inverted repeat to the fragment ends) were performed by incubation for 5 min at 55°C followed by purification of the tagmented DNA by the use of the brief Zymo protocol outlined in the Nextera protocol for use with a Clean and Concentrator-5 kit (Zymo Research, Orange, CA). Purified DNA was eluted from the column with 11 μl of nuclease-free water. Purified DNA (5 μl) was used as the template in a 50-μl volume for limited-cycle PCR (15 cycles) and processed as outlined in the Nextera protocol. Amplified DNA was purified using a Zymo Clean and Concentrator-5 kit according to the manufacturer's protocol, and the resulting DNA concentration was measured using a Qubit fluorometer (Invitrogen Corp., Carlsbad, CA). The fragment size distribution of the tagmented DNA was analyzed utilizing a 2100 Bioanalyzer with a 7500 DNA assay kit (Agilent Technologies, Santa Clara, CA).
Sequencing.
454 Titanium sequencing (with 9.6 million capture beads in total) was performed at the Institute for Genome Sciences at the Maryland School of Medicine. Samples were sequenced on half of a 454 sequencing plate.
Bioinformatic analysis.
Assembly of phage genomes from sequence libraries prepared using the Nextera system was performed using Geneious 5.0.4 bioinformatics software (Geneious version 5.1, 2010; A. J. Drummond). Assembly analyses included the nine known phages in the mock metagenome (see reference 3), four unknown cyanophages for which no reference genome sequence was available (see Table 2), and eight mycobacteriophage sequence libraries (see Fig. 5b) prepared using the standard Roche 454 sequence library preparation protocol. Details of the Roche protocol are provided by the manufacturer of the 454 instrument. Phage genomes prepared for sequencing using the LASL technique (11) were assembled using Geneious 5.4.5 software (2011; A. J. Drummond). After preparation performed using the Nextera system, sequences that contained any error in the expected 5′ transposon sequence were removed using a custom Perl script (http://sourceforge.net/projects/readfilter/). The transposase linker and adaptor sequences were removed using the Trim Vector feature in Geneious, and sequences were quality trimmed at a base call error probability limit of less than 0.01. Reads shorter than 100 bp after quality trimming were not included in the assembly. For the TUSD phages (cyanophage), host DNA contamination was screened out using cross_match (http://www.phrap.org/), with a value of 30 for the minimum match and 10 for the minimum score. The original and cleaned files were used for assembly to determine the importance of the transposon quality screening step for de novo assembly (see Table 2).
Table 2.
Results determined for de novo assemblies of 454 pyrosequencing reads from Nextera and LASL fragment libraries
| Phage | No. of reads |
No. of contigs |
N50 contig (bp) | N90 contig (bp) | Major contig(s) of alignmenta |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Before cleaning | After cleaning | Before cleaning | After cleaning | Size(s) (bp) | No. of reads | Avg coveragec (± SD) | Maximum coveragec | Pairwise identity (%) | GC (%) | |||
| Angelica | 4,908 | 3,968 | 11 | 7 | 28,452 | 18,196 | 28,452 | 1,677 | 22 (12) | 73 | 99.5 | 66.9 |
| 18,196 | 1,396 | 29 (15) | 73 | 93.1 | 65.8 | |||||||
| Athena | 18,599 | 15,198 | 186 | 5 | 69,410 | 69,410 | 69,410 | 15,180 | 84 (39) | 253 | 99.1 | 67.5 |
| Avrafan | 15,562 | 12,975 | 4 | 2 | 41,902 | 41,902 | 41,902 | 12,970 | 111 (60) | 349 | 99.4 | 66.6 |
| Blue7 | 32,010 | 25,495 | 115 | 1 | 52,233 | 52,233 | 52,233 | 25,490 | 165 (54) | 381 | 99.4 | 61.3 |
| Wee | 18,525 | 16,012 | 12 | 2 | 59,245 | 59,245 | 59,245 | 16,004 | 95 (39) | 273 | 99.4 | 61.8 |
| TUSD 1b | 13,763 | 9,942 | 272 | 158 | 37,944 | 378 | 50,727, 37,944 | 5,665, 3,435 | 36 (11), 29 (11) | 84, 70 | 99.4, 99.4 | 36.7, 37.3 |
| TUSD 14 | 15,345 | 7,568 | 1,139 | 201 | 497 | 350 | 38,178 | 6,628 | 55 (16) | 108 | 99.4 | 37.3 |
| TUSD 20 (Nextera) | 18,066 | 14,311 | 53 | 31 | 38,098 | 443 | 38,098 | 14,151 | 118 (31) | 210 | 99.3 | 37.3 |
| TUSD 20 (LASL) | 12,705 | 8,855 | – | 20 | 7,610 | 726 | 7,610 | 1,005 | 26 (13) | 67 | 99.0 | 38.0 |
| TUSD 21b (Nextera) | 16,061 | 11,184 | 88 | 5 | 36,669, 32,801 | 465, 315 | 36,669, 37,626 | 9,403, 1,754 | 81 (27), 15 (6) | 173, 35 | 99.1, 99.4 | 35.9, 37.3 |
| TUSD 21b (LASL) | 31,069 | 20,588 | 62 | 37,873, 32,801 | 12,916, 6,640 | 71 (188), 39 (27) | 2,326, 221 | 99.4, 98.9 | 37.2, 35.7 | |||
For genomes with two or more major contigs, the information on the second contig is also listed.
For TUSD phages, host DNA contamination was removed using cross_match.
Coverage is defined as number of bases per position in the consensus sequence.
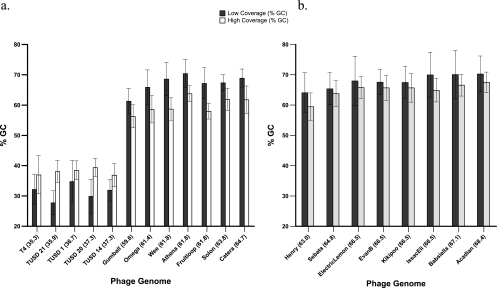
Fig. 5.
Average GC content (± SD) of regions with low or high coverage in comparisons of individual phage genomes. (a) Phage comprising the mock metagenome and unknown phage genomes prepared using the Nextera protocol. (b) Mycobacteriophage genomes prepared using the Roche GS FLX Titanium general library preparation method. Values in parentheses after the phage name indicate the average % GC content of the genome. Classification of the results into low- and high-coverage areas corresponded to the regions above or below 1.5 standard deviations from the average coverage. Genomes with at least 5 regions of low and high coverage of 50 bp or longer were included in the analysis.
Assembly and recruitment.
Recruitment of the mock-metagenome reads to a reference set of concatenated phage genome sequences was processed using Geneious software with the following custom parameters: word length, 10; index word length, 10; maximum gap size, 100; maximum gaps per read, 20%; maximum mismatches, 5%; maximum ambiguity, 16. Unassembled reads reflected sequences that did not recruit to any of the reference genomes in the concatenated reference sequence. Mock-metagenome sequences that recruited to each genome within the concatenated reference sequence were extracted and realigned individually to calculate coverage and read frequency statistics. De novo assemblies of the four unknown cyanophages and the mycobacteriophage sequence libraries were performed using the Geneious assembler using the “Highest Sensitivity/Slow” setting (word length, 10; index word length, 10; maximum gap size, 100; maximum gaps per read, 20%; maximum mismatches, 20%; maximum ambiguity, 16). Contig lengths were adjusted for genomes with redundancy at the terminal ends, denoting a circular genome or a genome with circular characteristics (sticky ends). N50 and N90 contig length values (defined as the smallest assembled contig representing greater than 50% or 90% of the combined contig lengths) were calculated for the assembled phage genomes. The major contig for each assembly was extracted as a BAM file, and a custom Perl script was used to generate the average percent GC and coverage over 50-bp sliding windows. Regions of each assembly with coverage above or below 1.5 standard deviations from the average coverage of a particular genome were further examined. These regions were extracted and analyzed for variations in GC content. Only regions ≥ 50 bp in length were included in calculating average GC content and standard deviation values.
Statistical analysis.
Linear regression analysis was performed using Aabel software (version 2; Gigawiz Ltd. Co., Tulsa, OK). Wilcoxon and Kruskal-Wallis tests were performed to determine whether there was a statistical difference between the GC contents of regions of high and low coverage for genomes prepared using Nextera and Roche library preparations using JMP (version 8.0.2; SAS, Cary, NC). Statistical tests where P values were <0.05 were considered significant.
RESULTS AND DISCUSSION
Nextera protocol.
For each of the samples, the recommended starting quantity of 50 ng of DNA was utilized in the transposase tagmentation reaction. The high-molecular-weight buffer was used to generate 500- to 1,100-bp fragments. After the limited-cycle PCR and cleanup, total DNA quantities of the samples ranged from 306.8 ng to 915.3 ng, with an average of 580.8 ng (Table 1). The majority of Nextera library fragments (mean = 59.7%) fell between 300 and 800 bp, slightly lower than the range predicted in the manufacturer's protocol. However, peak DNA abundance occurred at longer fragment lengths for samples with higher final DNA concentrations (e.g., Athena and TUSD 1; Table 1). This was likely a result of fragment “nesting,” which occurs as DNA concentrations increase during limited-cycle PCR; fragments form noncovalently bound chains through homologous base pairing of adaptor sequences. Bioanalyzer results from fragmented mycobacteriophage Athena gDNA represent a particularly good example of this phenomenon (see Fig. S1 in the supplemental material). It is likely that a majority of the long fragments actually represented shorter, nested fragments. While “nesting” makes accurate estimation of fragmentation length distribution difficult, it did not seem to have an effect on the quality of the 454 pyrosequencing results.
Table 1.
DNA yields and fragmentation results after completion of the Nextera protocol
| Phage | Typea | Final DNA concn (ng/μl) | Total DNA (ng)b | Concn fragments of 300–800 bpc (ng/μl) | % total DNA (300–800 bp)d | Fragment size of greatest abundancec (bp) | Avg read length for 454 sequences (bp)e |
|---|---|---|---|---|---|---|---|
| Angelica | M | 12 | 307 | 9 | 79 | 569 | 373 |
| Athena | M | 33 | 858 | 27 | 81 | 1,402 | 382 |
| Avrafan | M | 21 | 322 | 11 | 49 | 843 | 355 |
| Blue7 | M | 13 | 332 | 9 | 68 | 398 | 336 |
| Wee | M | 19 | 482 | 12 | 65 | 529 | 348 |
| TUSD 1 | Cy | 34 | 915 | 20 | 58 | 1,392 | 320 |
| TUSD 14 | Cy | 21 | 571 | 11 | 51 | 1,348 | 307 |
| TUSD 20 | Cy | 20 | 536 | 10 | 52 | 732 | 317 |
| TUSD 21 | Cy | 29 | 778 | 11 | 39 | 1,359 | 315 |
| Mock metagenome | 22 | 708 | 12 | 55 | 1,151 | 349 |
M, mycobacteriophage; Cy, cyanophage.
x ± SD = 581 ± 226 ng.
Data were determined using an Agilent Bioanalyzer.
x = 60%.
The average read lengths were calculated after removal of sequences of less than 100 bp.
Effects on sequence read length.
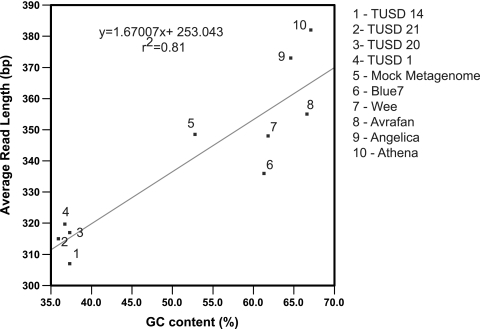
The average 454 sequence read length was most strongly associated with the GC content of the input DNA and not with the final DNA concentration after Nextera preparation. For the nine de novo-assembled phages and the nine phages in the mock metagenome, average read length and GC content were positively correlated (r2 = 0.81) (Fig. 2). This trend was also observed when genomes within the mock metagenome were analyzed individually (see Fig. S2 in the supplemental material). Transposase fragmentation is a possible explanation for this trend. Most transposases do not integrate randomly but choose sites based on a particular nucleotide frequency or DNA structure (7). The Nextera transposase is based on a mutated form of Tn5 that exhibits increased insertion frequency in comparison to wild-type Tn5 (22). The consensus target sequence of Tn5 gives a slight bias toward AT-rich sequences (10). Examination of read length histograms for nine phage genomes indicated that genomes with higher GC content tended to have a greater proportion of sequences in the third and fourth quartiles of read lengths, whereas genomes with lower GC content had a greater proportion of reads in the first and second read length quartiles, with the majority of reads occurring in the third quartile (see Fig. S3 in the supplemental material). However, analysis of raw reads indicated that only 1.5% to 4.5% of reads represented full fragments (i.e., fragments consisting of the span from the 454A adapter to the 454B adapter). So, the finding of possibly longer fragments coming from genomes with high GC content does not seem to explain the longer reads coming from these genomes. Instead, the differences in the distribution of read lengths for genomes with high and low GC content may be a function of the processivity of the 454 sequencing reaction, which can vary depending on the GC content of the fragment. As a surprising consequence, fragments with higher GC content resulted in longer sequence reads.
Fig. 2.
Linear regression analysis of the percent GC content of the largest contig versus average read length for nine unknown genomes and the mock metagenome. The percent GC content for the mock metagenome represents the collective percent GC for all nine reference genomes.
Quality screening of 454 sequence reads.
The importance of sequence quality in generating accurate alignments has been appreciated since the development of automated DNA sequencing (6). While trimming and removal of poor-quality sequences usually result in the loss of data, they also result in increased alignment accuracy (16). The 19-bp transposon sequence inserted during the Nextera tagmentation reaction allowed assessment of sequencing quality. A script was developed to remove sequences with any error in the transposon sequence, as such errors might have been indicative of poor overall read quality. After transposon sequence screening, the percentages of identical sites and pairwise identity values of short reads (<100 bp) recruiting to the T4, Catera, Gumball and P-SS2 reference genome sequences were similar to those of longer sequences (>500 bp). This result indicated that even the small fragments were of good quality. Thus, the shorter reads likely represented smaller fragments produced by Nextera library construction rather than poor-quality reads (see Table S1 in the supplemental material). Comparison of de novo assemblies showed a significant decrease in the number of contigs after quality screening despite the loss of reads from the screening step (Table 2). Therefore, it is recommended that a similar screening of sequences according to the quality of the transposase sequence be performed for all libraries that utilize a 454 Nextera kit.
De novo assembly of phage genomes.
In the genome assemblies with large numbers of contigs (e.g., the TUSD phages), the majority of contigs consisted of only a few reads (Table 2). The small contigs may have represented sequences that were too degenerate to assemble with the major contig or fragments at the extreme terminal ends of the genome, where sequencing coverage is expected to be low. Some of the small contigs from the TUSD genomes resulted from contaminating bacterial DNA, since many of these small contigs had strong hits with respect to known Prochlorococcus host strains. Nevertheless, the extent of coverage for the largest contig for each of the cyanophages (the TUSD phages) was greater than the 13-fold coverage needed for the generation of single-contig genomes (11). Phage TUSD 1 assembled into 158 separate contigs, with 130 of these contigs containing only two or three sequences. The two major contigs from TUSD 1 could not be connected due to either a gap in sequence coverage or a nonclonal phage lysate. A similar phenomenon was also seen with TUSD 21 and Angelica (Table 2). However, the two major contigs from Angelica were segments of the same genome, as verified by recruitment to the reference genome (see Table S2 in the supplemental material).
TUSD 20 and 21 phages were prepared for sequencing using both the LASL and Nextera protocols. For TUSD 20, LASL genome assembly resulted in shorter contig lengths than Nextera genome assembly, despite the generation of fewer contigs overall. For TUSD 21, the lengths of the two major contigs generated from the assembly of the LASL and from the Nextera preparations were similar. In the largest LASL contig, however, there was a ∼600-bp region exhibiting unusually high coverage (Table 2). The same region in the Nextera assembly was not found to have abnormal coverage. While it remains unclear why this region was so highly covered in the LASL assembly, it is possible that preferential amplification of this segment occurred during the linker amplification (Fig. 1). While the LASL technique has been shown to be an effective method for the preparation of sequence libraries (11), these results indicate that for cyanophages TUSD 20 and TUSD 21, genomic library preparation using Nextera resulted in better assemblies.
The best de novo genome assemblies occurred for mycobacteriophages Blue7, Avrafan, Athena, and Wee, for which five or fewer contigs were constructed (Table 2). Over the course of this study, full-length genome sequences of Athena, Avrafan, Blue7, and Wee were released through an independent project (http://phagesdb.org/). Consensus sequences from the Nextera libraries were compared to those genome sequences by the use of Blast-2 (3), and the comparison showed greater than 99% coverage and near-perfect identity between the reference genomes and the assemblies, with length differences occurring at the ends of genomes. For Blue7 and Wee, which had consensus sequences shorter than the genome length, small regions at the terminal ends were missing (see Table S2 in the supplemental material). Overall, these results indicate that Nextera library construction followed by 454 pyrosequencing appears to be a reasonable approach for de novo assembly of microbial genomes.
Mock metagenome analysis.
Metagenomic approaches have provided an unprecedented opportunity to explore the composition of viral assemblages within natural environments. However, accurate prediction of the frequencies of viral taxa and overall viral diversity depends on maintaining the relative ratios of viral genomes derived from DNA isolation through sequencing (4). Because amplification procedures can introduce bias into metagenomic sequencing data, their use should be minimized or at least carefully evaluated. For instance, multiple-displacement amplification approaches utilizing the phi29 DNA polymerase are known to result in chimeric sequences and preferential amplification of circular single-stranded DNA (ssDNA), which can skew the distribution of viral sequences within metagenomic libraries (4, 14, 20, 28). Ideally, procedures that amplify isolated DNA for library preparation should do so without altering the distribution of genotypes in the original sample.
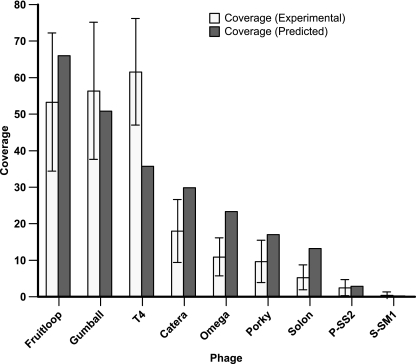
A mock viral metagenome sample was constructed from viral genomic DNA to test the applicability of Nextera library construction for shotgun metagenomic analysis of microbial community structure (Table 3). Of the 64,718 reads that remained after quality screening, only 0.4% (257 sequences) did not recruit to one of the phage genomes in the mock metagenome community. The experimental rank distribution of sequence coverage indicated that the order of the six least abundant phages was conserved, while the order of the three most abundant phages was shuffled in comparison to the predicted ranking, with phage T4 moving from the third to the first rank (Fig. 3). Discounting possible errors in DNA quantification during construction of the mock metagenome, it is possible that T4 genomic DNA, with its lower GC content, was preferentially amplified in the Nextera procedure. The ranking with respect to abundance of phages with similar levels of GC content (e.g., the mycobacteriophages) was preserved. Thus, for natural viral communities, the rank abundance of member viruses with similar levels of GC composition would be preserved in the Nextera procedure. In contrast, virus genomes with low GC content could be overrepresented in viral communities where viruses with high GC content are also present.
Fig. 3.
Comparison of the predicted sequence coverage of each member of the mock metagenome to the experimental coverage (± SD). The predicted coverage was normalized to the experimental average read length and the number of sequence reads obtained after quality screening.
The results of assembly of all sequences within the mock metagenome met expectations in that, with a decreasing rank abundance of a given virus, contig number increased and contig size decreased (Table 3). These expectations match the assumptions behind the approaches, such as that represented by Phage Communities from Contig Spectrum (PHACCS) software (4), that use whole-metagenome assembly statistics to estimate viral diversity. Whole-genome assembly was achieved only for three phages with sequence coverage greater than 50× (Table 3). The requirement of such high levels of coverage for whole-genome assembly in the mock metagenome is a consequence of the short read length of the 454 sequences as well as the lack of paired-end sequences, both of which are well known to improve genome assembly.
An important scientific goal of metagenomic analyses is to compare the composition and diversity characteristics of microbial communities across environments. However, a continuing challenge is that of minimizing the influence of methodological differences between studies on the outcome of comparative analyses. The impact of sample processing methodology on subsequent comparative analyses of microbial communities by the use of shotgun metagenome sequence data can be substantial. A recent study found that the rank frequency of microbial species within a simulated microbial community (as determined from sequence reads) varied significantly with the DNA extraction procedure (18). Across the experimental trials, the best agreement between replicated metagenome samples was seen for protocols that more readily standardized the treatment of samples, such as kit-based methods for extracting DNA. Because every methodological step in preparing an environmental sample for metagenomic sequencing (e.g., DNA isolation, shearing, ligation, and amplification) can potentially introduce bias into the frequency distribution of sequence reads within a metagenome library, comparative metagenomic analyses are advisable only for libraries obtained using identical sample-processing protocols. While use of the Nextera library construction did not result in a perfect frequency distribution of reads predicted for each genome, the kit does provide a readily available means for standardizing shotgun metagenome library construction across laboratories, thus constraining the overall methodological bias from field sample to sequence library. Ultimately, it is this quality of standardization that may be the most important contribution of Nextera to metagenomic studies.
Analysis of low- and high-coverage regions.
Assembly data from Nextera libraries revealed opposing trends in comparisons of sequence coverage and GC content. Genomes with high GC content tended to exhibit a decrease in mean coverage with increasing GC content (Fig. 4b), while genomes with low GC content tended to exhibit an increase in mean coverage with increasing GC content (Fig. 4c). However, the large range of coverage values for a given level of GC content suggests that there is not a strong correlation between percent GC and sequence coverage.
Fig. 4.
Box-and-whisker plots of the sequence coverage range as a function of phage genome GC content. Phage names are provided above each panel. (a) The Roche GS FLX Titanium general library preparation method. (b) The Nextera method with high-GC-content phage. (c) The Nextera method with low-GC-content phage. Whiskers represent the 5th and 95th percentiles; open diamonds represent mean values.
To further assess whether percent GC composition varied with the level of coverage, regions of each phage genome that fell 1.5 standard deviations (SD) away from the mean of the coverage values were extracted and analyzed. For genomes with intermediate coverage in the mock metagenome (Catera, Fruitloop, Gumball, Omega, and Solon), the level of GC content appeared to have an effect. Areas of high coverage had lower GC content than areas of low coverage (Fig. 5a). The same trend occurred for the de novo-assembled mycobacteriophage genomes Athena and Wee. An opposing trend was seen for genomes with low GC content, such as T4 and the TUSD phages. For all genomes, differences in GC content determinations between high- and low-coverage regions were found to be significant (Kruskal-Wallis test; P ≤ 0.03) (Fig. 5).
The coverage biases may have partly resulted from the PCR amplification step (1, 12). For example, recent reports have shown that GC-rich and GC-poor areas are generally underrepresented in Illumina sequencing data due to the PCR enrichment step (2, 13). Omitting the limited-cycle PCR step reduced GC coverage bias in Nextera sequence libraries of an E. coli genome (1). Therefore, it is possible that amplification biases from the Nextera limited-cycle PCR step or the 454 preparation protocol contributed to GC coverage bias. To address this issue, sequence data from mycobacteriophages prepared using a 454 pyrosequencing library preparation method developed by Roche (in which the only amplification step uses emulsion PCR [em-PCR]) were compared to data prepared using Nextera libraries. As with the Nextera libraries, the Roche libraries showed decreases in mean coverage with increasing GC content (Fig. 4a). Once again, a large range of coverage values within a given range of percent GC was observed. Lower-coverage regions tended to exhibit higher GC content results for genomes prepared using the Roche protocol compared to Nextera (Fig. 5b). However, this trend was not found to be statistically significant.
It is possible that events that occur during the 454 sequencing process itself, including the em-PCR step or even physical fragmentation that is not completely random (19, 26), may slightly contribute to coverage trends (Fig. 4a). However, our data suggest that the GC coverage trend observed in Nextera libraries was likely due to amplification bias that occurred during Nextera limited-cycle PCR rather than to the 454 sequencing technology. For metagenomic analyses in which read frequency is used as a measure of the prevalence of a biological feature (e.g., taxonomic origin or gene function), such biases can influence analytical outcomes.
From these experiments, it is evident that the Nextera 454 library preparation procedure can provide high-quality sequence data from small quantities of starting dsDNA, with a substantial reduction in processing time. While coverage may be influenced by GC content, this protocol was successful in the de novo assembly of phage genomes. In metagenomic studies, where small starting quantities of DNA generally make amplification procedures unavoidable, all extant library preparation procedures likely cause some measure of percent GC sampling bias in the resulting sequence library. To date, no other in vitro studies have systematically examined sources of bias in the preparation of viral metagenome libraries. While the Nextera protocol is not bias-free, it provides a rapid means of generating libraries as well as standardization and consistency in library construction that should improve comparative metagenomic analyses across and between laboratories (18).
Supplementary Material
ACKNOWLEDGMENTS
We thank Bonnie Poulos from the Sullivan laboratory for her work in providing and preparing the TUSD phages, funded by NSF grant DBI-0850105. We also thank Nicholas Caruccio from Epicentre for all of his assistance and Laura Marinelli, Divya Tumuluru, Greg Broussard, Ellie Pierce, Jeff Witters, Kathy Van Hoeck, and Mantha Makume for the isolation of mycobacteriophages Athena, Blue7, Acadian, Electric Lemon, Isaac Eli, Kikipoo, and Sebata, respectively.
This work was supported through grants from the National Science Foundation (MCB-0731916) and the U.S. Department of Agriculture Cooperative State Research, Education and Extension Service (2005-35107-15214). R.M. was supported through a graduate fellowship from the Institute for Soil and Environmental Quality.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 23 September 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Adey A., et al. 2010. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 11:R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aird D., et al. 2010. Analyzing and minimizing bias in Illumina sequencing libraries. Genome Biol. 11(Suppl. 1):P3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Angly F., et al. 2005. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics 6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breitbart M., et al. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99:14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chou H. H., Holmes M. H. 2001. DNA sequence quality trimming and vector removal. Bioinformatics (Oxford, England) 17:1093–1104 [DOI] [PubMed] [Google Scholar]
- 7. Craig N. L. 1997. Target site selection in transposition. Annu. Rev. Biochem. 66:437–474 [DOI] [PubMed] [Google Scholar]
- 8. Dean F. B., Nelson J. R., Giesler T. L., Lasken R. S. 2001. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11:1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delwart E. L. 2007. Viral metagenomics. Rev. Med. Virol. 17:115–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goryshin I. Y., Miller J. A., Kil Y. V., Lanzov V. A., Reznikoff W. S. 1998. Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. U. S. A. 95:10716–10721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henn M. R., et al. 2010. Analysis of high-throughput sequencing and annotation strategies for phage genomes. PLoS One 5:e9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Henry R., Oono K. 1991. Amplification of a GC-rich sequence from barley by a two-step polymerase chain reaction in glycerol. Plant Mol. Biol. Rep. 139–144 [Google Scholar]
- 13. Kozarewa I., et al. 2009. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods 6:291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lasken R. S., Stockwell T. B. 2007. Mechanism of chimera formation during the multiple displacement amplification reaction. BMC Biotechnol. 7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu C., et al. 2005. Elucidation of the small RNA component of the transcriptome. Science (New York, NY) 309:1567–1569 [DOI] [PubMed] [Google Scholar]
- 16. Malde K. 2008. The effect of sequence quality on sequence alignment. Bioinformatics (Oxford, England) 24:897–900 [DOI] [PubMed] [Google Scholar]
- 17. Mardis E. R. 2008. The impact of next-generation sequencing technology on genetics. Trends Genet. 24:133–141 [DOI] [PubMed] [Google Scholar]
- 18. Morgan J. L., Darling A. E., Eisen J. A. 2010. Metagenomic sequencing of an in vitro-simulated microbial community. PLoS One 5:e10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oefner P. J., et al. 1996. Efficient random subcloning of DNA sheared in a recirculating point-sink flow system. Nucleic Acids Res. 24:3879–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Polson S. W., Wilhelm S. W., Wommack K. E. 2011. Unraveling the viral tapestry (from inside the capsid out). ISME J. 5:165–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pop M., Salzberg S. L. 2008. Bioinformatics challenges of new sequencing technology. Trends Genet. 24:142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reznikoff W. S. 2003. Tn5 as a model for understanding DNA transposition. Mol. Microbiol. 47:1199–1206 [DOI] [PubMed] [Google Scholar]
- 23. Sato M., Ohtsuka M., Ohmi Y. 2005. Usefulness of repeated GenomiPhi, a phi29 DNA polymerase-based rolling circle amplification kit, for generation of large amounts of plasmid DNA. Biomol. Eng. 22:129–132 [DOI] [PubMed] [Google Scholar]
- 24. Schoenfeld T., et al. 2010. Functional viral metagenomics and the next generation of molecular tools. Trends Microbiol. 18:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schoenfeld T., et al. 2008. Assembly of viral metagenomes from Yellowstone hot springs. Appl. Environ. Microbiol. 74:4164–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schwartz S. L., Farman M. L. 2010. Systematic overrepresentation of DNA termini and underrepresentation of subterminal regions among sequencing templates prepared from hydrodynamically sheared linear DNA molecules. BMC Genomics 11:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tringe S. G., Rubin E. M. 2005. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genetics 6:805–814 [DOI] [PubMed] [Google Scholar]
- 28. Yilmaz S., Allgaier M., Hugenholtz P. 2010. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat. Methods 7:943–944 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.