Abstract
Saccharomyces cerevisiae generates complex biofilms called mats on low-density (0.3%) agar plates. The mats can be morphologically divided into two regions: (i) hub, the interior region characterized by the presence of wrinkles and channels, and (ii) rim, the smooth periphery. Formation of mats depends on the adhesin Flo11p, which is also required for invasive growth, a phenotype in which the S. cerevisiae yeasts grow as chains of cells that dig into standard-density (2%) agar plates. Although both invasive growth and mat formation depend on Flo11p, mutations that perturb the multivesicular body (MVB) protein sorting pathway inhibit mat formation in a FLO11-independent manner. These mutants, represented by vps27Δ, disrupt mat formation but do not affect invasive growth, FLO11 gene or protein expression, or Flo11p localization. In contrast, an overlapping subset of MVB mutants (represented by ESCRT [endosomal sorting complex required for transport] complex genes such as VPS25) interrupt the Rim101p signal transduction cascade, which is required for FLO11 expression, and thus block both invasive growth and mat formation. In addition, this report shows that mature Flo11p is covalently associated with the cell wall and shed into the extracellular matrix of the growing mat.
INTRODUCTION
Microbes exhibit multicellular behaviors such as swarming and the formation of colonies, fruiting bodies, and biofilms (1, 14, 34–36). All of these behaviors depend on cells interacting with one another and the local environment. The baker's yeast Saccharomyces cerevisiae is able to grow in a number of different multicellular forms, including pseudohyphae, floating biofilms on sherry wine, and biofilms on the surface of low-density agar plates (referred to herein as mats) (9, 30, 42). All of these growth forms are dependent on the presence of a glycosylphosphatidylinositol (GPI)-anchored, cell surface adhesion protein called Flo11p, which is similar to fungal adhesins found in a number of different yeasts, including several pathogens (38, 39).
This communication is focused on the Flo11p-dependent multicellular phenotypes of invasive growth and mat formation. During invasive growth, yeasts grow as chains of cells that invade into the relatively dry surface of 2% agar plates made with yeast extract-peptone-dextrose (YPD) medium (8). During mat formation, yeasts grow as biofilms that spread over the wet surface of 0.3% agar YPD plates. As the mats mature, they generate two morphologically distinct regions. The central region of the mat is called the hub and consists of aggregates of cells that adhere to the agar surface and one another and form channels and wrinkles that are hallmarks of biofilms. The outer region of the mat is called the rim, and it is smooth in appearance and consists of a dividing, spreading population of cells that are not particularly adherent to one another or the agar surface (30, 31).
The regulation of Flo11p and its impact on the yeast multicellular behaviors of invasive growth (which occurs in haploid yeast cells) and pseudohyphal growth (a related phenotype that occurs in diploid yeast cells) have been the subjects of numerous studies, many of which have been reviewed previously (8, 38). Several of these mutations that perturb FLO11 expression and affect invasive growth, such as mutations in glucose-sensing pathways and transcription factors that regulate inositol biosynthesis, also disrupt mat formation (29, 31).
In contrast, there are examples in the literature of mutations that cause defects in invasive growth but not mat formation, and vice versa. The ste12Δ mutation has only a very minor effect on mat formation but quite a strong effect on invasive growth (30). Conversely, a number of Hsp70-encoding genes, such as SSA1 and SSA2, have strong defects in mat formation but not invasive growth. These Hsp70 mutants also do not appear to affect Flo11 protein expression (22).
In this communication, it is examined whether the Rim101p signal transduction cascade, which is known to control invasive growth and FLO11 expression (3, 19), also regulates mat formation. The Rim101p signaling pathway is required for cells to respond to neutral or basic pH (6, 24) and is necessary for invasive growth. A model for the Rim101p pathway is that a plasma membrane receptor called Dfg16p detects extracellular signals, such as neutral pH, and is recruited to the endosome by the β-arrestin-like protein Rim8p (5, 12, 18, 41). This event recruits several of the ESCRT (endosomal sorting complex required for transport) complexes (I, II, and III), which are also required for proper protein sorting in the endosome. Snf7p of the ESCRT-III complex recruits the Rim13p protease via the Rim20p scaffolding protein, and Rim13p cleaves off the Rim101p C-terminal inhibitory domain to activate it.
The ESCRT complex subunits involved in Rim101p processing (5, 41) are part of a subset of vacuolar protein sorting (vps) components called class E vps proteins. The original vps mutants were grouped into 6 classes (A through F) on the basis of distinct vacuolar morphology defects (28). About 13 vps mutants belong to class E and are characterized by the formation of an aberrant prevacuolar compartment within the endosome, referred to as the class E compartment (28). The class E vps mutants perturb the ubiquitin-dependent sorting of proteins through the endosome to the vacuole by a pathway referred to as the multivesicular body (MVB) pathway (13, 25). However, only the ESCRT proteins affect Rim101p signaling (5, 41).
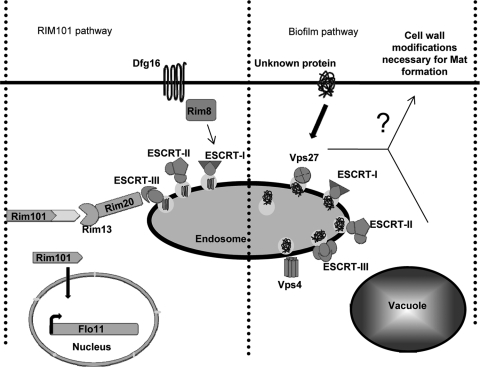
The steps of the MVB cascade involve (i) identification of the ubiquitinylated cargo by Vps27p and Hse1p; (ii) deformation of the endosomal membrane by the ESCRT-I complex (Vps37p, Vps28p, Vps23p) to allow subsequent steps of cargo intake; (iii) formation of invaginations by the ESCRT-II complex (Vps22p, Vps25p, Vps32p), leading to cargo protein engulfment; and finally, (iv) abscission by the ESCRT-III complex (Vps2p-Vps24p, Vps20p-Snf7p) to form intraluminal vesicles containing the cargo. The complex is disassembled by the ATPase Vps4p. Fusion of the limiting membrane of the endosome with the vacuole ultimately leads to degradation of the intraluminal vesicles and cargo (the MVB pathway is illustrated in Fig. 7) (26, 27, 41).
Fig. 7.
Two pathways affect mat formation through MVB. One pathway is the well-characterized Rim101p pathway, which uses components of the ESCRT-I, -II, and -III complexes to transduce the signal to activate Rim101p and FLO11 expression, which are necessary for both invasive growth and mat formation. The second pathway is the putative biofilm pathway, which is hypothesized to have a component that must be properly sorted by the MVB in order to function. The biofilm pathway is not necessary for FLO11 expression or invasive growth but is necessary for mat formation, presumably by altering the cell wall in some unknown way.
In this report, we reveal that several MVB mutants that are not part of ESCRT-I, -II, or -III affect mat formation but not invasive growth and can be used to genetically separate these phenotypes. Our results indicate the existence of two overlapping pathways that pass through the MVB and affect mat formation by FLO11-dependent and -independent mechanisms. The first pathway is the Rim101p pathway, and it affects invasive growth and mat formation by controlling FLO11 expression. The second pathway, which we tentatively call the biofilm pathway, requires the entire complement of class E vps components necessary for a properly functioning MVB and affects mat formation in a FLO11-independent manner.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All strains used in this study belong to the yeast strain background Σ1278 (30) (Table 1). The strains found in Table 3 are from a whole-genome deletion collection created in the Σ1278b background by Owen Ryan and colleagues in the laboratory of Charles Boone at the University of Toronto. A full characterization of the library and the phenotypes of all mutants regarding mat formation, invasive growth, and pseudohyphal growth will be published soon (O. Ryan et al., unpublished data). Mutants were generated by PCR-based gene disruption methods (20, 31). Primers are listed in Table 2. The RIM101-531 dominant active allele was also generated by PCR-based disruption of the C-terminal 95 codons of the RIM101 gene (see Table 2 for primers). Transformations were performed by the standard lithium acetate transformation method (37). The yeast strain L6906 (10) carries a hemagglutinin (HA)-tagged form of FLO11, with the HA tag between amino acids 30 and 31 (FLO11-HA30), and this was used for the immunofluorescence analyses. Primers PC675 and PC676 (Table 2) were used to insert an additional HA tag-encoding DNA sequence between codons encoding amino acids 1015 and 1016 of Flo11p (FLO11-HA30,1015) (16) by the method of Schneider et al. (33). All strains were maintained on standard YPD medium (37), and 250 μg/ml G418 was used for selection of transformants, with the exception of the RIM101-531 truncation transformant, which was selected on minimal medium lacking histidine (37). Strains grown on low-density agar plates (YPD with 0.3% agar) (30) for 5 days at 25°C were used for overlay adhesion assays, immunofluorescence, and Western blotting.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| L6906 | MATaura3-52 his3::hisG FLO11::HA30 | 11 |
| TRY181 | MATaura3-52 his3::hisG FLO11::HA30,1015 | This study |
| CPY74 | MATaura3-52 his3::hisG FLO11::HA30vps4::kanMX6 | This study |
| CPY15 | MATaura3-52 his3::hisG FLO11::HA30vps25::kanMX6 | This study |
| NY70 | MATaura3-52 his3::hisG FLO11::HA30,1015vps25::kanMX6 | This study |
| CPY160 | MATaura3-52 his3::hisG FLO11::HA30vps28::kanMX6 | This study |
| CPY24 | MATaura3-52 his3::hisG FLO11::HA30vps27::kanMX6 | This study |
| NY64 | MATaura3-52 his3::hisG FLO11::HA30,1015vps27::kanMX6 | This study |
| CPY105 | MATaura3-52 his3::hisG FLO11::HA30rim13::kanMX6 | This study |
| NY62 | MATaura3-52 his3::hisG FLO11::HA30,1015rim13::kanMX6 | This study |
| CPY115 | MATaura3-52 his3::hisG FLO11::HA30rim101::kanMX6 | This study |
| NY78 | MATaura3-52 his3::hisG FLO11::HA30,1015rim101::kanMX6 | This study |
| TRY120 | MATaura3-52 his3::hisG FLO11::HA30vps27::kanMX6 RIM101-531 | This study |
| NY60 | MATaura3-52 his3::hisG FLO11::HA30,1015vps27::kanMX6 RIM101-531 | This study |
| TRY118 | MATaura3-52 his3::hisG FLO11::HA30vps25::kanMX6 RIM101-531 | This study |
| NY58 | MATaura3-52 his3::hisG FLO11::HA30,1015vps25::kanMX6 RIM101-531 | This study |
| TRY124 | MATaura3-52 his3::hisG FLO11::HA30,1015rim13::RIM101-531 | This study |
| NY82 | MATaura3-52 his3::hisG FLO11::HA30,1015rim13::kanMX6 RIM101-531 | This study |
| CPY154 | MATaura3-52 his3::hisG FLO11::HA30vps20::kanMX6 | This study |
| CPY96 | MATaura3-52 his3::hisG FLO11::HA30rim9::kanMX6 | This study |
| CPY112 | MATaura3-52 his3::hisG FLO11::HA30rim101::kanMX6 | This study |
Table 3.
Mat and invasive growth phenotypes of vps mutants
| Mutant | Mat | Invasive growth | Class |
|---|---|---|---|
| vps1Δ | − | + | |
| vps2Δ/did4Δ | − | + | E |
| vps3Δ | − | + | |
| vps4Δ | − | + | E |
| vps8Δ | + | + | |
| vps13Δ | + | + | |
| vps15Δ | − | + | |
| vps17Δ | + | + | |
| vps20Δ | − | − | E |
| vps21Δ | + | ± | |
| vps22Δ/snf8Δ | − | − | E |
| vps23Δ | − | − | E |
| vps24Δ | − | + | E |
| vps25Δ | − | − | E |
| vps26Δ/pep4Δ | ± | + | |
| vps27Δ | − | + | E |
| vps28Δ | − | + | E |
| vps30Δ | + | + | |
| vps31Δ/bro1Δ | − | + | E |
| vps32Δ/snf7Δ | − | − | E |
| vps34Δ | − | − | |
| vps35Δ | ± | + | |
| vps36Δ | − | − | E |
| vps37Δ | ± | + | E |
| vps38Δ | − | + | |
| vps39Δ/vam6Δ | + | + | |
| vps41Δ | − | + | |
| vps43Δ/vam7Δ | ± | + | |
| vps44Δ/nhx1Δ | − | + | |
| vps46Δ/did2Δ | + | + | |
| vps51Δ | + | + | |
| vps52Δ | ± | + | |
| vps53Δ | ± | + | |
| vps54Δ | ± | + | |
| vps60Δ/mos10Δ | − | ± | E |
| vps62Δ | + | + | |
| vps64Δ | + | ± | |
| vps66Δ | ± | + | |
| vps68Δ | + | + |
Table 2.
Primers used in this studya
| Primer | Purpose | Sequence |
|---|---|---|
| TRO369 | Confirm disruptions | GCACGTCAAGACTGTCAAGG |
| TRO394 | Disrupt Vps4 | CCAACTTCTACGCCAAGTATCCTA |
| TRO395 | Disrupt Vps4 | CAATCCTGAAAGTGAAGAATCCA |
| TRO396 | Confirm vps4Δ | TAAGAGCAGTAAACCCGTTAGTGAC |
| TRO156 | Disrupt Vps25 | CAAATGATTACACCCCATGAA |
| TRO157 | Disrupt Vps25 | AAGGTTCAAGACTGGACCATG |
| TRO162 | Confirm vps25Δ | TTTTAGATATTTGCGTTAGCTAAGG |
| TRO379 | Disrupt Vps28 | CGGATCCTTCTAAATTGAGAAGAG |
| TRO380 | Disrupt Vps28 | TGGATCAAAGATGATAGTCGCAG |
| TRO381 | Confirm vps28Δ | TCCTTGCCGCCAATAATT |
| TRO266 | Disrupt Vps27 | CCGATTTTTTGGTAATATGTCAA |
| TRO267 | Disrupt Vps27 | AGCCAGGTGGTCAAAAAACA |
| TRO268 | Confirm vps27Δ | ACAAAAGCAAACTGTTCGGAG |
| TRO503 | Disrupt Rim13 | AGTATCTTTGAACCGCGCAG |
| TRO504 | Disrupt Rim13 | GGATGGTCGTTCATTATTTTTGAG |
| TRO505 | Confirm rim13Δ | CGTTACCTCCCACAAAACTTTTG |
| TRO482 | Disrupt Rim101 | GTCCAGCTCGGAGTTTCTAAA |
| TRO483 | Disrupt Rim101 | CGGGATCAACCGATCAAGATA |
| TRO484 | Confirm rim101Δ | ACTTTTCTCTGCCCAGTGACA |
| TRO516 | Generate RIM101-531 dominant allele | CAATGGCAGGTGGAACTTCATTGAAGCCTAACTGGGAATTTAGCCTGAACTGAGGCGCGCCACTTCTAAA |
| TRO517 | Generate RIM101-531 dominant allele | TCTTCAATCGCCAGCTTACTCATGATAATATCATTAGTACAGCTTTTTTGGAATTCGAGCTCGTTTAAAC |
| TRO518 | Confirm RIM101-531 | CCGCCTCTACAATCAAAGATACC |
| PC675 | Insert HA tag between residues 1015 and 1016 | GGATGCTCTCCAAAGACCCATTACAACTACTGTTCCATGTTCAACCAGGGAACAAAAGCTGG |
| PC676 | Insert HA tag between residues 1015 and 1016 | GGTAGGTGAAGTGGTTGTTGATT CCGAGGCGGTTTCGCTTGGACTCTGTAGGGCGAATTGG |
| TRO621 | Real-time PCR primers for FLO11 | CACTTTTGAAGTTTATGCCACACAAG |
| TRO622 | Real-time PCR primer for FLO11 | CTTGCATATTGAGCGGCACTAC |
| TRO632 | Real-time PCR primer for ACT1 | CTCCACCACTGCTGAAAGAGAA |
| TRO636 | Real-time PCR primer for ACT1 | CCAAAGCGACGTAACATAGCTTT |
TRO369 was used as a reverse primer in combination with the primers listed as confirming primers to confirm disruptions on the chromosome by PCR.
Invasive growth assay and overlay adhesion assay.
The invasive growth assay was performed as described previously (29). The overlay adhesion assay was performed as described previously (31).
rtRT-PCR.
Five-day-old mats were used to perform real-time reverse transcriptase PCR (rtRT-PCR). The cells from growing mats were collected from the surface of low-density agar YPD plates using a clean dry spatula and washed with ice-cold water. Total RNA was extracted as previously described (17). Contaminating DNA was removed with a Turbo DNA-free kit (Ambion) according to the manufacturer's protocol. rtRT-PCR was performed on a Bio-Rad iCycler real-time PCR machine using a Verso SYBR green two-step kit with random primers for the reverse transcription step according to the manufacturer's protocol. rtRT-PCR primers for FLO11 and ACT1 (reference gene) are listed in Table 2.
Immunofluorescence of Flo11-HA30 on the surface of cells from rim and hub.
The immunofluorescence assay was performed as described in reference 31, where cells were taken from the rim of the growing mats.
Cell fractionation.
Fractionation of cells carrying Flo11-HA30,1015 was carried out as follows. Mats were grown on low-density-agar YPD plates for 5 days at 25°C. Overlay adhesion assays were performed on the wild-type mats to separate rim and hub cells. Rim cells were washed off the plastic wrap and into a microcentrifuge tube with 1 ml of 50 mM Tris-HCl (pH 7.4) buffer. The adherent cells forming the central hub or the cells composing the entire mat from defective mutants were scraped from the agar using a clean dry spatula, paying attention to bring a minimum carryover of agar during this process. The hub (wild-type) or mutant cells were then suspended in 1 ml of 50 mM Tris-HCl (pH 7.4). Microcentrifuge tubes containing cells from all of these separate samples were then taped onto a roller barrel and washed for 20 min at 23°C. Twenty microliters of sample was removed to a separate tube to be used for normalization calculations for loading sodium dodecyl sulfate (SDS)-polyacrylamide gels (see “Normalization of fractionation samples for loading” below). The remaining cells from each sample were then pelleted, and the supernatant was removed to a separate tube. This supernatant represents proteins shed from the cell wall (S fraction), and proteins in this fraction were precipitated as described below (see “Precipitation of extracellular proteins from the mat” below). The cell pellet was resuspended in 0.8 ml of 50 mM Tris-HCl (pH 7.4) and ruptured using glass beads in the presence of protease inhibitors (protease inhibitor cocktail SE; EMD Chemicals Inc.) by vortexing for 1 min and cooling on ice for 1 min and by repeating this two-step cycle five times. The liquid above the glass bead layer was removed to a separate tube and centrifuged at ∼13,000 × g to pellet the cell wall and membranes. The supernatant (SF; representing the cytosolic or soluble fraction) was stored at −20°C. The membrane/cell wall pellet was resuspended in 100 μl of 50 mM Tris-HCl (pH 7.4) plus 2% SDS and boiled for 5 min, followed by centrifugation for 10 min. The supernatant containing membrane-bound and noncovalently attached cell wall proteins was removed to a fresh tube to create the membrane/noncovalent (M) fraction. The remaining cell wall pellet was boiled again for 10 min in 100 μl 50 mM Tris-HCl (pH 7.4) plus 2% SDS, followed by centrifugation. This second membrane supernatant was then combined with the first (membrane fraction) to obtain the total membrane/noncovalent (M) fraction. The final cell wall pellet was then resuspended in 100 μl of 50 mM Tris-HCl (pH 7.4) containing 2 units of β-1,3-glucanase (Quantazyme; MP Biomedicals) and 0.3 μl of β-mercaptoethanol, and the suspension was incubated for 2 h at 30°C, followed by centrifugation for 10 min at 13,000 rpm. The supernatant from the β-1,3-glucanase treatment represents the fraction of proteins that is covalently attached to the cell wall (C fraction). Proteins from both the membrane/noncovalent (M) and covalent (C) fractions were precipitated by adding 3 volumes of cold acetone and incubating at 4°C overnight. The samples were then centrifuged and dried in a SpeedVac apparatus, after which samples were resuspended in loading buffer. The fractions were analyzed by SDS-PAGE (4% stacking gel, 5% resolving gel), followed by Western blotting with an anti-HA antibody.
Normalization of fractionation samples for loading.
Ten microliters of cells from the washed mat samples was diluted into 490 μl of deflocculation buffer (50 mM EDTA) and sonicated with a Misonix Microson XL2000 ultrasonic homogenizer sonicator set on 4 for 5 pulses (∼5 s each). The cells were then enumerated with a hemocytometer.
Precipitation of extracellular proteins from the mat.
Precipitation of extracellular proteins from the mat was adapted from reference 4. Proteins from the extracellularly shed (S) fraction described above were precipitated from the Tris-HCl buffer by first adding 1/100 volume of 2% sodium deoxycholate (DOC) and incubating for 30 min at 4°C. A 1/10 volume of 100% trichloroacetic acid (TCA) was then added for overnight precipitation at 4°C.
RESULTS
Mutations in class E VPS mutants that block Rim101p processing disrupt invasive growth.
The class E VPS genes that encode members of the ESCRT-I, -II, or -III complex were hypothesized to regulate haploid invasive growth because they affect Rim101p processing, which is required for invasive growth and FLO11 expression in S. cerevisiae (3, 19, 41). We refer to these ESCRT-I, -II, or -III mutants as class E-1 mutants. Their orthologs have also been shown to affect filamentous growth in Candida albicans by affecting Rim101p processing (18, 41). In contrast, non-ESCRT-I, -II, or -III class E VPS mutants, such as vps27Δ or vps4Δ mutants (which we refer to as class E-2 mutants), do not affect Rim101p processing in S. cerevisiae or C. albicans (40) and were expected to not affect invasive growth in S. cerevisiae.
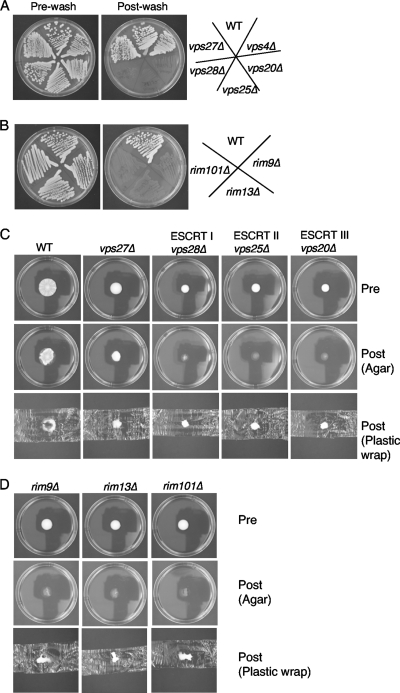
In the Σ1278b background of S. cerevisiae, class E-1 mutants representing three ESCRT complexes, vps28Δ (ESCRT-I), vp25Δ (ESCRT-II), and vps20Δ (ESCRT-III) mutants, show a strong defect in invasive growth (Fig. 1 A), while class E-2 mutants, such as those with vps27Δ and vps4Δ mutations, do not disrupt invasive growth.
Fig. 1.
Class E vps mutants that affect the Rim101p signaling pathway (class E-1) cause defects in mat formation and invasive growth, but class E vps mutants that do not affect the Rim101p pathway (class E-2) disrupt mat formation but not invasive growth. (A) The class E vps mutants vps27Δ, vps28Δ (ESCRT-I), vps25Δ (ESCRT-II), vps20Δ (ESCRT-III), and vps4Δ were subjected to the invasive growth assay (WT, wild type); (B) members of the Rim101p signaling pathway, rim101Δ, rim9Δ, and rim8Δ, were subjected to the invasive growth assay; (C and D) representative members of the class E-1 and E-2 vps mutants (C) and the Rim101p signal transduction pathway (D) were subjected to the overlay adhesion assay.
Class E-1 mutants exhibit stronger invasive growth defects than the rim101Δ mutant itself or mutants with mutations in upstream Rim101p processing components, such as rim13Δ or rim9Δ. While there is a thin layer of cells left behind in the rim101Δ, rim13Δ, and rim9Δ Rim101p pathway mutants, there are practically no cells left behind for the class E-1 mutants (Fig. 1A and B).
Both class E-1 and E-2 mutants perturb mat formation.
On the basis of the invasive growth assays whose results are shown in Fig. 1, it was predicted that the mutations that are known to perturb invasive growth (mutations in class E-1 mutants) would perturb mat formation. In particular, it was hypothesized that the class E-1 mutants would form defective biofilms that differ from the wild-type biofilm in three respects. (i) They would fail to form the wrinkles and channels that are hallmarks of the hub in the wild type (Fig. 1C). (ii) They would not spread over as large of a surface area of the agar plates as the wild type. (iii) They would not adhere to the agar surface when tested for adhesion. In contrast, the mutations that did not perturb invasive growth (those in the class E-2 mutants; Fig. 1A) were predicted to have little to no effect on mat formation.
However, when this was tested, it was discovered that both class E-1 and class E-2 mutants exhibit strong defects in mat formation that are similar to those of the Rim101p pathway mutants (Fig. 1C and D). Similar results have been found by Ryan et al. in the laboratory of Charlie Boone at the University of Toronto while screening the Σ1278b whole-genome deletion collection that they have generated (unpublished data). The vps4Δ mutant's biofilm resembles that of the vps27Δ strain (data not shown).
The class E-1 and class E-2 mutants spread poorly compared to wild type, and they did not generate noticeable patterns on low-density agar. In addition, they all exhibited defects in adhesion to agar, based on the overlay adhesion assay (31). This assay is performed by laying a piece of commercial plastic wrap on the agar over the growing cells and then removing it by lifting up on both sides. Cells that adhere to the agar surface stay behind, as seen for the wrinkled center (hub) of the wild-type mat (Fig. 1C). Cells that are not agar adherent are removed, as seen for the outer edge of the wild type (rim). The entire cell population of the Rim101p pathway mutants and the class E-1 mutants were removed by the plastic wrap (Fig. 1C and D and data not shown), a finding which is similar to what is seen for the flo11Δ mutant (31). The vps27Δ and vps4Δ mutants adhered to the agar surface slightly better than the other mutants (only vps27Δ is shown in Fig. 1C); however, the cells from these mutants that remained on the agar plate were poorly adherent compared to the hub cells from the wild type.
FLO11 expression is diminished in class E-1 mutants but not class E-2 mutants.
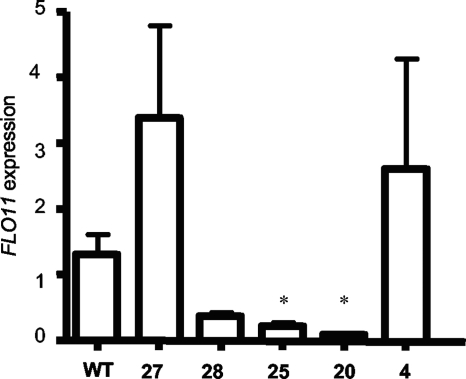
One reason for the difference between the class E-1 and class E-2 mutants might be that the class E-2 mutants exhibit diminished FLO11 gene expression during mat formation but not invasive growth. In order to test this, both groups of mutants were compared for FLO11 expression levels during mat formation by rtRT-PCR. This analysis revealed that the vps28Δ, vps25Δ, and vps20Δ mutants all expressed little FLO11 compared to the wild type (Fig. 2). In contrast, vps27Δ and vps4Δ mutants expressed either higher or similar levels of FLO11 compared to the wild type (Fig. 2).
Fig. 2.
FLO11 expression is greatly diminished in class E-1 mutants known to affect Rim101p processing, but class E-2 mutants like vps27Δ and vps4Δ mutants do not show a decrease in FLO11 expression. Fold change in FLO11 expression was measured by rtRT-PCR, and ACT1 was used as a reference gene. WT, wild type; 27, vps27Δ; 28, vps28; 25, vps25Δ; 20, vps20Δ; 4, vps4Δ. *, P < 0.05 compared to wild type.
Two pathways act through the endosome to affect mat formation.
A hypothesis to explain the different phenotypes between the class E-1 and class E-2 mutants is that there are two distinct, but overlapping, pathways that affect mat formation and act through the endosome. One pathway is the Rim101p signal transduction cascade (6), which requires specific ESCRT-I, -II, and -III components (18, 41) and is required for FLO11 expression (3) (Fig. 2) and therefore affects both invasive growth and mat formation (Fig. 1). The other pathway depends on a functional MVB pathway in general but has little to no effect on FLO11 expression and affects only mat formation and not invasive growth (Fig. 1 and 2).
The hypothesis presented above suggests that the whole MVB pathway is required for mat formation, but it is possible that there is a unique role for Vps27p and Vps4p. This was tested by examining the invasive growth and mat formation phenotypes of a collection of vps mutants in the Σ1278b background. Analysis of 9 additional class E vps mutants (did4Δ, snf8Δ, vps23Δ, vps24Δ, bro1Δ, snf7Δ, vps36Δ, vps37Δ, and mos10Δ) reveals that they all have defects in mat formation, although the defects in the vps37Δ and mos10Δ mutants are less pronounced (Table 3). Consistent with our above-described results, there is a correlation between ESCRT mutations known to perturb Rim101p signaling (class E-1 mutants) and defects in both invasive growth and mat formation. The vps37Δ mutant is an exception to this, as it is defective for mat formation but not invasive growth. However, the vps37Δ mutant gave mixed results regarding its role in Rim101p processing (32, 41). The results for the vps37Δ mutant notwithstanding, these results suggest that MVB trafficking is important for proper mat formation.
An alternative interpretation is that disruption of vacuolar function may be the root cause of the defect in mat formation. However, we have tested an additional 25 vps mutants that do not belong to class E. Of these non-class E vps mutants, 11 have no defect in mat formation, and 8 cause only a partial defect leaving the mutants with less well defined pattern formation but a clear rim and hub by the overlay adhesion assay. Thus, 19 out of 25 non-class E mutants exhibit only a partial defect or no defect in mat formation (Table 3). Only two genes represented among these 19 mutants, VPS21 and VPS62, have relatively close homologs in S. cerevisiae; therefore, for most of the 19 mutants, the lack of a strong defect in mat formation cannot be accounted for by redundant gene functions. In addition, a pep4Δ mutant, which disrupts vacuolar protease activity (2, 15), is also wild type for mat formation (data not shown).
Thus, there is a second pathway required for mat formation, which we will tentatively call the biofilm pathway, which is dependent on the MVB pathway and is hypothesized to act independently of the Rim101p pathway. If the biofilm pathway is really independent of the Rim101p pathway, then restoration of Rim101p transcription factor activity via a dominant allele of RIM101 should bypass upstream defects in the Rim101p pathway but not the biofilm pathway. The RIM101-531 dominant allele encodes a truncated form of Rim101p missing the inhibitory C-terminal tail following amino acid 531. This truncated protein is active even when upstream components of the signal transduction pathway are disrupted, including both class E-1 (i.e., vps28Δ, vps25Δ, and vps20Δ) and non-MVB (i.e., rim13Δ) components (19).
Addition of the RIM101-531 dominant active allele should have different predictable phenotypes in the non-MVB, class E-1, and class E-2 mutants. If RIM101-531 is expressed in a rim13Δ strain (non-MVB), then this should restore FLO11 expression, invasive growth, and mat formation since the rim13Δ mutation should block only Rim101p processing and not MVB trafficking. In contrast, the RIM101-531 allele in the vps25Δ mutant (class E-1) should suppress defects in FLO11 expression and invasive growth but not mat formation, since the RIM101-531 allele will not restore MVB sorting. Finally, the RIM101-531 allele should have no impact on the vps27Δ mutant (class E-2).
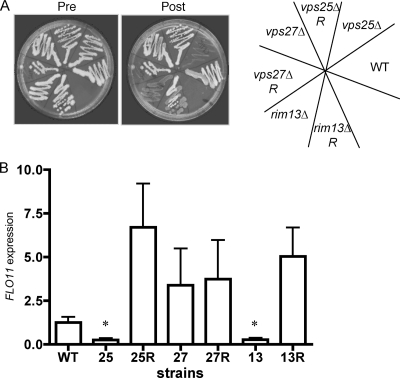
The RIM101-531 allele was introduced into the vps27Δ, vps25Δ, and rim13Δ mutants by deleting the C-terminal 95 codons on the chromosome by homologous recombination (see Materials and Methods). The resulting double mutants were examined for invasive growth, FLO11 expression, and mat formation. The rim13Δ RIM101-531 double mutant was fully restored for invasive growth compared to the rim13Δ single mutant, as was the vps25Δ RIM101-531 mutant. The vps27Δ RIM101-531 double mutant appears to be no different from the vps27Δ mutant (Fig. 3 A). Consistent with these results, FLO11 gene expression in growing mats measured by rtRT-PCR is restored in the vps25Δ RIM101-531 and rim13Δ RIM101-531 double mutants and is not significantly different between vps27Δ and vps27Δ RIM101-531 strains (Fig. 3B).
Fig. 3.
The RIM101-531 allele suppresses invasive growth and FLO11 expression defects in the vps25Δ and rim13Δ mutants. (A) Strains carrying the RIM101-531 allele were subjected to the invasive growth assay. Mutants with a capital R are double mutants carrying the named mutation and the RIM101-531 allele. (B) Fold change in FLO11 expression was measured by rtRT-PCR, and ACT1 was used as a reference gene. WT, wild type; 25, vps25Δ; 25R, vps25Δ RIM101-531; 27, vps27Δ; 27R, vps27Δ RIM101-531; 13, rim13Δ; 13R, rim13Δ RIM101-531. *, P < 0.05 compared to wild type.
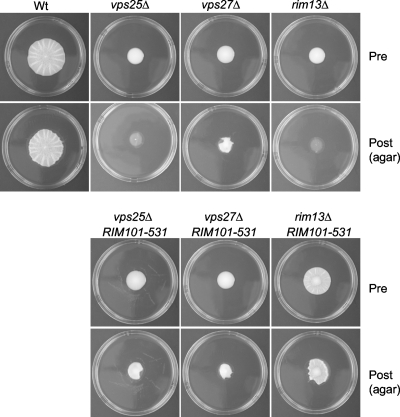
These mutants also behave as predicted in the mat formation assay. In the rim13Δ RIM101-531 double mutant, although mat formation is slightly reduced compared to the wild type, mat formation is restored compared to the rim13Δ parent strain. It exhibits increased spreading on low-density agar with formation of patterns such as a clear hub, and it adheres similarly to the wild type in the overlay adhesion assay, yielding a distinct hub and rim (Fig. 4; data for plastic not shown). The vps27Δ mutant is unaffected by the introduction of the RIM101-531 allele. In contrast, the vps25Δ RIM101-531 double mutant resembles a vps27Δ single mutant in the overlay adhesion assay (Fig. 4).
Fig. 4.
The RIM101-531 allele suppresses the mat formation defect in the rim13Δ mutant but not the vps25Δ or vps27Δ mutant. Pre, before the overlay adhesion assay; Post, agar after the overlay adhesion assay.
Expression of Flo11p is diminished in class E-1 mutants but is similar to that in wild type in class E-2 mutants.
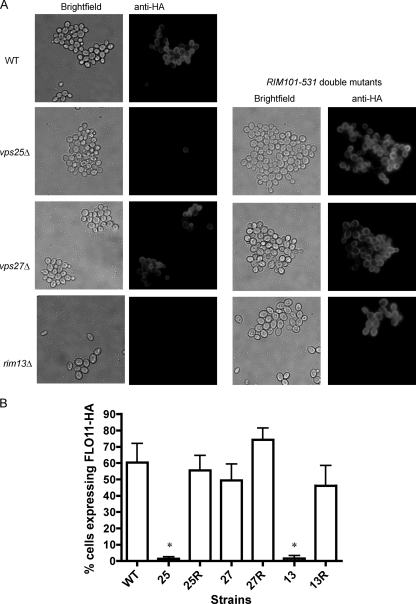
The above-described experiments support the hypothesis that there is a biofilm signaling pathway that depends on functional MVB trafficking and is necessary for mat formation but is independent of Rim101p signaling and FLO11 expression. However, since the MVB pathway affects protein trafficking within the cell, it seemed possible that the mat formation defects were due to poor expression or mislocalization of Flo11p. In order to test this, the percentages of cells expressing Flo11p on the cell surfaces within the mats of different strains were compared. Flo11p is expressed in a variegated manner in the Σ1278b strain, such that only ∼40 to 50% of the wild-type cells express the protein on the surface, as assessed by immunofluorescence (11, 31). Each of these strains carries on its chromosome an allele of FLO11 encoding a protein with an HA epitope tag located between amino acids 30 and 31 (FLO11-HA30). Cells were collected from growing mats in the wild-type strain and in the vps25Δ, vps27Δ, and rim13Δ strains plus their respective RIM101-531 double mutants and subjected to staining with anti-HA antibody to assess the percentages of cells expressing Flo11-HA30 on their surfaces. Consistent with FLO11 gene expression results (Fig. 3B), the vps25Δ strain expressed little Flo11-HA30, while the vps25Δ RIM101-531 strain expressed wild-type levels of the protein (Fig. 5). In contrast, the vps27Δ and vps27Δ RIM101-531 strains were similar to wild type. The rim13Δ mutant, like the vps25Δ strain, expressed much less Flo11-HA30 than the wild type, but the rim13Δ RIM101-531 double mutant was restored for Flo11-HA30 expression.
Fig. 5.
The RIM101-531 allele restores Flo11-HA30 expression in the vps25Δ and rim13Δ mutants. (A) Cells were subjected to secondary immunofluorescence with an anti-HA monoclonal primary antibody directed toward the HA tag in strains carrying Flo11-HA30. (B) Quantification of the percentage of cells expressing Flo11-HA30 from each strain. WT, wild type; 25, vps25Δ; 25R, vps25Δ RIM101-531; 27, vps27Δ; 27R, vps27Δ RIM101-531; 13, rim13Δ; 13R, rim13Δ RIM101-531. *, P < 0.05 compared to wild type.
Flo11p shedding and cell wall localization are not altered in class E-2 mutants.
Although Flo11-HA30 was clearly expressed on the cell surface of class E-2 mutants, it was recently reported that Flo11p is shed outside the cell wall and that this extracellular form is important for mat formation (16). A separate report by another group indicated that Flo11p is not covalently attached to the cell wall, unlike other canonical adhesins (7, 21), but is found in the membranes of yeast cells or is noncovalently associated with the cell wall (23). Thus, it seemed possible that although we do not see differences in Flo11-HA30 expression between wild-type and vps27Δ strains on the basis of immunofluorescence, the subcellular localization of Flo11-HA30 at the surface or outside the cells might be different. For example, perhaps the mutants shed all or most of their Flo11-HA30 or its association with the wall or membrane is altered.
In order to address the above-described concerns, we collected cells from the growing mats and subjected them to subcellular fractionation. Cells were collected from the wild type and vps25Δ (class E-1), vps27Δ (class E-2), and rim13Δ (non-MVB) mutants. The overlay adhesion assay was used to collect separate populations of rim cells from the wild type, and the hubs were scraped from the agar with a spatula. Whole mats from mat-defective vps mutants were collected by scrapping from the agar surface. The cells were then fractionated (see Materials and Methods for more details) to obtain shed (S), membrane-associated (M), and covalently attached cell wall (C) fractions. Protein fractions were then analyzed by SDS-PAGE and Western blotting against Flo11-HA. Loading was normalized to the number of cells represented in each sample from which proteins were extracted (see Materials and Methods for more details).
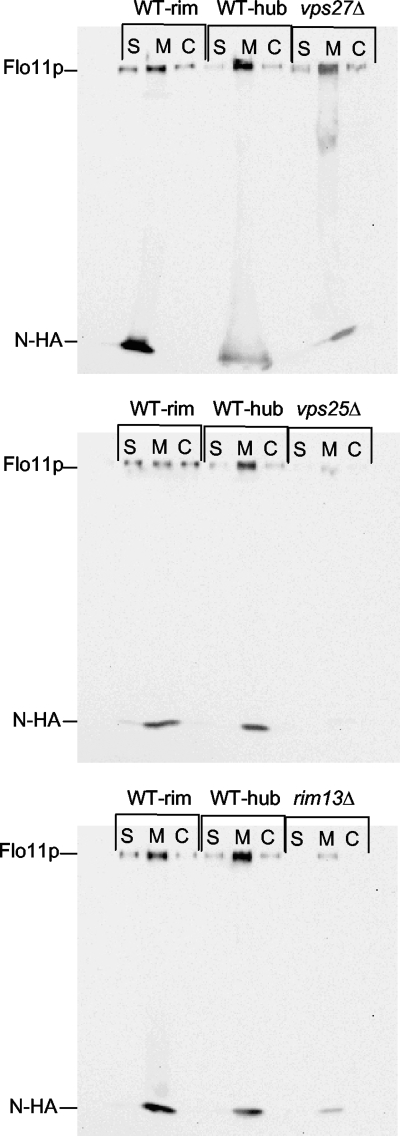
When this procedure was performed on the strains carrying Flo11-HA30, it was found that the expected high-molecular-mass Flo11p band (>260 kDa) seen by Karunanithi et al. (16) was seen only in the membrane fraction, and showed substantial degradation, even in the presence of protease inhibitors (data not shown). Our version of Flo11-HA was tagged between amino acids 30 and 31 (Flo11-HA30). Unlike ours, the Flo11-HA used by Karunanithi et al. (16) was tagged at amino acid residue 1015 (Flo11-HA1015). Therefore, another HA tag was added to FLO11-HA30 in our strain at residue 1015 to create doubly HA tagged Flo11-HA30,1015 strains, and the fractionation was repeated. In this case, we saw a band corresponding to Flo11p that ran at >260 kDa in the shed (S), membrane/noncovalent cell wall (M), and covalently attached cell wall (C) fractions (Fig. 6, Flo11p band). These data indicate that Flo11p is both shed outside the cell wall and covalently attached to the cell wall and is also found in the M fraction containing both membrane- and noncovalently cell wall-associated forms of the protein.
Fig. 6.
Flo11p is both shed from the cell wall and covalently attached to it and is expressed and localized similarly in wild-type and vps27Δ strains. Western blotting was performed on fractionated samples from wild-type, vps27Δ, vps25Δ, and rim13Δ strains carrying Flo11-HA30,1015. A high-molecular-mass Flo11p-HA30,1015 band (>260 kDa) was observed in wild-type and vps27Δ strains in all fractions, including shed (S), membrane bound/noncovalently cell wall associated (M), and covalently attached to cell wall (C) fractions. The vps25Δ and rim13Δ mutants show the absence of Flo11p-HA30,1015 in S and C fractions and considerably decreased signals in the M fraction. A small N-terminal fragment (17 kDa) referred to as N-HA was consistently observed in the M fraction.
A very-low-molecular-mass band is also present and is found primarily in the M fraction (Fig. 6, N-HA). Further analysis revealed this band to be ∼17 kDa (see Fig. S1, N-HA, in the supplemental material), although we do see faint amounts of an ∼37-kDa band as well. We suspect that the ∼17-kDa N-HA band seen in our Western blots corresponds to the N-terminal 33-kDa myc-tagged band of Flo11p reported by Karunanithi et al. (16) but may differ in size due to strain-associated differences in protease sites in Flo11p (see Discussion).
We have previously reported that the percentage of cells expressing Flo11-HA30 in the rim and hub is identical on the basis of immunofluorescence data (31). Consistent with these previous results, the Western blot analysis reveals no obvious or reproducible differences in the overall amounts or distribution of Flo11-HA30,1015 in the S, M, or C fraction of the rim or hub of the wild type (Fig. 6, WT-rim and WT-hub). This is despite the fact that there is a profound difference in the manner in which these cell populations adhere to agar in the overlay adhesion assay (Fig. 1C).
Finally, when Flo11-HA30,1015 expression and distribution are compared between the wild-type and mutant strains, there is a clear decrease in Flo11-HA30,1015 expression in all of the fractions in the vps25Δ and rim13Δ mutants, while there is no reproducible difference between wild-type and vps27Δ strains (Fig. 6). Thus, these results are once again consistent with those from the rtRT-PCR and immunofluorescence experiments (Fig. 3 and 5). Therefore, on the basis of three different measures of FLO11 gene or Flo11p protein expression (Fig. 2, 3, 5, and 6) it appears that the vps27Δ mutant does not differ from the wild type in Flo11p expression, distribution, or shedding. The vps27Δ mutant's failure to form a mat is likely attributable to some unidentified effector protein or molecule.
DISCUSSION
FLO11 is clearly necessary for mat formation; however, it is not sufficient for this phenotype. Martineau et al. (22) reported a similar finding in which they described several mutants with mutations in hsp70 homologs that exhibit defects in mat formation but not in Flo11p expression or invasive growth. We have found that class E-2 mutants cause defects in mat formation in a manner that is independent of Flo11p expression or localization. Thus, class E-2 mutants, along with the hsp70 mutants reported previously, reveal that the phenotypes of invasive growth and mat formation can be clearly separated at the genetic level.
The differences in the expression of FLO11 between the class E-1 and class E-2 mutants can be ascribed to the different roles of these two types of mutants in processing of the Rim101p transcription factor. Class E-1 mutants, such as vps28Δ (ESCRT-I), vps25Δ (ESCRT-II), and vps20Δ (ESCRT-III) mutants, are necessary for Rim101p processing (41), which is in turn necessary for FLO11 expression (Fig. 2, 3, and 5) (3). In contrast, the class E-2 mutants, such as the vps27Δ mutant, do not affect Rim101p processing (41) and therefore do not cause diminished FLO11 expression.
These data indicate that MVB sorting, the common process affected by both class E-1 and E-2 mutants, is required for mat formation but not invasive growth. This hypothesis is further strengthened by the fact that addition of a RIM101-531 dominant active allele to the vps25Δ mutant could rescue this class E-1 mutant's FLO11 expression and invasive growth phenotypes but not its mat formation defect (Fig. 3 and 4). In fact, the vps25Δ RIM101-531 double mutant strongly resembled the vps27Δ mutant in the overlay adhesion assay with its slightly adhesive cells (Fig. 4). Thus, even a constitutively active RIM101-531 allele cannot rescue mat formation as long as MVB sorting is compromised. As a control, it was found that the rim13Δ mutant, which is defective for Rim101p processing but not MVB function, was rescued for mat formation, invasive growth, and FLO11 expression by the RIM101-531 dominant active allele. Finally, our data support a model suggesting that class E vps mutants cause mat formation defects by affecting MVB sorting rather than vacuolar function, as numerous non-class E vps mutants have little or no defect in mat formation (Table 3).
Taken altogether, we present a model suggesting that there are two pathways passing through the endosome that affect mat formation (Fig. 7). One pathway, the Rim101p pathway, affects FLO11 expression, invasive growth, and mat formation, while the biofilm pathway, which is dependent on proper MVB sorting, is required for mat formation but not FLO11 expression or invasive growth.
It is suspected that the MVB mutations (class E-1 or class E-2) cause mislocalization of a component of the biofilm signaling pathway that is necessary for proper mat formation (Fig. 7). We further suspect that this pathway ultimately affects the cell wall in some way that strongly impacts mat formation in a Flo11p-independent manner but has only a very modest effect on invasive growth. In the future, we plan to identify and characterize the components of the biofilm signaling pathway.
On the basis of the rtRT-PCR data (Fig. 2 and 3B), one might get the impression that the class E-2 mutants, such as the vps27Δ mutant, actually overexpress FLO11 and the biofilm pathway represses FLO11. However, when Flo11-HA is examined in these mutants (Fig. 5), this does not appear to be the case. We suspect that the higher expression of FLO11 mRNA in the class E-2 mutants may be misleading due to the size of wild-type mats compared to mutant mats and the fact that there are more glucose-starved cells within wild-type mats that are no longer growing, thus giving a large hub population with diminished FLO11 expression (31).
What forms of Flo11p are found at the cell surface and shed extracellularly?
Karunanithi et al. (16) recently showed that Flo11-Myc30, HA1015 is proteolytically cleaved during its synthesis in a Kex2p-dependent manner and that this leads to the release of a 33-kDa fragment that includes the N terminus of the protein (16).
Surprisingly, we find that the N terminus of Flo11p is present in the cell wall since Flo11-HA30 is detected by immunofluorescence (Fig. 5). Thus, the form of Flo11p in the cell wall of yeast is present with an intact N terminus. However, the >260-kDa form of Flo11-HA30 is difficult to detect by Western blotting, even with the addition of protease inhibitors, and is seen almost exclusively in the membrane fraction (data not shown).
We suspect that release of proteases during cell fractionation may result in cleavage of the Flo11p N terminus, since a 17-kDa N-terminal fragment accumulates in the M fraction from Flo11-HA30,1015 strains (Fig. 6; see Fig. S1 in the supplemental material) and Flo11-HA30 strains (data not shown). This is similar to the findings of Karunanithi et al. (16), who found a 33-kDa Flo11p N-terminal fragment in the cell pellet, which would include membrane and noncovalently attached cell wall proteins. We did not detect the >260-kDa form of Flo11p in the shed (S) or covalent (C) fractions in Flo11-HA30 strains (data not shown), but we did find it in the S and C fractions in the Flo11-HA30,1015 strains (Fig. 6). The shed fraction is collected from intact cells, so the N terminus may be normally cleaved during shedding, as suggested by Karunanithi et al. (16). Since the >260-kDa form of Flo11-HA30 is also not observed in the covalent fractions, the covalent form may be similarly cleaved (and perhaps it is even a precursor to the shed form).
Mats are biofilms.
As a final point, the discovery by Karunanithi et al. (16) that mucin-like proteins such as Flo11p are shed extracellularly by Saccharomyces and our follow-up discovery that Flo11p is shed extracellularly in the mat in both the rim and hub (Fig. 6) suggest to us that Flo11p could itself be defined as part of an extracellular matrix (ECM). Flo11p greatly resembles the mucin proteins of mammals that make gel-like mucous layers. Thus, we believe that S. cerevisiae mats can rightly be described as biofilms that contain an ECM.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul J. Cullen for sending us the primer sequences for generating the HA-tagged version of Flo11p carrying the HA tag at residue 1015 and for sharing with us data and manuscripts regarding shed Flo11p long before they were published. We also appreciate Gerald R. Fink for his comments on the manuscript. Finally, we thank Charles Boone for generously sharing the whole-genome deletion collection in the Σ1278b strain background with us.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Allison C., Hughes C. 1991. Bacterial swarming: an example of prokaryotic differentiation and multicellular behaviour. Sci. Prog. 75:403–422 [PubMed] [Google Scholar]
- 2. Ammerer G., et al. 1986. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol. Cell. Biol. 6:2490–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrales R. R., Jimenez J., Ibeas J. I. 2008. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae. Genetics 178:145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bensadoun A., Weinstein D. 1976. Assay of proteins in the presence of interfering materials. Anal. Biochem. 70:241–250 [DOI] [PubMed] [Google Scholar]
- 5. Boysen J. H., Mitchell A. P. 2006. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol. Biol. Cell 17:1344–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davis D. 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 44:1–7 [DOI] [PubMed] [Google Scholar]
- 7. Frieman M. B., McCaffery J. M., Cormack B. P. 2002. Modular domain structure in the Candida glabrata adhesin Epa1p, a beta1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 46:479–492 [DOI] [PubMed] [Google Scholar]
- 8. Gancedo J. M. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:107–123 [DOI] [PubMed] [Google Scholar]
- 9. Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090 [DOI] [PubMed] [Google Scholar]
- 10. Guo B., Styles C. A., Feng Q., Fink G. R. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. U. S. A. 97:12158–12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halme A., Bumgarner S., Styles C., Fink G. R. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116:405–415 [DOI] [PubMed] [Google Scholar]
- 12. Herranz S., et al. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. U. S. A. 102:12141–12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hurley J. H., Emr S. D. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35:277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jelsbak L., Sogaard-Andersen L. 2003. Cell behavior and cell-cell communication during fruiting body morphogenesis in Myxococcus xanthus. J. Microbiol. Methods 55:829–839 [DOI] [PubMed] [Google Scholar]
- 15. Jones E. W. 1984. The synthesis and function of proteases in Saccharomyces: genetic approaches. Annu. Rev. Genet. 18:233–270 [DOI] [PubMed] [Google Scholar]
- 16. Karunanithi S., et al. 2010. Shedding of the mucin-like flocculin Flo11p reveals a new aspect of fungal adhesion regulation. Curr. Biol. 20:1389–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohrer K., Domdey H. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398–405 [DOI] [PubMed] [Google Scholar]
- 18. Kullas A. L., Li M., Davis D. A. 2004. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 3:1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li W., Mitchell A. P. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Longtine M. S., et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 21. Lu C. F., Kurjan J., Lipke P. N. 1994. A pathway for cell wall anchorage of Saccharomyces cerevisiae alpha-agglutinin. Mol. Cell. Biol. 14:4825–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martineau C. N., Beckerich J. M., Kabani M. 2007. Flo11p-independent control of “mat” formation by hsp70 molecular chaperones and nucleotide exchange factors in yeast. Genetics 177:1679–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martineau C. N., Melki R., Kabani M. 2010. Swa2p-dependent clathrin dynamics is critical for Flo11p processing and ‘mat’ formation in the yeast Saccharomyces cerevisiae. FEBS Lett. 584:1149–1155 [DOI] [PubMed] [Google Scholar]
- 24. Penalva M. A., Arst H. N., Jr 2004. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 58:425–451 [DOI] [PubMed] [Google Scholar]
- 25. Piper R. C., Cooper A. A., Yang H., Stevens T. H. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131:603–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raiborg C., Rusten T. E., Stenmark H. 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15:446–455 [DOI] [PubMed] [Google Scholar]
- 27. Raiborg C., Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458:445–452 [DOI] [PubMed] [Google Scholar]
- 28. Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reynolds T. B. 2006. The Opi1p transcription factor affects expression of FLO11, mat formation, and invasive growth in Saccharomyces cerevisiae. Eukaryot. Cell 5:1266–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reynolds T. B., Fink G. R. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878–881 [DOI] [PubMed] [Google Scholar]
- 31. Reynolds T. B., Jansen A., Peng X., Fink G. R. 2008. Mat formation in Saccharomyces cerevisiae requires nutrient and pH gradients. Eukaryot. Cell 7:122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rothfels K., et al. 2005. Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 25:6772–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schneider B. L., Seufert W., Steiner B., Yang Q. H., Futcher A. B. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265–1274 [DOI] [PubMed] [Google Scholar]
- 34. Shapiro J. A. 1995. The significances of bacterial colony patterns. Bioessays 17:597–607 [DOI] [PubMed] [Google Scholar]
- 35. Shapiro J. A. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81–104 [DOI] [PubMed] [Google Scholar]
- 36. Stoodley P., Sauer K., Davies D. G., Costerton J. W. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187–209 [DOI] [PubMed] [Google Scholar]
- 37. Styles C. 2002. How to set up a yeast laboratory. Methods Enzymol. 350:42–71 [DOI] [PubMed] [Google Scholar]
- 38. Verstrepen K. J., Klis F. M. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60:5–15 [DOI] [PubMed] [Google Scholar]
- 39. Verstrepen K. J., Reynolds T. B., Fink G. R. 2004. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2:533–540 [DOI] [PubMed] [Google Scholar]
- 40. Wolf J. M., Johnson D. J., Chmielewski D., Davis D. A. 2010. The Candida albicans ESCRT pathway makes Rim101-dependent and -independent contributions to pathogenesis. Eukaryot. Cell 9:1203–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu W., Smith F. J., Jr., Subaran R., Mitchell A. P. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 15:5528–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zara S., et al. 2005. FLO11-based model for air-liquid interfacial biofilm formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.