Abstract
Casein kinases regulate a wide range of cellular functions in eukaryotes, including phosphorylation of proteins that are substrates for degradation via the ubiquitin-proteasome system (UPS). Our previous study demonstrated that Fbp1, a component of the SCFFBP1 E3 ligase complex, was essential for Cryptococcus virulence. Because the Saccharomyces cerevisiae homolog of Fbp1, Grr1, requires casein kinase I (Yck1 and Yck2) to phosphorylate its substrates, we investigated the function of casein kinase I in Cryptococcus neoformans. In this report, we identified a C. neoformans casein kinase I protein homolog, Cck1. Similar to Fbp1, the expression of Cck1 is negatively regulated by glucose and during mating. cck1 null mutants showed significant virulence attenuation in a murine systemic infection model, but Cck1 was dispensable for the development of classical virulence factors (capsule, melanin, and growth at 37°C). cck1 mutants were hypersensitive to SDS treatment, indicating that Cck1 is required for cell integrity. The functional overlap between Cck1 and Fbp1 suggests that Cck1 may be required for the phosphorylation of Fbp1 substrates. Interestingly, the cck1 mutant also showed increased sensitivity to osmotic stress and oxidative stress, suggesting that Cck1 regulates both cell integrity and the cellular stress response. Our results show that Cck1 regulates the phosphorylation of both Mpk1 and Hog1 mitogen-activated protein kinases (MAPKs), demonstrating that Cck1 regulates cell integrity via the Mpk1 pathway and regulates cell adaptation to stresses via the Hog1 pathway. Overall, our study revealed that Cck1 plays important roles in regulating multiple signaling pathways and is required for fungal pathogenicity.
INTRODUCTION
Cryptococcus neoformans is a major human fungal pathogen and the causative agent of the often fatal cryptococcal meningoencephalitis, which is an AIDS-defining illness. Due to the medical significance and genetic tractability of C. neoformans, extensive studies have been conducted on the mechanism of its virulence. Several signaling pathways important for Cryptococcus virulence have been identified (1, 16, 38). However, fungal virulence is a complex trait, and additional virulence-determining mechanisms remain to be discovered.
The SCF (Skp1, Cullins, and F-box proteins) E3 ubiquitin ligase-mediated ubiquitin-proteasome system (UPS) is a principle intracellular mechanism for controlled protein degradation in eukaryotes and has recently emerged as an attractive drug target for human diseases, such as cancer (26, 35). Fungal SCF complexes have been reported to regulate a variety of cellular functions (17). Our previous studies identified an F-box protein, Fbp1, that is essential for fungal virulence despite its dispensability for the development of several classical virulence factors, including the production of melanin, capsule formation, and growth at 37°C (21). We hypothesize that Fbp1 may be part of a novel virulence mechanism and could have potential as a drug target. Identification of the substrates important for Fbp1-mediated virulence regulation could potentially be the key to understanding the molecular mechanism by which Fbp1 controls fungal virulence. Because only phosphorylated substrates can be targeted by the E3 ligase for ubiquitination and degradation, understanding these protein phosphorylation processes is important for characterizing the downstream substrates. Casein kinase I proteins are involved in the phosphorylation of many E3 ligase substrates, including the substrates of the well-studied SCF E3 ligase containing the Grr1 protein (SCFGrr1) in Saccharomyces cerevisiae (18). Two casein kinase I proteins in S. cerevisiae, Yck1 and Yck2, play important roles in the phosphorylation of the Grr1 substrates that are involved in glucose sensing (Mth1 and Std1) (27), amino acid signaling (Stp1, Stp2, and Ptr3) (22, 36), pheromone sensing (Ste2) (14), and uracil uptake (Fur4) (24, 25). Fbp1 is a homolog of Grr1, and its expression is also regulated by glucose (21). Therefore, we predict that a similar substrate phosphorylation mechanism exists to activate Fbp1-mediated protein turnover and that the casein kinase I family in C. neoformans is involved in the phosphorylation of Fbp1 substrates for degradation. If so, mutations of casein kinase I would abolish the phosphorylation of the Fbp1 substrates and prevent them from being recognized and ubiquitinated by the SCFFbp1 E3 ligase. Thus, casein kinase I mutants should exhibit phenotypes similar to those of fbp1 mutants since both types of mutations would cause the accumulation of Fbp1 substrates. The overlapping phenotypes can be used in genetic screening for potential substrates of Fbp1. Hence, we decided to study the function of the casein kinase I proteins in C. neoformans.
Casein kinase I (CKI) family members are serine/threonine protein kinases that function as regulators of signal transduction pathways in eukaryotic organisms from yeast to humans (18). There are seven CKI family members in human which are involved in Wnt signaling, circadian rhythms, nucleo-cytoplasmic shuttling of transcription factors, DNA repair, and DNA transcription (4, 32). In fungi, most studies of the casein kinase I family have been done in S. cerevisiae, where Yck1 and Yck2 share redundant functions in cell morphogenesis, nutrient sensing, septin assembly, and endocytic trafficking (33, 34). Deletion of both YCK1 and YCK2 is lethal in S. cerevisiae (34). The Candida casein kinase I protein Yck2 has been found to be important for the fungus to damage host epithelial cells and necessary for resistance to cell membrane stress (29). The Schizosaccharomyces pombe casein kinase I Cki1 was reported to function in the phosphorylation of phosphatidylinositol 4-phosphate 5-kinase to regulate the production of inositol polyphosphates (37). CKI in Neurospora crassa mediates the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the circadian negative-feedback loop (13). Despite the importance of the casein kinase I proteins in regulating fungal development, there is no report as yet on their functions in basidiomycetous fungi.
In this report, we have identified a protein that shares high sequence identity with both Yck1 and Yck2 in S. cerevisiae. Cryptococcus casein kinase 1 (Cck1) can complement the function of Yck1/Yck2 (Yck1/2) in an yck1 yck2ts mutant, suggesting that it is a bona fide casein kinase I protein. In a murine systemic infection model, the cck1 mutant developed normal virulence factors but showed attenuated virulence. Our study found that Cck1 is essential for cell integrity and sexual reproduction, functions shared by Fbp1. Meanwhile, the cck1 mutant also showed additional phenotypes that are not related to the Fbp1 function, indicating that Cck1 regulates multiple downstream targets. Epistasis analysis revealed that Cck1 regulates both the Mpk1 mitogen-activated protein kinase (MAPK) pathway that is known to regulate cell integrity in fungi and the Hog1 pathway that mediates fungal stress responses. Overall, our results demonstrated that the casein kinase I in C. neoformans regulates a wide range of cellular functions via multiple pathways. Our study also uncovered a new layer of regulation for cell integrity and fungal virulence.
MATERIALS AND METHODS
Strains, media, and growth conditions.
C. neoformans and S. cerevisiae strains used in this study are listed in Table 1. Strains were grown at 30°C on yeast extract-peptone-dextrose (YPD) agar medium and synthetic dextrose (SD) medium. V8 medium (pH 5.0) was used for mating assays. Modified MS medium (Murashige and Skoog medium) was used for mating and sporulation assays and was prepared as previously described (43). All other media were prepared as described previously (1, 12).
Table 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| C. neoformans strains | ||
| H99 | MATα | 30 |
| KN99a | MATa | 28 |
| CUX106 | MATα cck1::NEO | This study |
| CUX107 | MATacck1::NEO | This study |
| CUX111 | MATacck1::NEO CCK1::NAT | This study |
| CUX2 | MATα fbp1::NEO | This study |
| CUX3 | MATafbp1::NEO | This study |
| CUX130 | MATacck1::NEO fbp1::NEO | This study |
| CUX131 | MATacck1::NEO mpk1::NAT-150 | This study |
| CUX132 | MATα cck1::NEO pka1::URA | This study |
| KGCN861 | MATα mpk1::NAT-150 | J. Lodge |
| LVCN1073 | MATampk1::NAT-150 | J. Lodge |
| JKH7 | MATα pka1::URA5 ura5 | 15 |
| S. cerevisiae strains | ||
| YSB4742 | MATα his3Δ1 leu2Δ0 ura3Δ0 | ATCC yeast deletion collection |
| FM635 | MATα yck1-1::URA3 yck2-2ts | M. Johnston |
| YUX41 | MATα yck1-1::URA3 yck2-2tsPADH1-GFP::CCK1 | This study |
| YUX42 | MATα yck1-1::URA3 yck2-2tsPADH1-GFP | This study |
Detection of CCK1 gene expression using qRT-PCR.
We measured CCK1 mRNA levels via quantitative reverse transcription-PCR (qRT-PCR) in cells grown with or without glucose and throughout the mating process. Cultures of C. neoformans var. grubii wild-type (wt) strain H99 were grown overnight on YPD (2% glucose) or yeast extract-galactose (YPG; 2% galactose) liquid medium at 30°C with shaking. Collected cells were washed with distilled H2O (dH2O), resuspended in YPD or YPG medium, and incubated for 2 h. Cells collected from YPD medium were washed with dH2O and resuspended in YPG medium while cells collected from YPG medium were resuspended in YPD medium. Both cultures were incubated for 2 h. Total RNAs were prepared from cells with each treatment, and cDNA was synthesized as described below.
Mating was performed by mixing the H99 and KN99a strains, which were then cocultured on V8 medium (pH 5.0). Mating mixtures were collected from agar surfaces using cell scrapers after 6, 24, 48, 72, or 96 h of incubation. Collected cells were washed with dH2O, and pellets were used for total RNA extraction. Purified RNAs were quantified using a Nanodrop spectrometer (Thermo Scientific) and used as templates for PCR amplification with primers of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene to determine potential genomic DNA contamination. First-strand cDNAs were synthesized using a Superscript III cDNA synthesis kit (Invitrogen) following the manufacturer's instructions. Expression levels of CCK1 and GAPDH were analyzed using SYBR advantage QPCR premix reagents (Stratagene) with an Mx4000 QPCR system (Stratagene) as previously described (42). Gene expression levels were normalized using the endogenous control gene GAPDH, and the relative levels were determined using the comparative threshold cycle (CT) method (23).
Generation of cck1 null mutants and their complemented strains.
Mutants for CCK1 were generated in both H99 and KN99a strain backgrounds by overlap PCR as previously described (6). The 5′ and 3′ regions of the CCK1 gene were amplified from H99 genomic DNA with the primer pair CX322 and CX323 and the pair CX324 and CX325, respectively (Table 2 gives primer sequences). The dominant selectable markers (NEOr) were amplified with the M13 primers (M13F and M13R) from plasmid pJAF1 (9). Each target gene replacement cassette was generated by overlap PCR with primers CX322 and CX325 (Table 2). Purified overlap PCR products were precipitated onto 10 μl of gold microcarrier beads (0.6 μm; Bio-Rad), and strains H99 or KN99a were biolistically transformed as described previously (7). Stable transformants were selected on YPD medium containing G418 (200 mg/liter). To screen for mutants of the CCK1 gene, diagnostic PCR was performed by analyzing the 5′ junction of the disrupted mutant alleles with primers CX328 and JH8994 (Table 2). Positive transformants were identified by PCR screening with primers CX326 and CX327.
Table 2.
Primers used in this study
| Primer | Sequence (5′–3′) | Description or functiona |
|---|---|---|
| CX5 | GTAAAACGACGGCCAG | M13F |
| CX6 | CAGGAAACAGCTATGAC | M13R |
| CX44 | GAAATGTCATCGCCTGTGTGCCAA | FBP1 qRT-PCR |
| CX45 | TGTCAAACTTGATCCTGCGGAGCA | FBP1 qRT-PCR |
| CX374 | ATCGCCTCTGAAAACACTCTC | HOG1 qRT-PCR |
| CX375 | ACTCCAATCAAACACCTCGG | HOG1 qRT-PCR |
| CX411 | CTAGCCCGTTACCTCAATCTC | CCK1 qRT-PCR |
| CX412 | TTTCCCTTGTTCTCCTGGTG | CCK1 qRT-PCR |
| CX420 | GTATCCTAACGCATCTCCCTTG | MPK1 qRT-PCR |
| CX421 | GGAAACTTCTGACCTCTTCGAG | MPK1 qRT-PCR |
| CX49 | TGA GAA GGA CCC TGC CAA CA | GAPDH qRT-PCR |
| CX50 | ACT CCG GCT TGT AGG CAT CAA | GAPDH qRT-PCR |
| CX322 | TCCACCTCCCAAGTCGTCAATG | CCK1 KO left |
| CX323 | CTGGCCGTCGTTTTACTGAGTGTGGGCGATGCTAGTG | CCK1 KO left |
| CX324 | GTCATAGCTGTTTCCTGGCCATCTCTGGTACTGATGCTG | CCK1 KO right |
| CX325 | GCAAAAGAGCTTGCAGGTACAG | CCK1 KO right |
| CX326 | CTTCCAGCTCGAGCAACATCGT | CCK1 KO negative |
| CX327 | ACTCACGTCTTTGGGTCTCTGT | CCK1 KO negative |
| CX328 | CTTCATCTTGCCAAAGGGTCTC | CCK1 KO positive |
| CX336 | GGTGTAGTAGTGCTTCGTTGTG | CCK1 complementation |
| CX337 | CTGGCCGTCGTTTTAC CAACCGTAACTTCCACTCATGC | CCK1 complementation |
| CX426 | GGGGATCCGTCGACCTGCAGCCATGGCTACCACACACGTCATC | Yeast expression |
| CX427 | CGGAATTAGCTTGGCTGCAGCTATCGGAAGCAGCCGCAAG | Yeast expression |
| MPK1-A | GATCATGGCGATAGGGATAC | mpk1 mutant screen |
| MPK1-B | CTCTCATCAAGTCATCCTCC | mpk1 mutant screen |
| CNACT7 | TCCTCTCCTCCGACAACC | mpk1 mutant screen |
| JH8994 | TGTGGATGCTGGCGGAGGATA | Mutant screen |
KO, knockout.
To generate complemented strains of cck1 mutants, a genomic DNA fragment that contains a 1.5-kb upstream promoter region, the CCK1 ORF, and its 500-bp downstream region was amplified in a PCR using primers CX336 and CX337. This PCR fragment was fused with the nourseothricin resistance (NATr) selective marker gene at its C terminus in an overlap PCR using primers CX336 and M13R. The overlap PCR product was biolistically transformed in MATa cck1 mutant strains. Phenotypic assays were performed to identify transformants in which the cck1 phenotype was complemented.
Assays for virulence factors and cell integrity.
Assays for melanin and capsule production were performed as previously described (21). In brief, melanin production was tested on niger seed agar and incubated at 30°C or 37°C for 2 days, and pigmentation of fungal colonies was assessed and photographed. Capsule production was induced on Dulbecco modified Eagle's (DME) agar medium and incubated at 30°C for 3 days. Capsule size was visualized by India ink staining and observed with an Olympus CX41 microscope.
To assay for stress responses, each strain was incubated overnight at 30°C in YPD medium and subcultured in fresh YPD medium to an optical density at 600 nm (OD600) of ∼0.7. The cells were washed, resuspended, and serially diluted (1:10) in dH2O and spotted (5 μl) on YPD agar plates containing 1.5 M NaCl or 1.0 M KCl for osmotic shock or 2.5 mM H2O2 for oxidative stress. To test cell integrity, cells were also spotted on YPD agar plates containing 0.05% SDS, 0.5% Congo red, or 20 μg/ml calcofluor white (CFW). Plates were incubated at both 30°C and 37°C for 2 days and photographed. Trypan blue staining was performed as previously described (11, 21). The percentages of cells able to take up dye were calculated by counting at least 500 yeast cells for each strain.
Western blot analyses of Hog1 and Mpk1 phosphorylation.
Each strain was incubated overnight at 30°C in YPD medium and subcultured in fresh YPD medium for an additional 2 h. Cultures were then supplemented with 1.0 M NaCl, 0.05% SDS, or 2.5 mM H2O2 and incubated for another 10, 30, 60, or 120 min before they were rapidly frozen in a dry ice-ethanol bath. Protein extraction was performed as previously described (2). Protein concentrations were determined by a protein assay kit (Bio-Rad), and equal amounts of protein were loaded onto a 10% Tris-glycine gel. Separated proteins were transferred to Immuno-Blot PVDF (polyvinylidene difluoride) membrane (Bio-Rad). Membranes with proteins from NaCl- or H2O2-treated cells were incubated with primary mouse p38-MAPK-specific antibody (Cell Signaling Technology) for 2 h and with a secondary anti-mouse IgG horseradish peroxidase (HRP)-conjugated antibody. Membranes with proteins from cells treated by either SDS or H2O2 were incubated with a phospho-p44/42 MAPK rabbit antibody (Cell Signaling Technology) for 2 h and with a secondary anti-rabbit IgG HRP-conjugated antibody. All blots were developed using an ECL Plus Western Blotting Detection System (GE). Subsequently, every blot was stripped and further used for detection of actin, with a rabbit beta-actin monoclonal antibody (Genscript) as a loading control.
Virulence studies and fungal burden in infected organs.
Groups of 10 female A/JCr mice (NCI-Frederick) were intranasally infected with 105 yeast cells of each strain as previously described (5, 39). Over the course of the experiments, animals that appeared moribund or in pain were sacrificed by CO2 inhalation. Survival data from the murine experiments were statistically analyzed between paired groups using the log rank test of the Prism program, version 4.0 (GraphPad Software, San Diego, CA). P values of <0.001 were considered significant.
Infected animals were sacrificed at the endpoint of the experiment according to the University of Medicine and Dentistry of New Jersey (UMDNJ) IACUC-approved animal protocol. For mice infected by the cck1 mutant strain, the experiment was terminated at 75 days postinfection. To compare the fungal burden, lungs and brains from mice infected by H99, the cck1 mutant, or the complemented strain of the cck1 mutant were isolated at the endpoint of the experiment (around 20 days for mice infected by H99 or the complemented strain and 75 days postinfection for mice infected by the cck1 mutant). Infected lungs and brains were isolated and homogenized in 1× phosphate-buffered saline (PBS) using a homogenizer. Resuspensions were diluted, 100 μl of each dilution was spread on YPD medium with ampicillin, and colonies were counted after 3 days of incubation at 30°C.
RESULTS
Cck1 is a bona fide casein kinase I in C. neoformans and regulated by glucose.
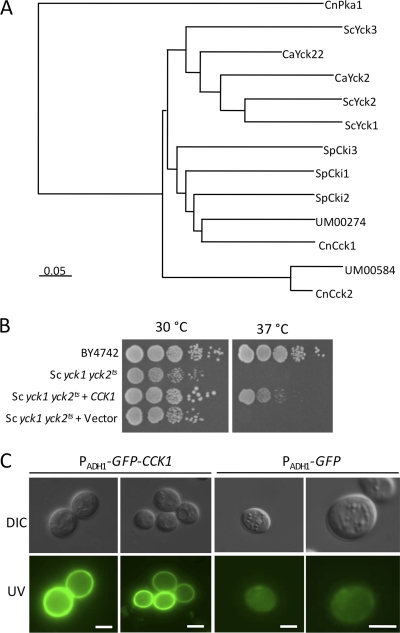
Casein kinase I proteins are conserved among eukaryotes from yeast to human and play an important role in ubiquitin-proteasome system (UPS)-mediated protein degradation by controlling the phosphorylation of substrates of E3 ligases. There are two proteins in the C. neoformans H99 genome database that share sequence similarity with casein kinase I in other organisms. One of them, Cck1 (CNAG_00556.2), shares high sequence similarity with the known casein kinase I proteins in S. cerevisiae, Candida albicans, and S. pombe. Cck2 (CNAG_07427.2) is more distinct from the casein kinase I proteins identified in these yeasts based on the phylogram (Fig. 1A).
Fig. 1.
Cck1 is a casein kinase I protein in C. neoformans. (A) A phylogenetic tree of the casein kinase I proteins in fungi. Full-length protein sequences from C. neoformans (CnCck1, CnCck2, and CnPka1), Ustilago maydis (UM00274 and UM00584), S. cerevisiae (ScYck1, ScYck2, and ScYck3), C. albicans (CaYck2 and CaYck22), and S. pombe (SpCki1, SpCki2, and SpCki3) were used for alignment. The phylogram was generated using ClustalX (version 2.1), viewed by TreeView. CnPka1 was used as an outgroup. (B) Expression of the Cryptococcus CCK1 in a Saccharomyces yck1 yck2ts mutant can partially complement the growth defect of this mutant. Tenfold serial dilutions of overnight cultures were made in dH2O, and 5 μl of each was plated. The plates were grown for 2 days at 30°C and 37°C. (C) The yck1 yck2ts mutant strains overexpressing GFP or a GFP-CCK1 fusion were grown on YPD medium for 24 h, and yeast cells were visualized by Nikon fluorescence microscopy. DIC, differential interference contrast. Scale bar, 5 μm.
In S. cerevisiae, Yck1 and Yck2 are membrane-bound proteins and have redundant functions in regulating cellular signaling. Mutation of both YCK1 and YCK2 was found to be lethal. The yck1 yck2ts mutant strain is conditionally lethal and fails to grow at an elevated temperature of 37°C (34). To examine whether Cck1 in C. neoformans functions as a casein kinase I, we overexpressed a green fluorescent protein (GFP)-Cck1 fusion protein under the control of the ADH1 promoter in an S. cerevisiae yck1 yck2ts mutant strain. Our results showed that Cck1 expression partially rescued the growth defect of the yck1 yck2ts mutant at 37°C, and the GFP-Cck1 protein was localized at the cell membrane, demonstrating that Cck1 is a bona fide casein kinase I protein (Fig. 1B and C).
Because the phosphorylation of substrates of Grr1, the F-box protein Fbp1 homolog in S. cerevisiae, requires casein kinase I proteins Yck1 and Yck2, we hypothesized that Cck1 may function in the same pathway to control the phosphorylation of Fbp1 substrates in C. neoformans. Our previous study has demonstrated that the expression of FBP1 was negatively regulated by glucose (21). Therefore, we checked the expression of Cck1 under glucose induction or repression conditions using qRT-PCR. Our results showed that the expression of CCK1 was also induced by glucose starvation but at a level much higher than that of FBP1 (Fig. 2A).
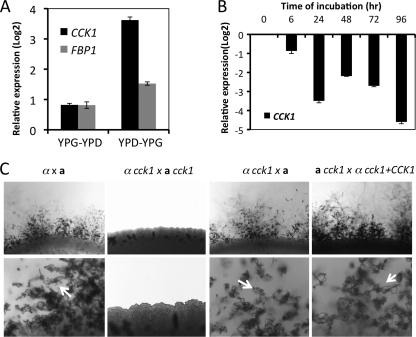
Fig. 2.
Expression of CCK1 and its role in fungal mating. (A) Expression of CCK1 under different glucose conditions. H99 cells were grown on YPG medium (YP with 2% galactose) or YPD (2% glucose) liquid medium overnight and were switched to YPD or YPG medium as described in Materials and Methods. Cells were collected, and gene expression was assayed by qRT-PCR as described in Materials and Methods. Values are expressed as relative expression (log 2) of the CCK1 gene or the FBP1 gene, normalized to the GAPDH gene endogenous reference. The error bars show standard deviations of three repeats. (B) Expression of CCK1 during mating on V8 medium. Mating cultures of H99 × KN99a were isolated from the plate surface after incubation for 6, 24, 48, 72, and 96 h. CCK1 expression was detected by qRT-PCR analysis relative to the zero-hour time point (H99 overnight liquid culture on YPD medium was considered time zero). (C) Mating assays for wild-type strains, cck1 mutants, and cck1 complemented strains were performed on MS medium. Mating structures at magnifications of ×40 (upper panel) and ×200 (lower panel) were photographed after 2 weeks of incubation in the dark at 25°C. Arrows indicate spore chains.
Cck1 regulates fungal mating.
To monitor the expression of CCK1 during the mating cycle, RNAs from mating mixtures between wild-type strains (α × a) were prepared at 0, 6, 24, 48, and 96 h postincubation. Our qRT-PCR results showed that the expression of CCK1 was negatively regulated by mating (Fig. 2B), similar to what we observed for FBP1 gene expression (21). The downregulation of expression at 24 h and 96 h postinoculation was significantly greater than at other time points.
To study the biological function of casein kinases in C. neoformans, CCK1 gene deletion mutants were generated in both α and a mating type backgrounds. We have tested the role of Cck1 in Cryptococcus sexual reproduction. Interestingly, the bilateral mating of cck1 mutants (cck1 × cck1) was sterile even though the cck1 unilateral mating (cck1 × wt) developed normal mating hyphae and still produced spores (Fig. 2C). These observations indicate that Cck1 may be required for the early development of the mating cycle. How Cck1 regulates mating remains unclear. Our previous study demonstrated that Fbp1 is dispensable for cell fusion and dikaryotic mating filament production but essential for sporulation due to its role in meiotic division (21). The different effects on fungal mating between Cck1 and Fbp1 suggest that Cck1 may regulate mating independent of the SCFFbp1 E3 ligase.
Cck1 is required for fungal virulence.
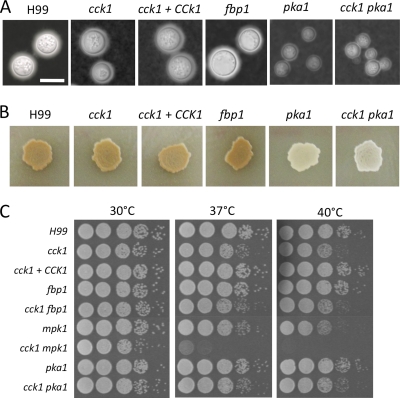
Fbp1 is essential for fungal virulence even though it is not required for the development of classical fungal virulence factors, including the production of melanin and capsule, as well as growth at 37°C. We have also examined the development of these virulence factors in cck1 mutants. Our results demonstrated that cck1 mutants produced normal melanin on niger seed agar and produced regular capsules on DME medium, indicating that Cck1 is also not important for the development of these virulence factors in vitro (Fig. 3). Interestingly, cck1 mutants did show a slower growth than the H99 strain when incubated at 37°C or 40°C, indicating a reduced fitness under thermal stress. The cyclic AMP (cAMP)-protein kinase A (PKA) pathway in C. neoformans plays a central role in the development of these virulence factors and fungal virulence (1, 16). Although cck1 mutants did not show an obvious defect in the development of melanin or capsule, we generated cck1 pka1 double mutants to examine a potential connection between Cck1 and cAMP-PKA signaling. Our results showed that the double mutant has significant defects in both melanin and capsule production, similar to the defects of the pka1 single mutant, confirming that Cck1 does not play a role in the cAMP pathway regulation.
Fig. 3.
Cck1 is not required for virulence factor development. (A) Cultures were grown overnight in YPD medium and diluted to an OD600 of 2.0. Tenfold serial dilutions were made in dH2O, and 5 μl of each was plated. The plates were grown for 2 days at 30°C, 37°C, and 40°C. (B) Capsule formation was assayed at 37°C on DME medium. Capsule production was visualized by India ink staining after cells were grown on DME medium for 3 days. Scale bar, 10 μm. (C) Melanin production was assayed on niger seed medium. Melanin levels produced by the strains were observed in photographs after incubation for 3 days at 37°C.
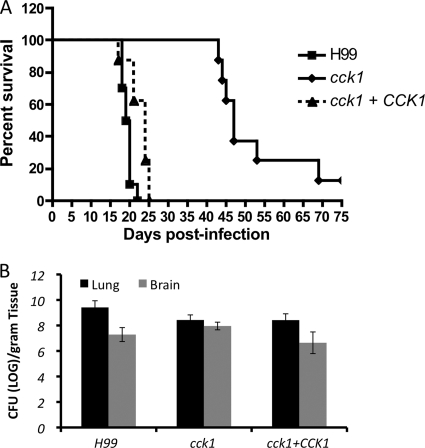
We then evaluated the potential involvement of Cck1 in fungal virulence by testing the cck1 mutant in a murine intranasal inhalation systemic infection model. Each mouse was inoculated with 1 × 105 yeast cells, and the survival of infected mice was analyzed. Our results showed that mice infected with the cck1 mutant had a significantly prolonged survival rate. Consistent with previous reports (21, 39), mice infected by H99 or the CCK1 complemented strain died around 20 days postinoculation while mice infected by the cck1 mutant did not show clear disease symptoms with a significant weight loss until 43 days. Seventy percent of the infected animals died between 45 days and 50 days, and the rest of the infected group survived for more than 60 days postinoculation (Fig. 4A). These results demonstrated that Cck1 is required for fungal virulence.
Fig. 4.
The cck1 mutant showed significant virulence attenuation in a murine infection model. (A) Survival curves of infected mice in a Cryptococcus murine inhalation model. Female A/JCr mice were inoculated intranasally with the following strains: H99, a cck1 mutant (CUX106), and its complemented strain (CUX111). Groups of 10 mice were infected with 1 × 105 yeast cells for each strain. (B) Lungs and brains from three mice infected with H99, the cck1 mutant, and the complemented strain were isolated at the end time point of infection. The numbers of yeast CFU per gram of fresh infected organ were measured in lung and brain homogenates. Each data point and error bar indicate the mean and standard deviation of the mean for values from three animals.
At the endpoint of the infection experiment, lungs and brains in mice infected by H99 or the cck1 mutant were recovered and homogenized. Fungal burdens from recovered organs were calculated by measuring the number of yeast CFU. The numbers of CFU per gram of fresh tissue infected by the H99 strain and the cck1 mutant were similar in both lungs and brains, suggesting that the mutant strain can still survive and proliferate in vivo, but the virulence was significantly attenuated (Fig. 4B).
Cck1 is involved in cell wall integrity by regulating the expression and phosphorylation of Mpk1.
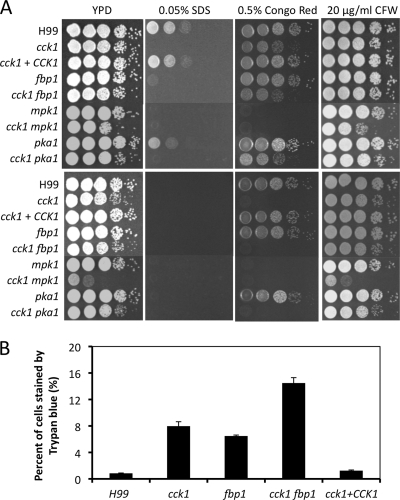
To understand why the cck1 mutant showed significant virulence attenuation, we evaluated other in vitro phenotypes of cck1 in addition to the virulence factors. Fbp1 is required for cell membrane integrity, which could be one factor that contributes to the avirulence of fbp1 mutants (21). To investigate whether Cck1 also plays a role in fungal cell wall integrity, we tested the growth of cck1 mutants on medium containing SDS, calcofluor white (CFW), or Congo red that are known to disrupt cell wall integrity (8, 40). Our results showed that cck1 mutants were also hypersensitive to 0.05% SDS at a level more severe than that of fbp1 mutants but grew normally on the medium containing 20 μg/ml CFW. In addition, cck1 mutants also showed a growth defect on medium with 0.5% Congo red, an indication that Cck1 is also important for maintaining cell wall integrity (Fig. 5). Fbp1 is not required for cell wall integrity since fbp1 mutants have no growth defect on medium with either CFW or Congo red. These results suggest that Cck1 has broader effects on cell wall integrity than Fbp1.
Fig. 5.
Cck1 is essential for cell integrity. (A) Tenfold serial dilutions of overnight cultures were made in dH2O, and 5 μl of each was plated. The plates were grown for 2 days at 30°C (top panel) and 37°C (bottom panel). H99 and mutant strains are indicated on the left, and the conditions are shown at the top. (B) Trypan blue staining to examine cell membrane integrity. Strains were inoculated in YPD medium for 48 h. Yeast cells were mixed with 0.4% trypan blue solution for 1 h before being visualized by microscope. The percentages of heavily stained cells from a total of over 500 yeast cells were calculated. The error bars indicate standard deviations.
Another indicator of cell membrane integrity is the level of trypan blue staining in yeast cells. Trypan blue stains only cells with membrane defects or dead cells. Wild-type H99 and cck1 mutants were stained with trypan blue after growth at 30°C for 48 h. Our results showed that while less than 2% of H99 cells were heavily stained, over 8% of cck1 mutant cells and over 6% of fbp1 mutant cells were stained under the same conditions (Fig. 5B). These results further indicate that both Cck1 and Fbp1 are important for maintaining cell membrane integrity.
To further dissect the role of Cck1 in cell wall integrity, we generated cck1 fbp1 double mutants. Epistasis analysis showed that the double mutant was sensitive to SDS and Congo red at a level similar to that of the cck1 single mutant while the double mutant showed more trypan blue-stained cells (13%), suggesting that Cck1 and Fbp1 have overlapping and distinct roles in the regulation of cell wall integrity (Fig. 5).
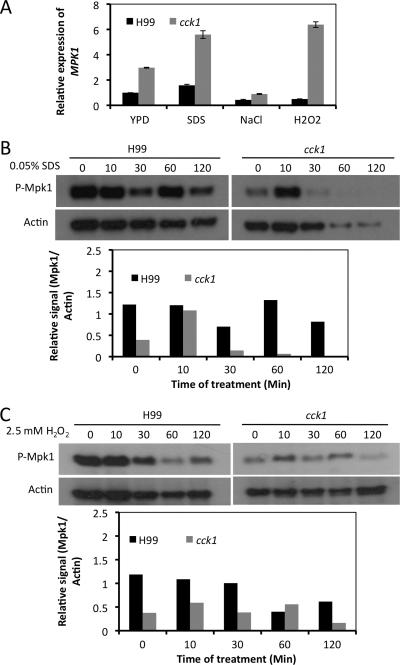
The Mpk1 MAP kinase pathway is the primary signaling pathway that regulates cell wall integrity in C. neoformans (11, 20). To understand whether Cck1 regulates cell wall integrity via the Mpk1 pathway, we measured the expression of MPK1 using qRT-PCR in both the wild-type strain and the cck1 mutant. When both strains were treated with 0.05% SDS for 60 min, MPK1 expression showed 3-fold increases in the cck1 mutant background, indicating that Cck1 negatively regulates MPK1 expression under the testing condition. We used samples treated with NaCl as a control since Mpk1 does not play a role in cell adaptation to osmotic shock. Our results showed that treatment of cells with NaCl did not cause the alteration of MPK1 expression, suggesting that the increased signals we observed were, indeed, due to SDS treatment (Fig. 6A).
Fig. 6.
Detection of the MPK1 gene expression and Mpk1 phosphorylation. Expression of MPK1 was measured by qRT-PCR. Fresh cultures of H99 and the cck1 mutant were supplemented with 1.0 M NaCl, 0.05% SDS, or 2.5 mM H2O2 and incubated for 60 min. Expression levels of MPK1 and GAPDH were analyzed by qRT-PCR as described in Materials and Methods. (B and C) Phosphorylation of Mpk1 under different conditions in a Western blot. Similar overnight cultures were prepared and induced with 0.05% SDS or 2.5 mM H2O2 for 10, 30, 60, and 120 min. The noninduced control for each was harvested before the addition of chemicals. The phosphorylation status of Mpk1 was monitored using a rabbit Mpk1 phosphorylation antibody in a Western blot. The same blots were restripped and probed with mouse beta-actin antibody as the protein loading control. The signal of each blot was quantified using the ImageJ program. The relative protein phosphorylation levels were normalized to the expression of actin protein, as shown in the graphs.
We also examined the phosphorylation of Mpk1 under the same conditions by Western blot analysis and found that Mpk1 phosphorylation is regulated by Cck1. In the wild-type background, the signal of Mpk1 phosphorylation was strong and persistent within a range of 120 min of the SDS treatment. On the other hand, in a cck1 mutant background, only a weak signal could be detected following the same treatment except for the sample that was treated with SDS for 10 min. A significantly increased signal was observed at that time point (Fig. 6B). These observations indicate that Cck1 regulates cell wall integrity by affecting both the expression and phosphorylation of Mpk1.
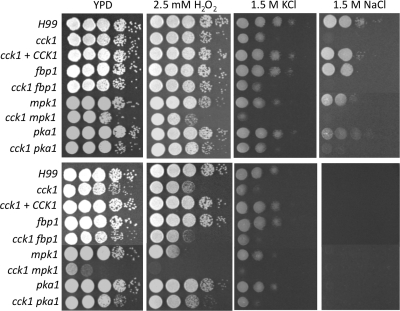
It has been reported that Mpk1 is required for Cryptococcus to respond to oxidative stress (10). We have also tested the potential role of Cck1 in the Mpk1-dependent cell response to oxidative stress by treating both wild-type and cck1 mutant cells with 2.5 mM H2O2. The cck1 mutant is more sensitive to the oxidative stress when incubated at 37°C (Fig. 7). By using qRT-PCR, significant induction of the MPK1 expression was observed compared to H99 under the same condition. The phosphorylation of Mpk1, however, was somewhat reduced in the cck1 mutant in our study (Fig. 6). Our results demonstrated that the sensitivity of cck1 mutants to oxidative stress is also dependent on Mpk1 signaling.
Fig. 7.
Cck1 is required for the osmotic stress response. Tenfold serial dilutions of overnight cultures were made in dH2O, and 5 μl of each was plated. The plates containing 2.5 mM H2O2 and 1.5 M KCl were grown for 2 days at 30°C (top panel) and at 37°C (bottom panel). The plates containing 1.5 M NaCl were grown for 5 days. H99 and mutant strains are indicated on the left, and the conditions are shown at the top.
We have also generated the cck1 mpk1 double mutants with genetic crosses between cck1 and mpk1 mutants. Epistasis analysis showed that the double mutant was sensitive to SDS and Congo red at levels similar to those of the cck1 or mpk1 single mutants but that there was increased sensitivity to the elevated temperature and treatment with CFW (Fig. 5). These results suggest that Cck1 functions upstream of Mpk1 and is partially involved in Mpk1 regulation. As we predicted, the pka1 mutant did not show a cell integrity defect while the cck1 pka1 double mutants showed the same sensitivity as that of the cck1 single mutant, thus excluding a potential functional interaction between Cck1 and the cAMP pathway (Fig. 5).
Cck1 regulates Hog1 phosphorylation but is not involved in HOG1 expression.
Growth of cck1 mutants was also tested on medium with a high salt concentration (1.5 M NaCl or 1.5 M KCl) at both 30°C and 37°C. The cck1 mutants were hypersensitive to 1.5 M KCl and could not grow on YPD medium with 1.5 M NaCl. These results indicate that Cck1 is required for cell adaptation to osmotic shock. In addition, the cck1 mutant was also found to be sensitive to H2O2 treatment at 37°C, an indication of its sensitivity to oxidative stress (Fig. 7). The hypersensitivity of the cck1 mutants to these stress conditions may also contribute to their hypovirulent phenotype in the murine model.
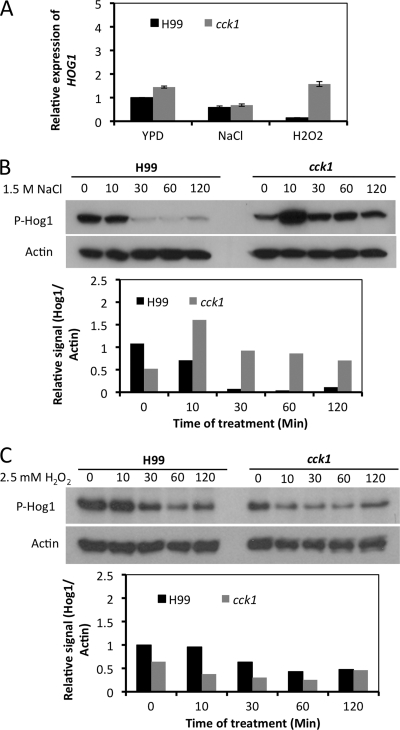
The Hog1 MAP kinase cascade is regulated by a histidine kinase two-component system and functions as a major regulator of the cellular response to high-osmotic stress and oxidative stress conditions (2). Because cck1 mutants showed hypersensitivity to NaCl and H2O2, we investigated whether Cck1 plays a role in the Hog1 MAPK signaling pathway by examining the expression and phosphorylation of Hog1 MAP kinase. qRT-PCR was performed to measure the expression of the HOG1 gene after cells were treated with 1 M NaCl or 2.5 mM H2O2 for 60 min. Our results showed that the HOG1 gene had similar expression levels, with changes less than 2-fold in either the wild-type or cck1 mutant background under all conditions tested, indicating that Cck1 does not play a role in regulating HOG1 expression (Fig. 8A).
Fig. 8.
The role of Cck1 in HOG1 gene expression and Hog1 phosphorylation. (A) Expression of HOG1 under osmotic stress and oxidative stress conditions was measured by qRT-PCR. Each fresh culture was supplemented with 1.0 M NaCl or 2.5 mM H2O2 and incubated for 60 min. Expression levels of HOG1 and GAPDH were analyzed by qRT-PCR as described in Materials and Methods. (B and C) Phosphorylation of Hog1 under different conditions in Western blot analysis. Similar overnight cultures were prepared and then induced with 1.0 M NaCl (B) or 2.5 mM H2O2 (C) for 0, 10, 30, 60, and 120 min. The dual phosphorylation status of Hog1 was monitored using a dual-phosphorylated p38 antibody (P-Hog1). The same blots were stripped and then probed with the mouse beta-actin antibody as the loading control. The relative signal of each blot was quantified using the ImageJ program. The relative protein phosphorylation levels were normalized to the expression of actin protein as shown in the graphs.
We also examined the phosphorylation of Hog1 protein by using the p38 MAPK phosphorylation antibody. In C. neoformans, Hog1 phosphorylation is repressed by treatment with NaCl (2). Our results showed that Hog1 is rapidly dephosphorylated in the wild-type strain background, consistent with previous reports (2, 3). Interestingly, the phosphorylation of Hog1 was stabilized in the cck1 mutant background when cells were treated with NaCl, even after treatment with NaCl for 120 min, indicating that Cck1 is required for Hog1 dephosphorylation in response to osmotic stress (Fig. 8B). On the other hand, no significant alteration of Hog1 phosphorylation was observed when cells were treated with 2.5 mM H2O2 (Fig. 8C), suggesting a less clear connection between Cck1 and Hog1 in the oxidative stress response.
DISCUSSION
Because Fbp1 is essential for fungal virulence, we are interested in understanding the rest of the pathway associated with the Fbp1 function as part of the UPS machinery. The homolog of Fbp1 in S. cerevisiae, Grr1, has been extensively studied for its role in the sensing of nutrients, such as glucose and amino acids (17). However, identification and characterization of Fbp1 substrates that are important for fungal virulence remain elusive. In general, phosphorylation is a necessary posttranslational modification that targets proteins for ubiquitination by the SCF E3 ligase of the UPS machinery. Casein kinase proteins are well conserved in eukaryotic organisms and play important roles in the phosphorylation of proteins, including those targeted by the ubiquitin-proteasome system for degradation. Because most Grr1 substrates reported are phosphorylated by the casein kinase I proteins Yck1 and Yck2 in S. cerevisiae and because the C. neoformans Fbp1 as a homolog of Grr1 also plays a role in glucose sensing, we hypothesize that casein kinase I proteins in C. neoformans play an important role in the phosphorylation of Fbp1 substrates, thus exhibiting shared function with Fbp1.
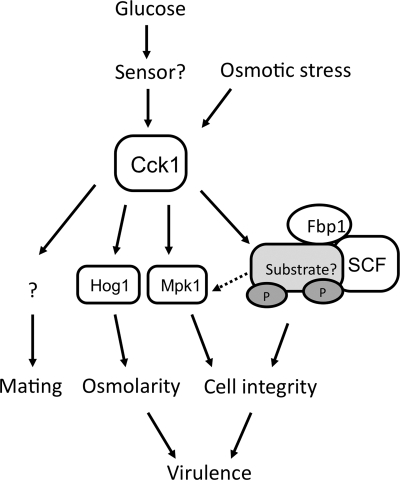
The main findings of this study were the identification of Cck1 as a casein kinase I protein in C. neoformans and of its role in regulating multiple downstream pathways to control fungal sexual reproduction, cell integrity, and fungal virulence. Our results also indicate that Cck1 shares overlapping functions with Fbp1. CCK1 and FBP1 showed similar expression patterns under several different conditions, and both of them were found to be important for cell integrity and fungal virulence. Because either dysfunction of the F-box protein Fbp1 or a defect in the substrate phosphorylation would result in the accumulation of Fbp1 substrates, our results suggest that Cck1 is involved in the Fbp1-mediated UPS and required for the phosphorylation of Fbp1 substrates that are important for cell integrity and fungal pathogenicity (Fig. 9).
Fig. 9.
The proposed model for Cck1 function. In our model, Cck1 regulates multiple downstream pathways and is essential for fungal virulence. Cck1 is regulated by glucose. It may participate in the phosphorylation of Fbp1 substrates to regulate cell integrity and fungal virulence. In addition, Cck1 may also play a more direct role in regulating Mpk1 signaling, or it regulates Mpk1 function via its target protein(s) in the Fbp1 pathway. Meanwhile, Cck1 also responds to stress signals and plays a role in the HOG pathway by regulating the phosphorylation of Hog1 protein and thus controls the cell adaptation to osmotic shock. Cck1 also regulates fungal mating, but its potential target(s) remains to be identified.
Although mice infected by the cck1 mutant showed significant virulence attenuation, the mutant strain still caused lethal infection. In contrast, the fbp1 mutant showed an avirulent phenotype (21). This difference can be explained by the possibility that additional casein kinase I proteins that may be able to partially compensate the loss of Cck1 exist in C. neoformans. The result that expression of CCK1 can only partially rescue the growth defect of the yck1 yck2ts mutant at 37°C also suggests that additional protein could be required for full complementation (Fig. 1). We have identified a Cck1 homolog in the H99 genome (Cck2), but its function remains to be understood. It is also possible that there are multiple substrates playing a collective role in fungal virulence and that Cck1 phosphorylates only some of them. Additional kinase proteins may be responsible for the phosphorylation of the other Fbp1 substrates important for fungal virulence. Either of these scenarios could lead to the more severe virulence attenuation in fbp1 mutants than in the cck1 mutant, as we observed. Comparison of the protein profiles in the fbp1 mutant and the cck1 mutant backgrounds under the same assay conditions should provide useful information to address these possibilities in the future.
How Fbp1 and Cck1 regulate fungal virulence remains to be understood. Our results demonstrated that Fbp1 and Cck1 are both important for cell integrity, suggesting that the cell integrity defect could be a potential cause of virulence attenuation. The Mpk1 MAPK pathway is known to regulate cell integrity in fungi and is activated by protein kinase C (PKC) signaling in C. neoformans (10, 11, 20). Our study connects Cck1 function with the Mpk1 pathway, suggesting that the Mpk1 MAPK cascade may have multiple upstream activators that regulate cell integrity. Because Pkc1 functions upstream of Mpk1 and because deletion of the PKC1 gene abolishes Mpk1 phosphorylation under several different stress conditions, as shown by a previous study (10), it is possible that Cck1 may regulate Pkc1 function to alter Mpk1 phosphorylation when cells are treated with SDS. Cck1 may also regulate Mpk1 function via its target protein in the Fbp1 pathway. Nevertheless, we expect that some proteins involved in cell membrane integrity could be substrates of Fbp1 and phosphorylated by Cck1 (Fig. 9). Searching for mutants that are sensitive or resistant to SDS treatment in a high-throughput setting could potentially provide useful information on the identification of potential substrates of Fbp1.
It is interesting that Cck1 also regulates the HOG pathway to influence cell responses to osmotic stress. Because Fbp1 does not play a role in cell adaptation to osmotic shock, the involvement of Cck1 in Hog1 phosphorylation indicates that Cck1 has additional functions beyond phosphorylating Fbp1 substrates. In addition to involvement in the osmotic stress response, Hog1 also negatively regulates the capsule size by inhibiting the cAMP-PKA pathway in C. neoformans var. grubii (2), but we found that Cck1 does not play a role in capsule formation in our study. Because a two-component system is the major upstream regulator of Hog1 expression and phosphorylation and because the Cryptococcus two-component system is composed of seven cell membrane-associated histidine kinases (Tco1 to Tco7) that sense extracellular signals (3, 19), it is possible that Cck1 may be partially or conditionally involved in HOG pathway regulation by controlling the turnover of some Tco proteins.
Both the Mpk1 MAPK pathway and the Hog1 MAPK pathway have been reported to participate in the oxidative stress response (2, 10). Therefore, we also examined the potential role of Cck1 in oxidative stress. Our studies showed that cck1 mutants were indeed more sensitive to treatment with H2O2 but only at the elevated temperature of 37°C. The phosphorylation of neither Mpk1 nor Hog1 was significantly altered in the cck1 mutant strain following treatment with 2.5 mM H2O2, suggesting a potential minor role of Cck1 in the oxidative stress.
The requirement for Cck1 in fungal sexual reproduction also indicates that Cck1 has other downstream targets independent of Fbp1 function since Fbp1 regulates only fungal sporulation (21). In S. cerevisiae, Yck1 and Yck2 are required for the turnover of the pheromone receptor Ste2 (14). Yck1 has also been reported to phosphorylate Ste50 (41). Because both Ste2 and Ste50 are involved in the mating pathway (31), this connection suggests that Yck1 may also play a role in mating regulation. It is possible that Cck1 phosphorylates pheromone receptors in C. neoformans in an Fbp1-independent E3 ligase pathway. It will be important to understand how Cck1 regulates mating of C. neoformans, a project for future study.
In summary, we identified a casein kinase I protein, Cck1, in C. neoformans that regulates a range of cellular functions via multiple downstream pathways, including the Fbp1-mediated UPS machinery. The consensus phenotypes observed in both fbp1 and cck1 mutants provide valuable information in our search for the substrates of Fbp1 that are important for fungal virulence.
ACKNOWLEDGMENTS
We thank Issar Smith and Carol Newlon for critical reading of the manuscript and valuable comments for the study. We thank Jenny Lodge for providing C. neoformans mpk1 and pkc1 mutant strains and Mark Johnston for S. cerevisiae strains including the yck1 yck2ts mutant for the study. We thank Neeraj Chauhan for providing the p38-MAPK phosphorylation and the p44/42 phosphorylation antibodies for this study. We also acknowledge use of the C. neoformans genome sequences at the Broad Institute.
This study was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions and the National Natural Science Foundation of China (number 30870107) grants to L.J. and by the UMDNJ institutional startup fund to C.X.
Footnotes
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Alspaugh J. A., Perfect J. R., Heitman J. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bahn Y. S., Kojima K., Cox G. M., Heitman J. 2005. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16:2285–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bahn Y. S., Kojima K., Cox G. M., Heitman J. 2006. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17:3122–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheong J. K., Virshup D. M. 2011. Casein kinase 1: complexity in the family. Int. J. Biochem. Cell Biol. 43:465–469 [DOI] [PubMed] [Google Scholar]
- 5. Cox G. M., Mukherjee J., Cole G. T., Casadevall A., Perfect J. R. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davidson R. C., et al. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607–2615 [DOI] [PubMed] [Google Scholar]
- 7. Davidson R. C., et al. 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29:38–48 [DOI] [PubMed] [Google Scholar]
- 8. Elorza M. V., Rico H., Sentandreu R. 1983. Calcofluor white alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J. Gen. Microbiol. 129:1577–1582 [DOI] [PubMed] [Google Scholar]
- 9. Fraser J. A., Subaran R. L., Nichols C. B., Heitman J. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2:1036–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gerik K. J., Bhimireddy S. R., Ryerse J. S., Specht C. A., Lodge J. K. 2008. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 7:1685–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerik K. J., et al. 2005. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58:393–408 [DOI] [PubMed] [Google Scholar]
- 12. Granger D. L., Perfect J. R., Durack D. T. 1985. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J. Clin. Invest. 76:508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He Q., Cha J., Lee H. C., Yang Y., Liu Y. 2006. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 20:2552–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hicke L., Zanolari B., Riezman H. 1998. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 141:349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hicks J. K., Bahn Y. S., Heitman J. 2005. Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans. Eukaryot. Cell 4:1971–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Idnurm A., et al. 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3:753–764 [DOI] [PubMed] [Google Scholar]
- 17. Jonkers W., Rep M. 2009. Lessons from fungal F-box proteins. Eukaryot. Cell 8:677–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knippschild U., et al. 2005. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell. Signal. 17:675–689 [DOI] [PubMed] [Google Scholar]
- 19. Kojima K., Bahn Y. S., Heitman J. 2006. Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology 152:591–604 [DOI] [PubMed] [Google Scholar]
- 20. Kraus P. R., Fox D. S., Cox G. M., Heitman J. 2003. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 48:1377–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu T. B., et al. 2011. The F-Box protein Fbp1 regulates sexual reproduction and virulence in Cryptococcus neoformans. Eukaryot. Cell 10:791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Z., Thornton J., Spirek M., Butow R. A. 2008. Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol. Cell. Biol. 28:551–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 24. Marchal C., Haguenauer-Tsapis R., Urban-Grimal D. 1998. A PEST-like sequence mediates phosphorylation and efficient ubiquitination of yeast uracil permease. Mol. Cell. Biol. 18:314–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marchal C., Haguenauer-Tsapis R., Urban-Grimal D. 2000. Casein kinase I-dependent phosphorylation within a PEST sequence and ubiquitination at nearby lysines signal endocytosis of yeast uracil permease. J. Biol. Chem. 275:23608–23614 [DOI] [PubMed] [Google Scholar]
- 26. Mitsiades C. S., Mitsiades N., Hideshima T., Richardson P. G., Anderson K. C. 2005. Proteasome inhibitors as therapeutics. Essays Biochem. 41:205–218 [DOI] [PubMed] [Google Scholar]
- 27. Moriya H., Johnston M. 2004. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. U. S. A. 101:1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nielsen K., et al. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park H., et al. 2009. Transcriptional responses of Candida albicans to epithelial and endothelial cells. Eukaryot. Cell 8:1498–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perfect J. R., Schell W. A., Rinaldi M. G. 1993. Uncommon invasive fungal pathogens in the acquired immunodeficiency syndrome. J. Med. Vet. Mycol. 31:175–179 [DOI] [PubMed] [Google Scholar]
- 31. Posas F., Witten E. A., Saito H. 1998. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 18:5788–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Price M. A. 2006. CKI, there's more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 20:399–410 [DOI] [PubMed] [Google Scholar]
- 33. Robinson L. C., et al. 1999. The Yck2 yeast casein kinase 1 isoform shows cell cycle-specific localization to sites of polarized growth and is required for proper septin organization. Mol. Biol. Cell 10:1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robinson L. C., Menold M. M., Garrett S., Culbertson M. R. 1993. Casein kinase I-like protein kinases encoded by YCK1 and YCK2 are required for yeast morphogenesis. Mol. Cell. Biol. 13:2870–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sakamoto K. M. 2002. Ubiquitin-dependent proteolysis: its role in human diseases and the design of therapeutic strategies. Mol. Genet. Metab. 77:44–56 [DOI] [PubMed] [Google Scholar]
- 36. Spielewoy N., Flick K., Kalashnikova T. I., Walker J. R., Wittenberg C. 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 24:8994–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vancurova I., Choi J. H., Lin H., Kuret J., Vancura A. 1999. Regulation of phosphatidylinositol 4-phosphate 5-kinase from Schizosaccharomyces pombe by casein kinase I. J. Biol. Chem. 274:1147–1155 [DOI] [PubMed] [Google Scholar]
- 38. Wang P., Heitman J. 1999. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 2:358–362 [DOI] [PubMed] [Google Scholar]
- 39. Wang Y., et al. 2011. Two major inositol transporters and their role in cryptococcal virulence. Eukaryot. Cell 10:618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wood P. J., Fulcher R. G. 1983. Dye interactions. A basis for specific detection and histochemistry of polysaccharides. J. Histochem. Cytochem. 31:823–826 [DOI] [PubMed] [Google Scholar]
- 41. Wu C., et al. 2003. Phosphorylation of the MAPKKK regulator Ste50p in Saccharomyces cerevisiae: a casein kinase I phosphorylation site is required for proper mating function. Eukaryot. Cell 2:949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xue C., et al. 2010. Role of an expanded inositol transporter repertoire in Cryptococcus neoformans sexual reproduction and virulence. mBio 1(1):e00084–10 doi:10.1128/mBio.00084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xue C., Tada Y., Dong X., Heitman J. 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1:263–273 [DOI] [PubMed] [Google Scholar]