Abstract
Wsc proteins have been identified in fungi and are believed to be stress sensors in the cell wall integrity (CWI) signaling pathway. In this study, we characterized the sensor orthologs WscA and WscB in Aspergillus nidulans. Using hemagglutinin-tagged WscA and WscB, we showed both Wsc proteins to be N- and O-glycosylated and localized in the cell wall and membrane, implying that they are potential cell surface sensors. The wscA disruptant (ΔwscA) strain was characterized by reduced colony and conidia formation and a high frequency of swollen hyphae under hypo-osmotic conditions. The deficient phenotype of the ΔwscA strain was facilitated by acidification, but not by alkalization or antifungal agents. In contrast, osmotic stabilization restored the normal phenotype in the ΔwscA strain. A similar inhibition was observed in the wscB disruptant strain, but to a lesser extent. In addition, a double wscA and wscB disruptant (ΔwscA ΔwscB) strain was viable, but its growth was inhibited to a greater degree, indicating that the functions of the products of these genes are redundant. Transcription of α-1,3-glucan synthase genes (agsA and agsB) was significantly altered in the wscA disruptant strain, resulting in an increase in the amount of alkali-soluble cell wall glucan compared to that in the wild-type (wt) strain. An increase in mitogen-activated protein kinase (MpkA) phosphorylation was observed as a result of wsc disruption. Moreover, the transient transcriptional upregulation of the agsB gene via MpkA signaling was observed in the ΔwscA ΔwscB strain to the same degree as in the wt strain. These results indicate that A. nidulans Wsc proteins have a different sensing spectrum and downstream signaling pathway than those in the yeast Saccharomyces cerevisiae and that they play an important role in CWI under hypo-osmotic and acidic pH conditions.
INTRODUCTION
Signal transduction plays an important role in sensing environmental stimuli and the subsequent regulation of gene expression required for appropriate cell development and morphology. In the yeast Saccharomyces cerevisiae, the cell wall integrity (CWI) signaling pathway has been identified as a major regulator of cell wall biogenesis in adaptation to environmental stresses such as heat shock and hypo-osmolarity (23, 24). Recent genomics studies revealed that the core component of this pathway is highly conserved in yeast and other fungal genomes (31, 38).
The CWI pathway has been well studied in S. cerevisiae (23, 24). The five plasma membrane sensor proteins, Wsc1, Wsc2, Wsc3, Mid2, and Mtl1, act as mechanosensors and detect cell wall perturbations caused by stressful events (35, 48). Wsc1 and Mid2 act as the main sensor proteins in this system. The activated sensors initiate the signaling cascades of downstream cytoplasmic transducers (23, 24). The sensor proteins interact with GDP/GTP exchange factors (Rom1 and Rom2) that activate a small G protein (Rho1). Rho1 GTPase activity in turn activates a mitogen-activated protein kinase (MAPK) module through sequential phosphorylation. The MAPK module consists of MAPKKK (Bck1), MAPKKs (Mkk1 and Mkk2), and MAPK (Slt2 [Mpk1]). Eventually, the phosphorylation relay activates the transcriptional factors Rlm1 and SBF (composed of Swi4 and Swi6), which regulate the transcriptional levels of cell wall-related genes.
A BLAST search using the sequences of these functionally characterized CWI components revealed that their orthologous genes, except for the stress sensor genes MID2 and MTL1, are conserved in the filamentous fungi Aspergillus fumigatus, A. nidulans, and A. niger (8). Several downstream signal-transducing orthologous genes, including rhoA, pkcA, mpkA, and rlmA, that are orthologs of the RHO1, PKC1, MPK1, and RLM1 genes, respectively, have been demonstrated to play essential roles in cell polarity, differentiation, and CWI in Aspergillus species. In A. nidulans, the Rho family GTPase RhoA was shown to be involved in establishing polarity, branching, and cell wall synthesis (13). It was suggested that the protein kinase C (PKC)-encoding gene pkcA plays an essential role in the maintenance of cell integrity and polarized growth in A. nidulans (18, 41, 46). The A. nidulans MpkA protein is involved in germination of conidial spores and polarized growth (4), and transcription of the mpkA gene seems to be autoregulated by the CWI pathway via MpkA (8). The transcription of most cell wall-related genes except for the α-1,3-glucan synthase genes agsA and agsB is independent of RlmA, unlike the case in the yeast model (8).
Although the downstream signal transducers and transcription factors involved in CWI signaling have been characterized in Aspergillus species, the functional roles of the upstream cell wall sensor WSC1-3 orthologs have not been elucidated in filamentous fungi. The Wsc family of sensor proteins has been characterized in S. cerevisiae and its close relative Kluyveromyces lactis (24, 40). A structural feature of the Wsc proteins is the presence of a cysteine-rich domain (also referred to as a WSC domain), a serine/threonine-rich region, a transmembrane region, and a highly charged C-terminal cytoplasmic region. The WSC domain contains up to eight conserved cysteine residues that may form S-S bonds and is believed to mediate noncovalent binding with cell wall glucans. The β-1,3-exoglucanase of Trichoderma harzianum also contains two copies of the WSC domain that may bind glucan chains (5). A GFP fusion localization study revealed that Wsc1 resides in membrane patches within the plasma membrane in both S. cerevisiae and K. lactis (39, 44). Recently, single-molecule atomic force microscopy revealed that Wsc1 behaves like a linear nanospring that is capable of resisting a high level of mechanical force and of responding to cell surface stress (6). The WSC domain of Wsc1 is required for clustering stimulated by stressful conditions (16). Based on the results of GFP fusion localization and single-molecule microscopy, it was proposed that the function of Wsc1 is coupled to its local enrichment within membrane patches called the “Wsc1 sensosome.”
During the course of our previous study on posttranslational modification by O-glycosylation in A. nidulans, we identified the gene for the Wsc family sensor ortholog wscA (11). The wscA gene was used to investigate the substrate specificities of the protein O-mannosyltransferases PmtA, PmtB, and PmtC in A. nidulans, since the conserved serine/threonine-rich region was shown to be highly O-mannosylated by the O-mannosyltransferases Pmt2 and Pmt4 in S. cerevisiae (26). O-mannosylation is believed to confer the rod-like structure upon the sensors by extending and stiffening the polypeptide. Our results also suggested that A. nidulans WscA is O-mannosylated in A. nidulans by PmtA and PmtC, but not by PmtB, and that the O-glycan attachment has a significant impact on the stability and degradation of WscA (11, 20). Protein O-glycosylation plays an essential role in fungal species, as demonstrated by the observation that the pmtA disruptant exhibits abnormal cell morphology and altered cell wall composition (10, 32). Notably, disruption of pmtC leads to a higher level of growth repression than disruption of the other pmt genes. The hypha in the pmtC disruptant is swollen and frequently branched, and cells lose the ability to form conidia under normal growth conditions. Accordingly, impairment of Pmt substrates, including WscA, may result in a crucial defect in the hyphal structure, germination, and CWI in A. nidulans.
In this study, we sought further understanding of how the substrates of Pmt function in Aspergillus species. We also identified and characterized the stress sensor ortholog wscB and examined its role in influencing cell morphology and cell wall biogenesis in A. nidulans. Using phenotypic and transcriptional analysis, we showed that WscA and WscB are involved in CWI in A. nidulans subjected to hypo-osmotic and acidic pH conditions.
MATERIALS AND METHODS
A. nidulans strains, media, and culture conditions.
The Aspergillus strains used in this study are listed in Table 1. Fungi were grown at 30°C in YG medium (0.5% [wt/vol] yeast extract, 1% [wt/vol] glucose) or MM medium (1% [wt/vol] glucose, 0.6% [wt/vol] NaNO3, 0.052% [wt/vol] KCl, 0.052% [wt/vol] MgSO4·7H2O, 0.152% [wt/vol] KH2PO4, 0.211% [wt/vol] arginine, 5 μg/ml of biotin, and Hunter's trace elements [pH 6.5]). Media were adjusted to the required pH with HCl and NaOH. To express genes under the control of the alcA promoter, 100 mM threonine and 0.1% fructose were added as carbon sources instead of glucose in MM medium. For the cultivation of pyrG-negative strains, 0.056% (wt/vol) uracil and 0.122% (wt/vol) uridine were added to MM medium.
Table 1.
A. nidulans strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| AKU89 | biA1 argB2 akuB::aurA | 11 |
| ΔwscA | biA1 argB2 akuB::aurA wscA::ptrA | This study |
| ΔwscB | biA1 argB2 akuB::aurA wscB::ptrA | This study |
| ΔwscA ΔwscB | biA1 argB2 akuB::aurA wscA::ptrA wscB::argB | This study |
| AΔwscA | biA1 argB2 akuB::aurA wscA::ptrA pyrG::wscA | This study |
| BΔwscB | biA1 argB2 akuB::aurA wscB::ptrA pyrG::wscB | This study |
| AKU89-wscA-HA | biA1 argB2 akuB::aurA alc(P)-wscA::3HA-bip(T)ptrA | This study |
| AKU89-wscB-HA | biA1 argB2 akuB::aurA alc(P)-wscB::3HA-bip(T)ptrA | This study |
| ΔmpkA | biA1 argB2 akuB::aurA mpkA::argB | This study |
Construction of wscA and wscB disruptants.
A. nidulans AKU89 served as the wild-type (wt) strain in this study (11). The wscA and wscB genes were disrupted in wt A. nidulans by insertion of the pyrithiamine resistance gene (ptrA). The gene replacement cassette encompassing 1 kb at the 5′ end of wscA, 2 kb of ptrA, and 1 kb at the 3′ end of wscA was constructed by recombinant PCR using the primer pairs F5660-1015/R5660-2042, F5660-ptrA/R5660-ptrA, and F5660-2939/R5660-3992, respectively (see Fig. S1A and Table S2 in the supplemental material). For amplification of the ptrA gene, pGEM-T Easy ptrA was used as the template DNA. The resultant DNA fragment amplified with primers F5660-1015 and R5660-3992 was used to transform wt A. nidulans. The gene replacement cassette encompassing 1.4 kb at the 5′ end of wscB, 2 kb of ptrA, and 1.5 kb at the 3′ end of wscB was constructed by recombinant PCR using the primer pairs F4674-599/R4674-1990, F4674-ptrA/R4674-ptrA, and F4674-3065/R4674-4505, respectively (see Fig. S1B and Table S2 in the supplemental material), and the resultant DNA fragment amplified with primers F4674-599 and R4674-4505 was used to transform wt A. nidulans. Standard transformation procedures for A. nidulans were used (53). For the selection of transformants, MM agar plates supplemented with 0.1 μg/ml of pyrithiamine were used. Disruption of wscA and wscB was confirmed by Southern blot analysis (see Fig. S1A and B in the supplemental material). A. nidulans genomic DNA was prepared as previously described (32). The digoxigenin (DIG)-labeled probes were constructed using primer sets F5660-2939/R5660-3992 for wscA and F4674-3065/R4674-4505 for wscB and a DIG labeling kit (Roche) according to the manufacturer's protocols.
To construct a wscA and wscB double disruptant strain, the wscB gene was disrupted in a wscA disruptant strain using argB insertion. The gene replacement cassette encompassing 1.4 kb of the 5′ end of wscB, 1.8 kb of argB, and 1.5 kb of the 3′ end of wscB was constructed by recombinant PCR using the primer pairs F4674-599/RargB4674-1990, F4674-argB/R4674-argB, and FargB4674-3065/R4674-4505, respectively (see Table S2 in the supplemental material), and the resultant DNA fragment amplified with primers F4674-599 and R4674-4505 was used to transform a wscA disruptant strain. The transformants were selected on MM agar plates without arginine. Introduction of the argB gene into the wscB locus was confirmed by PCR using the primer pair F-AnwscB/R4674-4505 (see Fig. S1C in the supplemental material).
Complementation of wscA and wscB deletion strains.
For analysis of complementation of the wscA disruptant with wt wscA, a gene replacement cassette encompassing 1.5 kb of the 5′ end of pyrG, 2.9 kb of wt wscA, and 1.2 kb of the 3′ end of pyrG was constructed by recombinant PCR using the primer pairs F1-AnpyrG/R1-AnpyrG, F-AnwscA/R-AnwscA, and F2-AnpyrG/R2-AnpyrG, respectively (see Table S2 in the supplemental material). The resultant DNA fragment amplified with primers F1-AnpyrG and R2-AnpyrG was used to transform the wscA disruptant. Transformants were selected on MM agar with 10 mM arginine, 5 mM 5-fluoroortic acid, 5 mM uridine, and 5 mM uracil. Introduction of the wt wscA gene into the wscA disruptant at the pyrG locus was confirmed by PCR using the primer pair F3-AnpyrG/R2-AnpyrG (see Fig. S1D in the supplemental material).
For analysis of complementation of the A. nidulans wscB disruptant with wt wscB, the plasmid pGTΔsal-pyrG::wscB was constructed as follows. A 3.1-kb DNA fragment of wscB was amplified by PCR using F-AnwscB and R-AnwscB and ligated into the SalI site of pGTΔsal-pyrG (11) (see Table S2 in the supplemental material), yielding pGTΔsal-pyrG::wscB. Using pGTΔsal-pyrG::wscB as a template, we amplified the DNA fragment carrying 1.3 kb of the 5′ end of pyrG, 3.1 kb of wscB, and 1.7 kb of the 3′ end of pyrG with primers F1-AnpyrG and R2-AnpyrG and used it to transform the wscB disruptant. Introduction of the wt wscB gene into the wscB disruptant at the pyrG locus was confirmed by PCR using the primer pair F3-AnpyrG/R2-AnpyrG (see Fig. S1D in the supplemental material).
Analysis of conidiation efficiency.
Approximately 105 conidia were spread onto an 84-mm agar plate containing YG medium or YG medium with 0.6 M KCl. After 5 days of incubation at 30°C, a 1-cm2 agar block containing newly formed conidia was suspended in 0.01% (wt/vol) Tween 20 solution and the conidia were counted using a hemocytometer. The mean number of conidia formed was determined from the results of 10 agar blocks from two independent plates.
Analysis of glucan and chitin content.
Strains were grown for 24 h at 30°C with shaking at 120 rpm in flasks containing YG liquid medium. Analysis of the glucan and chitin contents was carried out according to previously described methods (3, 37). Briefly, cells were dissolved in 250 mM phosphate buffer (pH 7.0) and placed on ice for 30 min. The cells were then disrupted using a French press (Ohtake Seisakusho, Tokyo, Japan) and centrifuged at 27,300 × g for 10 min at 4°C. The pellet was washed with 10 ml of phosphate buffer. The centrifugation and washing steps were repeated five times, and the resulting pellet was dissolved in 10 ml of ultrapure water. The centrifugation and washing steps using ultrapure water were repeated five times. The resulting pellet was freeze dried and used as a cell wall. Ten milligrams of cell wall pellet was dissolved in 1 ml of formic acid and incubated for 20 min at 100°C. After centrifugation at 20,000 × g for 5 min, the glucan content of the supernatant was determined as the total fraction using the phenol-sulfuric acid method (3). To obtain alkali-soluble and alkali-insoluble fractions, 10 mg of cell wall pellet was dissolved in 1 ml of ultrapure water and incubated for 30 min at 100°C. After centrifugation at 20,000 × g for 5 min, the pellet was dissolved in 1 ml of 1 M KOH and incubated for 30 min at 70°C. After centrifugation at 20,000 × g for 5 min, the supernatant was used as the alkali-soluble fraction, while the pellet was dissolved in 1 ml of formic acid and incubated for 20 min at 100°C and then centrifuged at 20,000 × g for 5 min. The content of α-1,3-glucan and β-1,3-glucan in the alkali-soluble and alkali-insoluble fractions, respectively, was confirmed by dot blot assay using MOPC-104E (Sigma) and β-1,3-glucan monoclonal antibodies (Biosupplies) (data not shown).
For chitin quantification, 10 mg of cell wall was digested with 2 mg/ml of Yatalase (Takara) in 250 mM phosphate buffer (pH 7.0) for 16 h at 30°C. After centrifugation at 20,000 × g for 5 min, the supernatant was used to quantify chitin using the Morgan-Elson method (37). The mean glucan and chitin contents were determined from analysis of three independent cultures.
Analysis of glycerol content.
Strains were grown for 19 h at 30°C with shaking at 120 rpm in flasks containing YG liquid medium with or without 0.6 M KCl. The collected mycelium was not washed because Witteveen et al. reported that the washing process resulted in considerable losses of the intracellular polyols (51). The glycerol content was measured as described previously (28). Briefly, 100 mg of freeze-dried mycelium was mechanically broken using a multibead shocker instrument with 500 μl of ultrapure water (2,000 rpm, 90 sec; Yasui Kikai) for three rounds, each round comprised of 3 cycles of 30 s each. The homogenates were centrifuged at 1,500 × g for 8 min. The supernatant was transferred to a new tube. The remaining pellet was resuspended in 400 μl of ultrapure water and subjected to two more disruptions by the multibead shocker. These supernatants were collected and heated at 95°C for 20 min to remove proteins and were centrifuged at 19,060 × g for 10 min. The supernatant was subjected to high-performance liquid chromatography (HPLC) using a CARBOSep COREGEL-87P column (Transgenomic) and a refractive index detector. The ultrapure water was used as a mobile phase at a flow rate of 1 ml per min. The column temperature was maintained at 85°C. The standards for polyols, including those for trehalose, glucose, mannitol, and glycerol, were used.
Microscopy.
To observe aerial hyphae, we inoculated conidia on a slide culture of YG agar medium. After incubation at 30°C for 48 h, the aerial hyphae were observed, using an inverted IX70 light microscope (Olympus). To observe submerged A. nidulans hyphae, we inoculated 2 × 108 conidia into 100 ml of YG liquid medium or YG liquid medium with 0.6 M KCl and incubated them with shaking at 120 rpm at 30°C. After 24 h, the mycelia were transferred to a 12-well plate and incubated with 10 ng/ml of fluorescent brightener 28 (calcofluor white; Sigma) for 10 min. The mycelia were mounted on the slide glass and observed using a FluoView FV10i confocal laser scanning microscope (Olympus). Image acquisition was performed using the Z-stack mode.
Construction of A. nidulans strains expressing HA-tagged WscA or WscB.
For the expression of hemagglutinin (HA)-tagged WscA and WscB in wt A. nidulans, plasmids pAGTB-wscA-HA-ptrA and pAGTB-wscB-HA-ptrA, which contained a wscA::HA or wscB::HA fusion between the alcA promoter and bipA terminator, were constructed as follows. HA was amplified using the F1-3HA/R1-3HA primer set and ligated into the SalI and PstI sites of pAST30-Tbip carrying Palc and Tbip (see Table S2 in the supplemental material), yielding pAGTB-HA. The wscA and wscB genes were amplified using primer sets F1-5660.2-exp/R1-5660.2-exp and F1-4674.2-exp/R1-4674.2-exp, respectively (see Table S2 in the supplemental material), and ligated into the NcoI and SalI sites of pAGTB-HA, yielding pAGTB-wscA-HA and pAGTB-wscB-HA. The ptrA gene was obtained by digesting pGEM-T Easy ptrA with HindIII and ligating pAGTB-wscA-HA and pAGTB-wscB-HA into the HindIII site, yielding pAGTB-wscA-HA-ptrA and pAGTB-wscB-HA-ptrA. The integrative plasmids were then transformed into wt A. nidulans. Southern blot analysis was used to confirm the pyrithiamine-resistant transformants carrying wscA::HA (AKU89-wscA-HA) and wscB::HA (AKU89-wscB-HA). The DIG-labeled probes were constructed using primer set F1-5660.2-exp/R1-5660.2-exp for AKU89-wscA-HA and F1-4674.2-exp/R1-4674.2-exp for AKU89-wscB-HA using a DIG-labeling kit (Roche) according to the manufacturer's protocols.
Preparation of A. nidulans extracts and immunoblot analysis.
To investigate the localization of HA-tagged WscA and WscB, total, membrane, microsomal, and cytosolic fractions were prepared. The A. nidulans membrane fraction was prepared according to the method of Yamazaki et al., with some modifications (52). The conidia (2 × 108) of AKU89-wscA-HA and AKU89-wscB-HA were inoculated into 100 ml of MM medium with 100 mM threonine and 0.1% fructose as carbon sources instead of glucose. After cultivation at 30°C with shaking at 120 rpm for 24 h, mycelia were harvested by filtration. Approximately 0.1 mg of cell pellets was mechanically broken using a multibead shocker instrument (2,500 rpm, 10 sec), and proteins were extracted with 2 ml of LY buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA) supplemented with 4% (vol/vol) Complete protease inhibitor cocktail (Roche) using a multibead shocker (2,000 rpm, 60 sec). The homogeneous suspension was centrifuged at 2,430 × g for 5 min at 4°C, and the supernatant was recentrifuged twice under the same conditions. The resulting supernatant represented the total fraction, and this supernatant was centrifuged at 21,880 × g for 15 min at 4°C. The pellet was washed with LY buffer. The centrifugation and wash steps were repeated three times, and the pellet was dissolved in 200 μl of LY buffer and used as the membrane fraction.
The supernatant remaining after the pellet was removed was centrifuged at 100,000 × g for 2 h, and the resulting supernatant was used as the cytoplasmic fraction, while the pellet was used as the microsomal fraction. Each fraction was precipitated with trichloroacetic acid (TCA) and centrifuged, and the pellet was washed three times with ice-cold acetone. The pellet was then dissolved in SDS-PAGE sample buffer. After separation of proteins on 10% SDS-polyacrylamide gels, WscA-HA and WscB-HA were detected using immunoblot analysis with anti-HA monoclonal antibody (Sigma). Proteins were visualized with nitroblue tetrazolium–bromochloroindolylphosphate (Roche) according to the manufacturer's instructions.
To determine the levels of MpkA and actin expression and the level of MpkA phosphorylation, we inoculated 2 × 108 conidia of each strain into 100 ml of YG medium or YG medium with 0.6 M KCl. After cultivation at 30°C with shaking at 120 rpm for 16 h, mycelia were harvested by filtration. Total protein was extracted and used for immunoblot analysis according to the method described above. MpkA, actin, and phosphorylated MpkA were detected using anti-p44/42 MAPK (L34F12) (Cell Signaling Technology), anti-actin monoclonal antibody clone C4 (MP Biomedicals), and anti-phospho-p44/42 MAPK (Cell Signaling Technology), respectively.
N-Glycosidase treatment of WscA-HA and WscB-HA and immunoblot analysis.
Membrane fraction proteins isolated as described above were treated with N-glycosidase F (Roche) according to the manufacturer's protocol. Briefly, the membrane fraction was denatured at 95°C for 5 min with 1% SDS and treated with N-glycosidase in 200 mM sodium phosphate (pH 8.0), 10 mM EDTA, 1% 2-mercaptoethanol, 0.5% Nonidet P-40, and 0.1% SDS at 37°C for 12 h. The proteins were then precipitated with TCA, washed three times with acetone, and subjected to SDS-PAGE and immunoblot analysis using anti-HA monoclonal antibody (Sigma).
TFMS treatment of WscA-HA and WscB-HA and immunoblot analysis.
The freeze-dried mycelia (20 mg) were broken with metal corn by the multibead shocker at 2,000 rpm for 10 sec. The resultant cells were dissolved in TNE buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100) as described previously (52) and further broken by a multibead shocker with 0.15 g of 0.5-mm beads at 2,000 rpm for 60 sec. To remove debris, we centrifuged the mixture at 19,060 × g for 5 min. The supernatant containing 1 mg protein was transferred to a glass bottle and freeze dried. The glass bottle was sealed with a Teflon-coated butyl rubber stopper and purged with nitrogen gas. Five hundred μl of trifluoromethanesulfonic acid (TFMS; Sigma) was added, and the mixture was incubated on ice for 40 min. It was neutralized by the addition of 5 ml of 1 M Tris solution. After TCA precipitation, the proteins were collected by centrifugation at 2,430 × g for 10 min, washed by ethanol, and subjected to SDS-PAGE and immunoblot analysis using anti-HA monoclonal antibody (Sigma).
Indirect immunofluorescence microscopy.
Fixed cells were prepared and processed as described by Harris et al. and Takeshita et al. except that Yatalase (30 mg/ml; Takara) and cellulase R10 (30 mg/ml; Yakult Pharmaceutical) were used for cell lysis (15, 45). A rabbit anti-HA antibody (Sigma) at 1:500 dilution and a mouse anti-actin monoclonal antibody C4 clone (MP Biomedical) at 1:200 dilution were used as primary antibodies. Cy3-conjugated anti-mouse IgG antibody (Sigma) at 1:1,000 dilution and Alexa Fluor 488-conjugated anti-rabbit antibody (Invitrogen) at 1:1,000 were used as secondary antibodies. The chitin was stained with calcofluor white. The pictures were taken by a FluoView FV10i confocal laser scanning microscope (Olympus).
Preparation of A. nidulans total RNA.
For investigation of the transcription of cell wall-related genes, the conidia of wt and wscA and wscB disruptants (2 × 108) were grown in 100 ml of YG liquid medium or YG liquid medium with 0.6 M KCl in flasks for 18 h at 30°C with shaking at 120 rpm. The mycelia were collected and frozen at −80°C. The frozen mycelia were disrupted using a multibead shocker instrument (2,000 rpm, 10 sec). One milliliter of RNAiso (Takara) was added, and the mycelia were again homogenized using a multibead shocker (2,000 rpm, 5 sec). Chloroform treatment was performed according to the manufacturer's protocol. The resulting RNA was precipitated with isopropanol and rinsed with 70% ethanol, and the pellet was air dried and then dissolved in diethylpyrocarbonate-treated water. Finally, DNase treatment was performed using RQ1 RNase-Free DNase (Promega) according to the manufacturer's protocol.
For analysis of agsB transcription after micafungin treatment, 2 × 108 conidia from the wt and wsc disruptant strains were inoculated into 100 ml of YG medium in a flask and incubated with shaking at 120 rpm at 30°C. After 18 h, micafungin was added to a final concentration of 10 ng/ml. Mycelia were collected at 0, 30, 60, and 120 min and immediately frozen at −80°C. Total RNA was extracted according to the method described above.
Real-time RT-PCR analysis.
cDNAs were synthesized using a PrimeScript Perfect real-time reagent kit (Takara) according to the manufacturer's protocol using 400 ng of total RNA as the template. Real-time reverse transcription (RT)-PCR analysis was performed using a LightCycler Quick 330 system (Roche) with SYBR Premix DimerEraser (perfect real time) reagent (Takara). The following primers were used: wscA-RT-F and wscA-RT-R for wscA, wscB-RT-F and wscB-RT-R for wscB, gpdA-RT-F and gpdA-RT-R for gpdA, agsA-RT-F and agsA-RT-R for agsA, agsB-RT-F and agsB-RT-R for agsB, gelA-RT-F and gelA-RT-R for gelA, csmA-RT-F and csmA-RT-R for csmA, chsB-RT-F and chsB-RT-R for chsB, fksA-RT-F and fksA-RT-R for fksA, rhoA-RT-F and rhoA-RT-R for rhoA, and histone-RT-F and histone-RT-R for the histone H2B gene (see Table S2 in the supplemental material). The histone H2B gene was used to standardize the mRNA levels of the target genes.
RESULTS
In silico identification of A. nidulans WscA and WscB.
Previously, WscA (encoded by ANID_05660.1) was identified as an ortholog of Wsc1 in S. cerevisiae (11). In this study, BLASTP analysis using the amino acid sequences of S. cerevisiae Wsc sensor proteins as the search queries identified three Wsc family sensor candidates, including WscA, ANID_04674.1, and ANID_06927.1. The amino acid sequences of these putative Wsc family proteins were analyzed using Pfam and TMHMM Server v. 2.0. Although WscA and ANID_04674.1 showed significant structural homology to Wsc1-4 from S. cerevisiae with regard to the domain constitution and N-terminal signal sequence, WSC motif, serine/threonine-rich region, transmembrane domain, and the highly charged C-terminal cytoplasmic region, in silico analysis indicated that ANID_06927.1 does not possess a region after the serine/threonine-rich domain, including a transmembrane domain that is important for membrane localization (Fig. 1). Thus, ANID_04674.1 was termed WscB as a putative stress sensor. Alignments of the WscA, WscB, and Wsc1 sequences from S. cerevisiae and K. lactis showed that the WSC domain (C1-X-S-X12–16-Φ-Q-S-X3-C2-X3-C3-X5–8-A-L(I)-X5–6-C4-Φ-C5-X12–17-C6-X3-C7-X-G-Φ-X4-C8-G-X6(30)-VY) found in Wsc proteins from S. cerevisiae is highly conserved in WscA and WscB, with the exception of several aromatic amino acids (designated by the symbol Φ) (48) (Fig. 1). Although the conserved KXYQ sequence in the C terminus of S. cerevisiae Wsc proteins was not found in A. nidulans WscA and WscB, the conserved DXXD sequence was found (48). ANID_06927.1 does not possess both KXYQ and DXXD sequences. The primary difference was in the length of the serine/threonine-rich region, which was shorter in WscA and WscB than in S. cerevisiae Wsc1 (81 amino acids [aa] in A. nidulans WscA and 73 aa in WscB). In addition, three putative N-glycosylation sites were found at N135, N176, and N258 in WscA and N41, N250, and N257 in WscB using the NetNGlyc 1.0 server.
Fig. 1.
Sequence comparison of the Wsc proteins. ClustalW multiple-sequence alignment of the deduced amino acid sequences of WscA, WscB, and ANID_06927.1 from A. nidulans (AnWscA, AnWscB, and ANID_06927.1), Wsc1 from K. lactis (KlWsc1), and Wsc1 from S. cerevisiae (ScWsc1) was performed using BioEdit software (47). Putative signal sequences and transmembrane regions were identified by the SignalP 3.0 server and TMHMM Server v. 2.0 and are shown in bold (2, 21). The conserved cysteine motifs (C1-X-S-X12–16-Φ-Q-S-X3-C2-X3-C3-X5–8-A-L(I)-X5–6-C4-Φ-C5-X12–17-C6-X3-C7-X-G-Φ-X4-C8-G-X6(30)-VY) are highlighted with a gray background (48). Aromatic amino acids are designated by the symbol Φ. The putative N-glycosylation sites identified by the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) are highlighted with a black background. The conserved sequences in the C terminus, KXYQ and DXXD, are boxed (48).
The A. nidulans Wsc sequences had 19.4% to 35.8% amino acid identity with the functionally characterized S. cerevisiae Wsc1-4 proteins (see Table S1 in the supplemental material). Phylogenetic analysis showed that WscA and WscB can be classified into the Wsc1 and Wsc4 branches, respectively, with relatively high bootstrap values (data not shown). Wsc1 localizes in the plasma membrane and acts as a main sensor transducer for the CWI signaling pathway in S. cerevisiae (12, 25, 30). On the other hand, Wsc4 localizes in the endoplasmic reticulum (ER) and is involved in the translocation of soluble secretory proteins and the insertion of membrane proteins into the ER membrane. Wsc4 may also have a role in the stress response, but it has only partial functional overlap with Wsc1-3 (27, 54).
Localization and glycosylation of HA-tagged WscA and WscB.
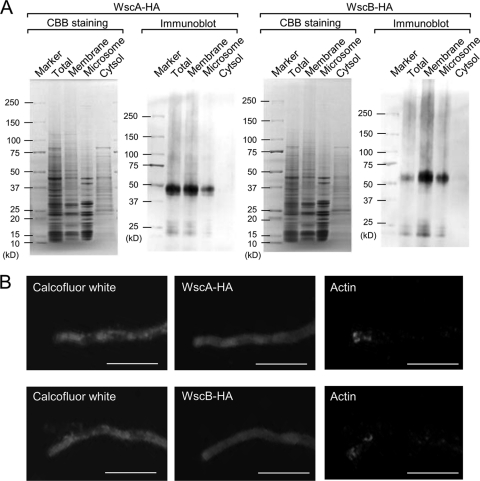
WscA and WscB fused with a triple HA tag were expressed in wt A. nidulans under the control of the alcA promoter, and their localization was investigated by immunoblotting (Fig. 2 A) and indirect immunostaining (Fig. 2B) using an anti-HA antibody. Both proteins were detected in the total, membrane, and microsomal fractions, but not in the cytoplasmic fraction (Fig. 2A). In addition, indirect immunostaining of WscA-HA and WscB-HA using an anti-HA antibody did not show any fluorescence intensity in intracellular structures, including the ER (Fig. 2B), indicating that Wsc proteins are localized in the cytoplasmic membrane. This result was in agreement with the prediction that WscA and WscB function as cytoplasmic membrane-spanning sensors. The WSC domain is believed to interact with cell wall carbohydrates. The molecular masses of WscA-HA and WscB-HA as determined by SDS-PAGE were 43 to 50 kDa and 55 to 63 kDa, respectively (Fig. 2A). Because the predicted molecular masses of WscA-HA and WscB-HA are 30 and 32 kDa, respectively (see Table S1 in the supplemental material), these two proteins appeared to be posttranslationally modified.
Fig. 2.
WscA and WscB are cytoplasmic membrane proteins. (A) WscA-HA and WscB-HA strain cell extracts were subjected to immunoblot analysis. (B) Localization of actin and WscA-HA and WscB-HA proteins in a hyphal tip of A. nidulans by indirect immunostaining. The horizontal microscopic pictures were taken from the same position. Anti-actin antibody was used as a positive control for this study. These hyphae were stained simultaneously with calcofluor white. The white bars represent 20 μm.
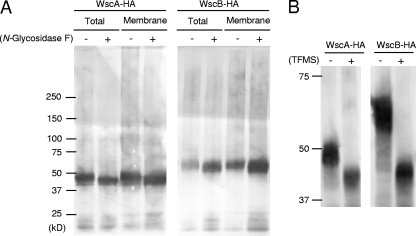
Since WscA and WscB possess putative N-glycosylation sites, we examined the proteins for the presence of N-glycans. Total and membrane fraction proteins were treated with N-glycosidase, and HA-Wsc proteins were subsequently analyzed by SDS-PAGE and immunoblotting (Fig. 3 A). After the N-glycosidase treatment, the apparent molecular masses of WscA-HA and WscB-HA decreased to 43 to 48 kDa and 48 to 63 kDa, respectively. The size differences before and after N-glycosidase treatment were consistent with the existence of three N-glycosylation sites with the typical N-glycan structure [Man(5-12)GlcNAc2] that has been described for Aspergillus glycoproteins (1, 19, 49, 50). Thus, our results suggest that both WscA and WscB are N-glycosylated in A. nidulans. Because WscA and WscB also have O-glycosylation sites, the remaining difference between the observed and native protein molecular masses of 30 kDa and 32 kDa, respectively, seems to be due to modification by O-linked glycosylation (11).
Fig. 3.
N- and O-glycosylation of WscA-HA and WscB-HA. The cell extracts from A. nidulans AKU89-wscA-HA and AKU89-wscB-HA were treated with N-glycosidase (A) and TFMS (B) and subjected to immunoblot analysis using anti-HA antibody.
We demonstrated previously that WscA-HA is O-glycosylated by the O-mannosyltransferases PmtA and PmtC (11). To further confirm the O-glycosylation, we treated the WscA-HA and WscB-HA proteins expressed in A. nidulans (Fig. 3B) with TFMS, which removed both N- and O-glycans, resulting in a significant decrease in the molecular masses of Wsc proteins. The extents of reduction in molecular mass of Wsc proteins by treatment with TFMS are much larger than those by treatment with N-glycosidase. Thus, these two proteins were also shown to be O-glycosylated.
Deletion of wscA and wscB results in sensitivity to hypo-osmolarity and low pH.
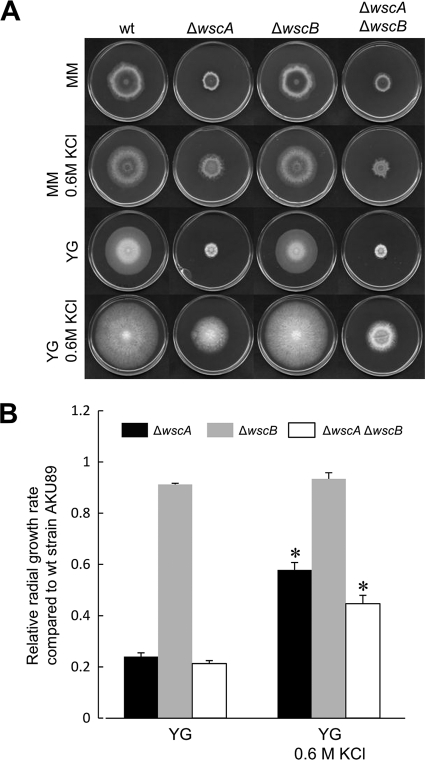
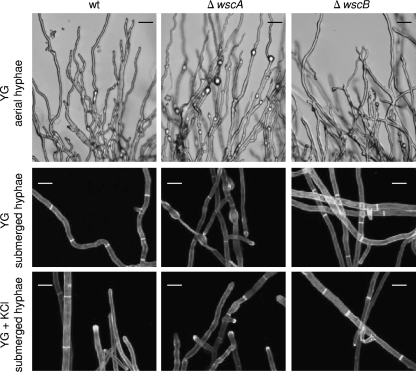
To explore the functional roles of wscA and wscB, we generated wsc deletion strains (ΔwscA, ΔwscB, and ΔwscA ΔwscB) (see Fig. S1 in the supplemental material). The ΔwscA strain colonies were significantly smaller in size on YG and MM media than the wt strain colonies (Fig. 4 A). The ΔwscA strain conidiophores were of similar structure to those of the wt strain on YG medium (data not shown). However, a high frequency of swollen structures was observed in both the aerial and submerged hyphae of the ΔwscA strain grown on YG medium (Fig. 5). The deficient phenotype of the ΔwscA strain was osmoremediable. The diameter of ΔwscA colonies became similar to that of wt colonies upon the addition of KCl to the media as an osmotic stabilizer. The radial growth rate of the ΔwscA strain relative to that of the wt strain was remedied from 0.24 to 0.58 by the addition of KCl to the YG medium (Fig. 4B). The balloon structures of submerged hyphae in ΔwscA were still present, but they were less prominent when ΔwscA was grown on YG medium with KCl than when grown on YG alone (Fig. 5). On the other hand, the ΔwscB strain formed slightly smaller colonies than did the wt strain. The relative radial growth rates of the ΔwscB strain in YG medium and YG medium with KCl were reduced to 0.91 and 0.93, respectively (Fig. 4B). A double disruption of the wscA and wscB genes (ΔwscA ΔwscB) inhibited growth to a greater degree than did the single disruptions (Fig. 4B). These results indicate that the functions of WscA and WscB are redundant.
Fig. 4.
Colony formation of the wt strain AKU89 and wsc disruptants. (A) A. nidulans strains were grown in YG and MM media with or without 0.6 M KCl at 30°C for 5 days. (B) For comparison of the growth rate in YG medium with or without 0.6 M KCl, the radial growth rate (cm/day) of each wsc disruptant was divided by that of wt strain AKU89. *, statistically significant difference relative to the result obtained using YG medium (P < 0.05).
Fig. 5.
Hyphal morphology of the wt and wsc disruptant strains. The wt strain AKU89 and disruptant strains ΔwscA and ΔwscB were grown at 30°C in YG medium with or without 0.6 M KCl. The dark bars represent 10 μm, and the white bars represent 25 μm.
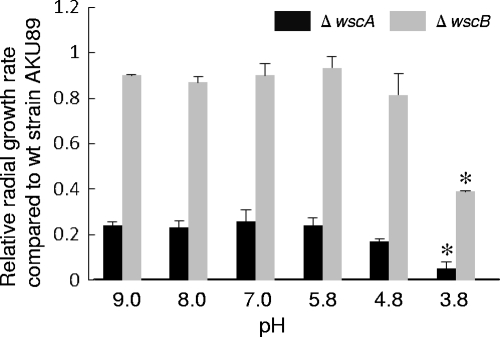
The defect in colony formation was facilitated under low-pH conditions (pH 3.8) in the ΔwscA and ΔwscB strains (Fig. 6). The relative radial growth rates of both the ΔwscA and ΔwscB strains were reduced when cells were cultivated on acidified YG medium (pH 3.8) but not when they were cultured on an alkalinized medium. Colony formation was also compared in the presence of several growth inhibitors (caffeine, calcofluor white, hygromycin B, Congo red, and micafungin). The growth of the ΔwscA and ΔwscB strains did not decrease relative to that of the wt strain when conidia were inoculated into YG medium with growth inhibitors at different concentrations (data not shown).
Fig. 6.
Relative sensitivity to pH change. A. nidulans strains were grown on YG medium at various pHs (3.8, 4.8, 5.8, 7.0, 8.0, and 9.0). The radial growth rate (cm/day) of each wsc disruptant was divided by that of wt strain AKU89. *, statistically significant difference (P < 0.05) relative to results using standard YG medium (pH 5.8).
Effect of wsc disruptions on conidia formation.
To examine the effects of wsc deletions on conidiation, mutants were cultivated on YG agar plates or YG agar plates with 0.6 M KCl for 5 days, at which time the numbers of conidia were counted. The number of conidia/cm2 in the ΔwscA strain was significantly reduced, to 3% and 2% of the number produced by the wt strain cultured on both YG and YG with KCl, respectively (Fig. 7). Thus, the addition of KCl did not improve conidiation in the ΔwscA strain. On the other hand, the number of conidiospores produced by the ΔwscB strain was reduced to 71% and 59% of the number produced by the wt strain grown on YG and YG with 0.6 M KCl, respectively. Therefore, conidiation in the ΔwscB strain was inhibited by both hypo-osmotic and hyperosmotic conditions.
Fig. 7.
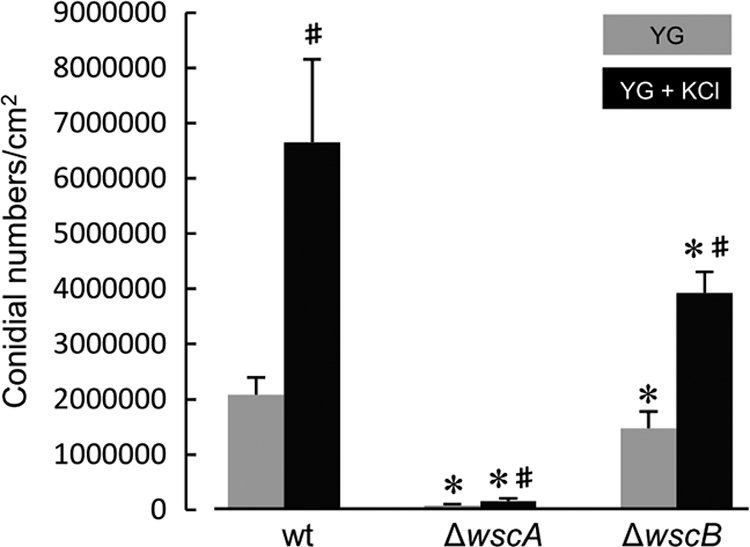
Conidiation of wsc disruptants was reduced. Formation of conidia on YG agar plates with or without 0.6 M KCl was measured after 5 days of cultivation at 30°C. All results were expressed as means with standard deviations. *, statistically significant difference (P < 0.05) relative to results for wt strain AKU89. #, statistically significant difference (P < 0.05) relative to the result obtained using YG medium.
Effect of wsc disruptions on cell wall components.
The CWI pathway is known to regulate cell wall-related genes during cell wall biosynthesis. We therefore compared the cell wall contents of the wt, ΔwscA, ΔwscB, and ΔwscA ΔwscB strains (Table 2). The alkali-soluble fraction containing α-glucan and soluble β-1,3-/1,6-glucan and the alkali-insoluble fraction containing β-1,3-/1,6-glucan covalently linked to chitin were quantified after 24 h of cultivation in YG liquid medium (3, 7, 22). In the ΔwscA and ΔwscA ΔwscB strains, the amount of α-glucan and soluble β-1,3/1,6-glucan increased to 130% and 125%, respectively, of that in the wt strain. The alkali-insoluble fraction containing insoluble β-1,3/1,6-glucan is believed to be responsible for fungal cell wall rigidity (7). However, we found no significant difference in cell wall composition between the wt and wsc disruptants with regard to the alkali-insoluble fraction and chitin.
Table 2.
Cell wall compositions of A. nidulans wild-type strain AKU89 and wsc disruptants
| Strain | Total sugara (%) | Alkali-soluble fractiona (%) | Alkali-insoluble fractiona (%) | Total GlcNAca (%) |
|---|---|---|---|---|
| AKU89 | 475 ± 71 (100) | 254 ± 9 (100) | 176 ± 30 (100) | 309 ± 8 (100) |
| ΔwscA | 537 ± 84 (114) | 329 ± 15 (130) | 176 ± 12 (100) | 297 ± 24 (96) |
| ΔwscB | 441 ± 97 (93) | 278 ± 40 (109) | 179 ± 40 (102) | 298 ± 22 (96) |
| ΔwscA ΔwscB | 536 ± 95 (113) | 317 ± 40 (125) | 160 ± 10 (91) | 291 ± 20 (94) |
Measurements are in μg per mg of cell wall.
Glycerol concentration in wt and wsc disruptants.
Because polyol concentrations in the cells were associated with stress tolerance (42), we determined them in the wt and wsc disruptants. HPLC analysis revealed that significant amounts of glycerol are present but only trace amounts of trehalose, glucose, and mannitol are present in the wt and wsc disruptants. The glycerol concentration was significantly decreased in the wscA disruptant and the wscA and wscB double disruptant compared to that in the wt strain in the YG medium (Fig. 8). In addition, the reduced glycerol concentration was retrieved by the addition of 0.6 M KCl into the YG medium and a statistically significant difference in glycerol content among the wt and the wsc disruptant was not shown. This result was consistent with the results that the hyphal growth rates of the wscA disruptant and the wscA and wscB double disruptant were significantly reduced in the YG medium and that the reduced growth was retrieved by the addition of 0.6 M KCl.
Fig. 8.
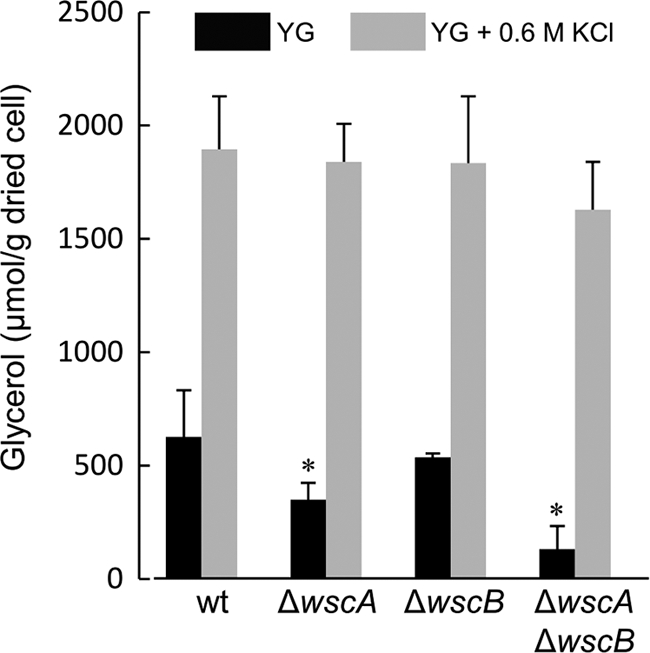
Glycerol concentrations in mycelia. A. nidulans strains were cultivated in YG medium or YG medium with 0.6 M KCl. *, statistically significant difference (P < 0.05) relative to the results for the wt strain.
Transcriptional analysis of wscA, wscB, gpdA, and cell wall-related genes.
Since disruption of wscA altered the cell wall composition, leading to an increase in the amount of the alkali-soluble fraction containing α-glucan and soluble β-1,3/1,6-glucan in YG medium, we hypothesized that WscA may be involved mainly in the transcriptional regulation of cell wall biogenesis-related genes under hypo-osmotic conditions. To elucidate the regulation of these genes in the wsc disruptants, we compared the transcription of wscA, wscB, gpdA (glyceraldehyde-3-phosphate dehydrogenase), and cell wall-related genes between the wt and wsc disruptant strains using real time RT-PCR (Fig. 9). We used RNA templates extracted from log-phase cells cultured for 18 h in YG liquid medium or YG liquid medium supplemented with 0.6 M KCl. Since osmotic stabilization restored the phenotype of ΔwscA, we hypothesized that the regulation of these genes is affected by the addition of 0.6 M KCl.
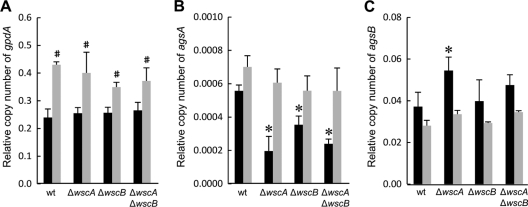
Fig. 9.
Transcription levels of the gpdA (A), agsA (B), and agsB (C) genes in the wt and wsc disruptant strains cultured in YG medium with (black) or without (gray) 0.6 M KCl. Strains were cultured at 30°C for 18 h. Transcription of the indicated genes was determined by real-time RT-PCR, and the mRNA levels shown are relative to the levels of histone H2B mRNA. #, statistically significant difference (P < 0.05) relative to the results obtained using YG medium. *, statistically significant difference (P < 0.05) relative to results for the wt strain cultured in YG medium.
The complete disappearance of wscA and wscB transcription was confirmed for each wsc disruptant strain, and similar levels of mRNA for these two genes were detected in the wt and wsc disruptant strains in YG liquid medium (data not shown). The transcriptional response of the wt and wsc disruptant strains to the saline environment was confirmed by examining the transcription of the gpdA gene, which is known to be transcriptionally upregulated under high osmotic conditions (33, 34). The gpdA gene has a constitutive promoter, but its expression was upregulated by the addition of NaCl, KCl, and sorbitol. Transcription of gpdA increased 1.4- to 1.8-fold in both the wt strain and wsc disruptants upon the addition of KCl (Fig. 9A). Thus, the response to high osmotic conditions seemed to be active in all four strains.
Next, we examined the transcription of several cell wall biogenesis-related genes, including α-1,3-glucan synthase genes (agsA and agsB), the β-1,3-glucan synthase gene (fksA), the β-1,3-glucanosyl transferase gene (gelA), and chitin synthase genes (csmA and chsB). Among the genes examined, agsA and agsB had significantly altered transcription in the wsc disruptant strains compared to that in the wt strain only when the cells were cultured in YG medium (Fig. 9B and C). The transcription of agsA decreased in all the wsc disruptant strains compared to that in the wt strain. The reduction in transcription was proportional to the degree of deficiency of the wsc disruptant strain phenotypes. On the other hand, the transcription of agsB increased in the ΔwscA strain compared to that in the wt strain. This result agrees with the observation that changes in the cell wall composition of the ΔwscA strain led to an increase in the amount of α-glucan extracted in the alkali-soluble fraction (Table 2). The differences observed among the wt, ΔwscA, ΔwscB, and ΔwscA ΔwscB strains cultured in YG medium were remedied when the cells were cultured in YG supplemented with 0.6 M KCl, as transcription of agsA and agsB was similar in the wt, ΔwscA, ΔwscB, and ΔwscA ΔwscB strains. This result was consistent with the determination that the reduced colony formation and abnormal hyphal structures found in ΔwscA could be reversed in the presence of KCl.
Effect of wsc disruptions on phosphorylation of MpkA.
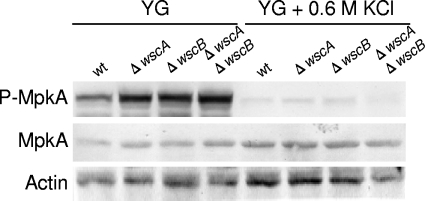
Because the mitogen-activated protein kinase MpkA is a response factor for the CWI signaling pathway, we examined the level of MpkA phosphorylation in the wt and wsc disruptant strains. Interestingly, in YG medium, phosphorylation of MpkA in the wsc disruptants increased even in the ΔwscA ΔwscB strain (Fig. 10). Thus, both Wsc proteins are not essential for the phosphorylation of MpkA. On the other hand, in the presence of 0.6 M KCl, the level of MpkA expression was similar among strains, but its phosphorylation was significantly repressed in the wt and wsc disruptant strains (Fig. 10), indicating that the remedial effect of KCl was due to a reduction in MpkA signaling under the hyperosmotic condition.
Fig. 10.
Phosphorylation of MpkA in the wt and wsc disruptants cultured in YG medium with or without 0.6 M KCl. The levels of MpkA phosphorylation (P-MpkA) and MpkA and actin expression were determined using immunoblot analysis. Actin served as the loading control.
Effect of micafungin treatment on transcription of agsB and phosphorylation of MpkA.
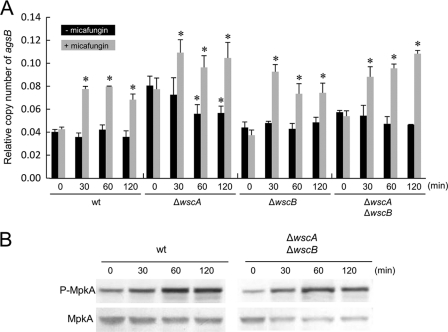
It has been shown that micafungin treatment transiently upregulates the transcription of agsB via MpkA-RlmA signaling in A. nidulans (8). Therefore, we compared this transcriptional response in the wt and wsc disruptant strains to obtain direct evidence that WscA and WscB are placed upstream of MpkA-RlmA signaling. However, we found that exposure to micafungin activated the transcription of agsB in both the wt strain and the wsc diruptants (Fig. 11 A), indicating that the Wsc proteins are not involved in sensing stress associated with exposure to micafungin. A consistent increase in the level of MpkA phosphorylation after micafungin treatment was observed in the ΔwscA ΔwscB strain, to the same degree as that observed in the wt strain (Fig. 11B). The result was also consistent with the observation that the ΔwscA and ΔwscB strains are not hypersensitive to micafungin, in contrast to the wt strain.
Fig. 11.
Transient transcriptional upregulation of agsB (A) and phosphorylation of MpkA (B) following exposure to micafungin in the wt and wsc disruptant strains. Strains were cultured at 30°C for 24 h, at which time micafungin was added to the medium and transcription of the agsB and histone H2B genes and the level of MpkA phosphorylation were monitored at 0, 30, 60, and 120 min. The y axis shows the level of mRNA relative to that of the histone H2B gene (A). *, statistically significant difference (P < 0.05) relative to the result obtained at 0 min.
DISCUSSION
In a previous study, we hypothesized that impairment of WscA is involved in producing some of the morphological deficiencies observed in pmtA and pmtC disruptants (11). In the present study, we observed several overlapping deficiencies between the wsc and pmt disruptants. The ΔwscA strain had a phenotype similar to that of the pmtA and pmtC disruptants, such as swollen hyphal structures, reduced conidiation, and reduced colony formation when cells were cultured under hypo-osmotic conditions. Osmotic stabilization of the medium remedied these defects in all wscA, pmtA, and pmtC disruptants. In contrast, disruption of pmtB had no significant effect on colony formation. The phenotypes we observed are in close agreement with the determination that O-mannosylation of WscA is catalyzed by PmtA and PmtC, but not by PmtB.
In our previous study on protein O-mannosylation in A. nidulans (11), N-glycosidase and TFMS treatments suggested that WscA and WscB are modified by both O- and N-glycosylation. S. cerevisiae Wsc1 also possesses two potential N-glycosylation sites; however, it is posttranslationally modified only by O-glycosylation (35). Both O- and N-glycosylation of the S. cerevisiae Mid2 sensor protein have been reported (17). O-mannosylation determines the stability of Mid2. On the other hand, N-glycosylation near the N-terminal end (N35) is required for proper function of Mid2. It has been suggested that the N-glycan may be directly involved in Mid2 sensing.
The sensing spectrum in A. nidulans seems to be different from that in the well-studied S. cerevisiae Wsc proteins. In our study, both the ΔwscA and ΔwscB strains exhibited hypersensitivity to stress associated with acidic but not alkaline conditions, indicating that WscA and WscB sense the perturbations in the cell wall caused by exposure to low pH. In contrast, S. cerevisiae Wsc1 is involved in the response to stress associated with exposure to alkaline conditions (43). Interestingly, phenotypic analysis suggested that WscA and WscB are not involved in sensing stress associated with exposure to antifungal agents, including caffeine, calcofluor white, hygromycin B, Congo red, and micafungin. In S. cerevisiae, Wsc1, but not Wsc2, Wsc3, or Mid2, mediates caspofungin-induced PKC pathway activation (36). Caspofungin is an echinocandin class β-1,3-glucan synthase inhibitor similar to micafungin. In A. nidulans, the downstream components of the CWI signaling pathway are involved in tolerance to antifungal agents. For example, the dominant negative rhoAE40I allele confers hypersensitivity to calcofluor white and caspofungin (13), while the ΔmpkA strain exhibits sensitivity to calcofluor white and micafungin, and the ΔrlmA strain is sensitive to calcofluor white (8). Consistently, the transcriptional level of rhoA was unchanged among wt and wsc disruptants (data not shown). Moreover, micafungin treatment transiently upregulates the transcription of agsB via MpkA-RlmA signaling in A. nidulans (8). In this study, a double disruptant strain of wscA and wscB demonstrated a transient upregulation of agsB and increased phosphorylation of MpkA after treatment with micafungin, similar to the wt strain. This suggests that WscA and WscB are not required for MpkA signaling and either that the sensor responsible for responding to stress associated with micafungin exposure has not been identified or that micafungin independently activates the downstream pathway of CWI signaling.
Although the BLASTP search did not identify any orthologous MID2 sensor genes in the Aspergillus genomes, a S. cerevisiae Cwh43 homolog was found (8). Cwh43 is a putative sensor/transporter protein that acts upstream of the BCK2 branch of the PKC1-dependent cell wall integrity pathway and is involved in cell wall biogenesis (29). The principal structural feature of Cwh43 is the presence of 14 to 16 transmembrane segments and several putative glycosylation and phosphorylation sites.
The colony formation and abnormal hyphal structures observed with the wsc disruptant strains were remedied when cells were cultured under high osmotic conditions. Therefore, the osmoresponsive activation pathway is retained in the wt and both wsc disruptants, indicating that WscA and WscB are dispensable in osmoadaptation. The osmoinducible gpdA gene was consistently upregulated in the wsc disruptant strains, and the level of MpkA phosphorylation was equally depressed under high osmotic conditions in the wt strain and all of the wsc disruptants. Osmotic signals are known to activate the high-osmolarity glycerol (HOG) pathway. Genome sequencing analyses have revealed that A. nidulans has genes orthologous to all the genes of the HOG response MAPK pathway of S. cerevisiae (14). Activation of the A. nidulans HOG pathway depends solely on the two-component signaling system, and MAPKK activation mechanisms in the A. nidulans HOG pathway differ from those in the yeast model (9). Although abnormal colony size was remedied by the addition of KCl in our study, conidiation efficiency was not recovered in the ΔwscA and ΔwscB strains. Wsc proteins seem to be required for the conidiation event even under high-osmolarity conditions.
In A. nidulans, the putative stress sensors WscA and WscB are involved in CWI under hypo-osmotic and acidic pH conditions. However, direct evidence of transcriptional response to cell wall-related stress has not been obtained. Further study is therefore needed to fully elucidate the stress sensor functions of these Wsc family proteins in A. nidulans.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by a Ministry of Education, Science, Sports and Culture Grant-in-Aid for Scientific Research (C) (21580096).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Aleshin A. E., Firsov L. M., Honzatko R. B. 1994. Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution. J. Biol. Chem. 269:15631–15639 [PubMed] [Google Scholar]
- 2. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 3. Borgia P. T., Dodge C. L. 1992. Characterization of Aspergillus nidulans mutants deficient in cell wall chitin or glucan. J. Bacteriol. 174:377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bussink H. J., Osmani S. A. 1999. A mitogen-activated protein kinase (MPKA) is involved in polarized growth in the filamentous fungus, Aspergillus nidulans. FEMS Microbiol. Lett. 173:117–125 [DOI] [PubMed] [Google Scholar]
- 5. Cohen-Kupiec R., Broglie K. E., Friesem D., Broglie R. M., Chet I. 1999. Molecular characterization of a novel beta-1,3-exoglucanase related to mycoparasitism of Trichoderma harzianum. Gene 226:147–154 [DOI] [PubMed] [Google Scholar]
- 6. Dupres V., et al. 2009. The yeast Wsc1 cell surface sensor behaves like a nanospring in vivo. Nat. Chem. Biol. 5:857–862 [DOI] [PubMed] [Google Scholar]
- 7. Fontaine T., et al. 2000. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 275:27594–27607 [DOI] [PubMed] [Google Scholar]
- 8. Fujioka T., et al. 2007. MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 6:1497–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furukawa K., Hoshi Y., Maeda T., Nakajima T., Abe K. 2005. Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol. Microbiol. 56:1246–1261 [DOI] [PubMed] [Google Scholar]
- 10. Goto M. 2007. Protein O-glycosylation in fungi: diverse structures and multiple functions. Biosci. Biotechnol. Biochem. 71:1415–1427 [DOI] [PubMed] [Google Scholar]
- 11. Goto M., et al. 2009. Protein O-mannosyltransferases B and C support hyphal development and differentiation in Aspergillus nidulans. Eukaryot. Cell 8:1465–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gualtieri T., Ragni E., Mizzi L., Fascio U., Popolo L. 2004. The cell wall sensor Wsc1p is involved in reorganization of actin cytoskeleton in response to hypo-osmotic shock in Saccharomyces cerevisiae. Yeast 21:1107–1120 [DOI] [PubMed] [Google Scholar]
- 13. Guest G. M., Lin X., Momany M. 2004. Aspergillus nidulans RhoA is involved in polar growth, branching, and cell wall synthesis. Fungal Genet. Biol. 41:13–22 [DOI] [PubMed] [Google Scholar]
- 14. Han K. H., Prade R. A. 2002. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol. Microbiol. 43:1065–1078 [DOI] [PubMed] [Google Scholar]
- 15. Harris S. D., Morrell J. L., Hamer J. E. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heinisch J. J., Dupres V., Wilk S., Jendretzki A., Dufrene Y. F. 2010. Single-molecule atomic force microscopy reveals clustering of the yeast plasma-membrane sensor Wsc1. PLoS One 5:e11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hutzler F., Gerstl R., Lommel M., Strahl S. 2008. Protein N-glycosylation determines functionality of the Saccharomyces cerevisiae cell wall integrity sensor Mid2p. Mol. Microbiol. 68:1438–1449 [DOI] [PubMed] [Google Scholar]
- 18. Ichinomiya M., Uchida H., Koshi Y., Ohta A., Horiuchi H. 2007. A protein kinase C-encoding gene, pkcA, is essential to the viability of the filamentous fungus Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71:2787–2799 [DOI] [PubMed] [Google Scholar]
- 19. Kainz E., et al. 2008. N-glycan modification in Aspergillus species. Appl. Environ. Microbiol. 74:1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kriangkripipat T., Momany M. 2009. Aspergillus nidulans protein O-mannosyltransferases play roles in cell wall integrity and developmental patterning. Eukaryot. Cell 8:1475–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 22. Lee H. H., Park J. S., Chae S. K., Maeng P. J., Park H. M. 2002. Aspergillus nidulans sod(VI)C1 mutation causes defects in cell wall biogenesis and protein secretion. FEMS Microbiol. Lett. 208:253–257 [DOI] [PubMed] [Google Scholar]
- 23. Lesage G., Bussey H. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levin D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lodder A. L., Lee T. K., Ballester R. 1999. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 152:1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lommel M., Bagnat M., Strahl S. 2004. Aberrant processing of the WSC family and Mid2p cell surface sensors results in cell death of Saccharomyces cerevisiae O-mannosylation mutants. Mol. Cell. Biol. 24:46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mamoun C. B., Beckerich J. M., Gaillardin C., Kepes F. 1999. Disruption of YHC8, a member of the TSR1 gene family, reveals its direct involvement in yeast protein translocation. J. Biol. Chem. 274:11296–11302 [DOI] [PubMed] [Google Scholar]
- 28. Managbanag J. R., Torzilli A. P. 2002. An analysis of trehalose, glycerol, and mannitol accumulation during heat and salt stress in a salt marsh isolate of Aureobasidium pullulans. Mycologia 94:384–391 [PubMed] [Google Scholar]
- 29. Martin-Yken H., et al. 2001. Saccharomyces cerevisiae YCRO17c/CWH43 encodes a putative sensor/transporter protein upstream of the BCK2 branch of the PKC1-dependent cell wall integrity pathway. Yeast 18:827–840 [DOI] [PubMed] [Google Scholar]
- 30. Nanduri J., Tartakoff A. M. 2001. The arrest of secretion response in yeast: signaling from the secretory path to the nucleus via Wsc proteins and Pkc1p. Mol. Cell 8:281–289 [DOI] [PubMed] [Google Scholar]
- 31. Nikolaou E., et al. 2009. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol. Biol. 9:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oka T., Hamaguchi T., Sameshima Y., Goto M., Furukawa K. 2004. Molecular characterization of protein O-mannosyltransferase and its involvement in cell-wall synthesis in Aspergillus nidulans. Microbiology 150:1973–1982 [DOI] [PubMed] [Google Scholar]
- 33. Punt P. J., et al. 1990. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101–109 [DOI] [PubMed] [Google Scholar]
- 34. Punt P. J., Zegers N. D., Busscher M., Pouwels P. H., van den Hondel C. A. 1991. Intracellular and extracellular production of proteins in Aspergillus under the control of expression signals of the highly expressed Aspergillus nidulans gpdA gene. J. Biotechnol. 17:19–33 [DOI] [PubMed] [Google Scholar]
- 35. Rajavel M., Philip B., Buehrer B. M., Errede B., Levin D. E. 1999. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3969–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reinoso-Martín C., Schuller C., Schuetzer-Muehlbauer M., Kuchler K. 2003. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2:1200–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reissig J. L., Storminger J. L., Leloir L. F. 1955. A modified colorimetric method for the estimation of N-acetylamino sugars. J. Biol. Chem. 217:959–966 [PubMed] [Google Scholar]
- 38. Rispail N., et al. 2009. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 46:287–298 [DOI] [PubMed] [Google Scholar]
- 39. Rodicio R., Buchwald U., Schmitz H. P., Heinisch J. J. 2008. Dissecting sensor functions in cell wall integrity signaling in Kluyveromyces lactis. Fungal Genet. Biol. 45:422–435 [DOI] [PubMed] [Google Scholar]
- 40. Rodicio R., Heinisch J. J. 2010. Together we are strong—cell wall integrity sensors in yeasts. Yeast 27:531–540 [DOI] [PubMed] [Google Scholar]
- 41. Ronen R., et al. 2007. The Aspergillus nidulans pkcA gene is involved in polarized growth, morphogenesis and maintenance of cell wall integrity. Curr. Genet. 51:321–329 [DOI] [PubMed] [Google Scholar]
- 42. Ruijter G. J., et al. 2003. Mannitol is required for stress tolerance in Aspergillus niger conidiospores. Eukaryot. Cell 2:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Serrano R., Martin H., Casamayor A., Arino J. 2006. Signaling alkaline pH stress in the yeast Saccharomyces cerevisiae through the Wsc1 cell surface sensor and the Slt2 MAPK pathway. J. Biol. Chem. 281:39785–39795 [DOI] [PubMed] [Google Scholar]
- 44. Straede A., Heinisch J. J. 2007. Functional analyses of the extra- and intracellular domains of the yeast cell wall integrity sensors Mid2 and Wsc1. FEBS Lett. 581:4495–4500 [DOI] [PubMed] [Google Scholar]
- 45. Takeshita N., Ohta A., Horiuchi H. 2002. csmA, a gene encoding a class V chitin synthase with a myosin motor-like domain of Aspergillus nidulans, is translated as a single polypeptide and regulated in response to osmotic conditions. Biochem. Biophys. Res. Commun. 298:103–109 [DOI] [PubMed] [Google Scholar]
- 46. Teepe A. G., Loprete D. M., He Z., Hoggard T. A., Hill T. W. 2007. The protein kinase C orthologue PkcA plays a role in cell wall integrity and polarized growth in Aspergillus nidulans. Fungal Genet. Biol. 44:554–562 [DOI] [PubMed] [Google Scholar]
- 47. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Verna J., Lodder A., Lee K., Vagts A., Ballester R. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 94:13804–13809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wallis G. L., Easton R. L., Jolly K., Hemming F. W., Peberdy J. F. 2001. Galactofuranoic-oligomannose N-linked glycans of alpha-galactosidase A from Aspergillus niger. Eur. J. Biochem. 268:4134–4143 [DOI] [PubMed] [Google Scholar]
- 50. Wallis G. L., Hemming F. W., Peberdy J. F. 1999. Investigation of the glycosyltransferase enzymes involved in the initial stages of the N-linked protein glycosylation pathway in Aspergillus niger. Biochim. Biophys. Acta 1426:91–98 [DOI] [PubMed] [Google Scholar]
- 51. Witteveen C. F. B., Weber F., Busink R., Visser J. 1994. Isolation and characterization of two xylitol dehydrogenases from Aspergillus niger. Microbiology 140:1679–1685 [Google Scholar]
- 52. Yamazaki H., Tanaka A., Kaneko J., Ohta A., Horiuchi H. 2008. Aspergillus nidulans ChiA is a glycosylphosphatidylinositol (GPI)-anchored chitinase specifically localized at polarized growth sites. Fungal Genet. Biol. 45:963–972 [DOI] [PubMed] [Google Scholar]
- 53. Yelton M. M., Hamer J. E., Timberlake W. E. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. U. S. A. 81:1470–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zu T., Verna J., Ballester R. 2001. Mutations in WSC genes for putative stress receptors result in sensitivity to multiple stress conditions and impairment of Rlm1-dependent gene expression in Saccharomyces cerevisiae. Mol. Genet. Genomics 266:142–155 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.