Abstract
Microbial infections delay wound healing, but the effect of the composition of the wound microbiome on healing parameters is unknown. To better understand bacterial communities in chronic wounds, we analyzed debridement samples from lower-extremity venous insufficiency ulcers using the following: conventional anaerobic and aerobic bacterial cultures; the Ibis T5000 universal biosensor (Abbott Molecular); and 16S 454 FLX titanium series pyrosequencing (Roche). Wound debridement samples were obtained from 10 patients monitored clinically for at least 6 months, at which point 5 of the 10 sampled wounds had healed. Pyrosequencing data revealed significantly higher bacterial abundance and diversity in wounds that had not healed at 6 months. Additionally, Actinomycetales was increased in wounds that had not healed, and Pseudomonadaceae was increased in wounds that had healed by the 6-month follow-up. Baseline wound surface area, duration, or analysis by Ibis or conventional culture did not reveal significant differences between wounds that healed after 6 months and those that did not. Thus, pyrosequencing identified distinctive baseline characteristics of wounds that did not heal by the 6-month follow-up, furthering our understanding of potentially unique microbiome characteristics of chronic wounds.
INTRODUCTION
Chronic wounds represent an increasing public health burden, driven by an aging population and the growing incidence of diabetes mellitus and obesity worldwide (34). Wounds that do not progress through the normal stages of healing have diverse etiologies and usually occur in patients with predisposing conditions, such as venous disease, arterial and vascular compromise, neuropathy, and immunosuppression, making the host more susceptible to pathogenic factors, including microbial colonization and the infection of the wound (11, 27). Microbial infections are recognized as one of the many destructive processes that delay wound healing, particularly in the form of biofilms. Biofilms are composed of microbes embedded in a polymeric matrix that protects them from antimicrobials and allows them to resist host defenses. The study of biofilms has introduced a new paradigm of chronic microbial infections: instead of single free-floating (planktonic) microbes causing disease patterns that can be reproduced by following Koch's postulates, biofilms are metabolically quiescent, sessile, polymicrobial communities in which synergistic relationships between microbes can alter virulence and pathogenicity (2, 9, 17). Thus, understanding the effect of biofilms on wound healing requires the study of the variation of the polymicrobial communities (microbiome) in relation to wound-healing parameters.
Conventional clinical diagnostic culture methods are biased largely toward the 2% of microorganisms that are able to grow rapidly in standard clinical culture media and are presumed to be significant (5). Genomic tools such as nucleic acid amplification, DNA sequencing, and the development of microbial ribosomal clone libraries have greatly expanded our view of wound microbial flora. As the genomic tools used to obtain the molecular signature of complex microbiomes are relatively new, more information is required to establish which microbes are deleterious and impair healing, which are benign colonizers, and which may even facilitate healing processes (22). In spite of the lack of understanding of the microbial composition of chronic wounds, topical and systemic antibacterial agents are used liberally in wound care, possibly promoting the development of resistant bacterial strains and/or killing potentially beneficial bacteria. Therefore, although microbial colonization may be only one aspect of the complex process of delayed healing, it is critical to understand the relationship between the presence of microbes and chronic wounds to further develop evidence-based treatment strategies.
Few studies have compared conventional diagnostic techniques to newly emerging diagnostic methods. This study therefore uses two molecular diagnostic modalities, the Ibis T5000 universal biosensor (Abbott Molecular) and tag-encoded FLX amplicon pyrosequencing (Research and Testing Laboratory), in addition to conventional anaerobic and aerobic bacterial cultures, to identify microorganisms in debridement samples from the wounds of patients with chronic venous insufficiency ulcers (CVU). CVUs account for 70 to 90% of chronic leg wounds (34). We chose to focus on this type of wound to standardize the etiology and body location of wounds being studied. After sampling, patients were followed clinically for at least 6 months to enable the comparison of microbial communities as determined by the different analytic methods in relation to wound-healing parameters, including time to healing, baseline duration of the wound, and baseline wound surface area.
The Ibis T5000 universal biosensor has several potential advantages over conventional culture as a diagnostic modality in the clinical microbiology laboratory, including the rapid and high-throughput testing of samples with a low per-sample analysis cost (8). To our knowledge, Ibis has not previously been compared to the conventional culture of chronic wounds. Ibis uses multiple PCR primer pairs to amplify selected regions of the genome of microbes and then uses high-performance electrospray ionization (ESI) time-of-flight (TOF) mass spectrometry (MS) to determine molecular sizes of the denatured amplicons, which then can be used to determine their base compositions. The base compositions from multiple primer pairs are used to triangulate the identity of the organisms present in the sample. The identity of microbes present at ∼3% or above the total microbial population in a sample can be deduced (8).
In contrast, pyrosequencing uses a single set of PCR primers targeting a highly conserved area of the 16S ribosomal subunit gene flanking variable areas. The sequencing of the PCR products then is used to identify microbes as well as their relative abundance within the given sample (4, 5, 15). In this study, the identity of microbes present at 0.02% of the total population in a sample were deduced by pyrosequencing, thus providing a comprehensive view of the microbial communities of a sample. Both of the molecular approaches make possible the detection of microbes without the time requirement for growth and environmental selection pressures inherent in the culturing process. The comparison of the different molecular approaches was performed in this study to determine the utility of the increased depth of identification provided by pyrosequencing as well as the comparison of the different approaches to the detection of microbes in wounds to further understand the unique microbiome characteristics of nonhealing wounds.
MATERIALS AND METHODS
Patient recruitment.
Patients were recruited from the Akron General Multidisciplinary Wound Center. Inclusion criteria included an age of more than 18 years and at least one venous insufficiency wound present for at least 1 month and requiring regular debridement as part of standard-of-care treatment. Patients were classified as having a venous insufficiency wound if their wound was located in the gaiter area of the leg (area extending from the midcalf to approximately one inch above the malleolus) and the patient had a history of lower leg edema that improved with leg elevation, or if the patient had other cutaneous findings of venous disease (venous blush or flare, varicose veins, or venous valvular incompetence). If a patient had more than one qualifying wound, one wound was randomly selected for the study. Also, the patient or legally authorized representative had to be able to read and sign informed consent. Patients with wounds that did not fit criteria for a venous insufficiency wound as defined above or expressed unwillingness to perform requirements of the study were excluded.
Sample collection.
During the initial study visit, informed consent was obtained and information was collected from the patient, including age, gender, patient-reported duration of the wound (in months), number of wounds, stage of wound, undermined wound margin, time since last antibiotic administration (topical or systemic), and other medication usage and comorbidities. Additionally, skin assessment, wound measurements, and photography of the affected area were conducted. Wound size was determined by measuring the maximum length of the wound and multiplying it by the maximum width perpendicular to that length, a method that closely approximates planimetric techniques (19). Debridement samples were collected after the wound was washed with sterile isotonic sodium chloride (saline) and anesthetized with topical 4% lidocaine under occlusion with sterile gauze for 5 min. Using the method established by Frankel et al. (13), a sterile 3-mm curette was used to collect debridement samples. Debridement samples were systematically collected from the most proximal wound edge, the most distal wound edge, and center of the wound for consistency in wound sampling. Approximately 100 mg of debrided tissue was collected and stored at −80°C to be processed for DNA extraction. No change in the patient's treatment was made as part of participation in this study.
Bacterial culture.
During debridement, samples were collected for aerobic and anaerobic bacterial culture, obtaining swabs of both the debrided tissue and base of the wound in the same areas of debridement (Copan Diagnostics ESwab 480C). The swab was placed in the ESwab transport medium and immediately delivered for aerobic and anaerobic bacterial culture in the Clinical Laboratory Improvement Amendments (CLIA) certified laboratory of University Hospitals Case Medical Center. Cultures were prepared by plating on BBL Trypticase soy agar with 5% sheep blood, MacConkey II, chocolate II, and CDC anaerobe 5% sheep blood agar. Cultures were incubated at 35°C aerobically and anaerobically (Remel AnaeroPack System) and deemed negative after 48 h of aerobic growth and 72 h of anaerobic growth.
DNA extraction.
Debridement samples were collected as detailed above into a sterile 2-ml tube and stored at −80°C. DNA extraction was performed using the MasterPure yeast DNA purification kit (no. MPY80200; Epicentre Biotechnolgies) as outlined in the manufacturer's protocol. Samples were further prepared for extraction by vortexing for 15 min in the kit's yeast cell lysis solution, heating to 65°C for 15 min, and then repeating 15 min of vortexing and heating once. Additionally, DNA samples diluted to 200 μg/ml in Tris-EDTA buffer were incubated with RNase (5 mg/ml stock at 1 μl per 100-μl sample) for 30 min at 37°C. Phenol-chloroform–isoamyl alcohol was added at equal volumes to the DNA sample, vortexed for 10 s, and centrifuged briefly to pellet debris. DNA was precipitated from the aqueous phase by adding an equal volume of 100% ethanol, incubating for 1 h at −80°C, and centrifuging at 14,000 rpm in an Eppendorf 5417C at 4°C for 10 min. The DNA pellet was washed with 500 μl of 70% ethanol and resuspended in 40 μl H2O.
bTEFAP sequence-processing pipeline and analysis. (i) Massively parallel bTEFAP.
Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) was performed using standard research methods as previously described (6) and was based upon 16S primers Gray28F 5′-GAGTTTGATCNTGGCTCAG and Gray519R 5′-GTNTTACNGCGGCKGCTG. Tag-encoded FLX amplicon pyrosequencing analyses utilized the Roche 454 FLX instrument with titanium reagents, titanium procedures (Roche), a one-step PCR, a mixture of Hot Start and HotStar high-fidelity Taq polymerases, and amplicons originating and extending from 28F for bacterial diversity based upon procedures developed by and performed at the Research and Testing Laboratory (RTL; Lubbock, TX) based upon RTL protocols.
(ii) Bacterial diversity data analysis.
Following sequencing, data were depleted of all reads <250 bp in length after the trimming of low-quality sequence ends, primers, and tags as well as any nonbacterial ribosome sequences and definite chimeras using custom software as previously described (16). To determine the identity of bacteria in the remaining sequences, a distributed BLAST algorithm (Kraken BLAST) was run against a database of high-quality 16S bacterial sequences derived from NCBI (1 August 2010 version). Using a.NET and C# analysis pipeline (Research and Testing Laboratory), the resulting BLASTn outputs were compiled and validated, and data reduction analysis was performed as described previously (26, 39). Based upon the BLASTn-derived sequence identity score (the percentage of alignment of the query sequence with a given database sequence), taxonomic levels were assigned based on the following: sequence identity scores of >97% (<3% divergence) were resolved at the species level, ≤97 and >95% at the genus level, ≤95 and >90% at the family level, ≤90 and >85% at the order level, ≤85 and >80% at the class level, and ≤80 and >77% at the phylum level. The percent abundance of each microbe then was determined for each sample based on the identity and numbers of sequence reads. Low relative percentages of organisms were not considered; only organisms with >0.20% identity scores were reported. Evaluations presented at each taxonomic level, including percent compilations, represent all sequences resolved to their primary identification or their closest relative taxonomic designation allowing any unknown or novel organisms to be characterized (35). Unifrac-like (20) and diversity index analyses based upon Mothur (33) were performed as described previously (15). Further statistical analyses, including the determination of the kappa statistic, McNemar's test, and Wilcoxon signed-rank test, were performed using JMP 9.0.0 software (SAS Institute, Inc.).
Ibis T5000 assay.
For the Ibis T5000 assay, an aliquot of each sample was loaded into each of the 16 wells of a 96-well bacterial artificial chromosome (BAC) detection PCR plate (PN 05N13-01; Abbott Molecular). Each of the 16 wells for each specimen contained a different broad-range primer set (14, 24). An internal calibrant consisting of a synthetic nucleic acid template was included in each assay, controlling for false negatives (e.g., from PCR inhibitors) and enabling a semiquantitative analysis of the amount of template DNA present. PCR amplification was carried out per Ecker et al. (7). The PCR products then were desalted in a 96-well plate format and sequentially electrosprayed into the TOF MS. The spectral signals were processed to determine the masses of each strand of each of the PCR products with sufficient accuracy so that the base composition of each amplicon could be deduced. Using combined base compositions from multiple PCRs, the identities of the microbes and a semiquantitative determination of their relative concentrations in the starting sample were established by using a proprietary algorithm to interface with the Ibis database of known organisms (8). Microbes were reported that were not present in the negative control, identified at >0.65 confidence, and detected by more than one primer detection.
RESULTS
Patient information is summarized in Table 1. Samples were collected from 10 patients, with ages ranging from 42 to 88 (average age, 68) years, and three were women. Patients were monitored for at least 6 months. Second samples were obtained from patients 1, 3, 4, and 8 within 10 to 14 weeks of the first collection for a total of 14 samples. Patients 5, 6, 7, 9, and 10 had healed before 5 months, and patients 1, 2, 3, 4, and 8 had not healed after more than 6 months. As the second samples were obtained only from nonhealed wounds, the data from the second samples was not included in comparisons of nonhealed and healed wounds. Patients had wounds of the same etiology and were treated at the same wound center, and thus they received the same standard of care. Antimicrobial use was not otherwise controlled in this study, and all but one of the patients had been using either a topical or a systemic antibiotic or antiseptic agent at the time of initial sample collection, as shown in Table 1. Two patients had type II diabetes mellitus (patients 5 and 7), both of whom controlled the condition with diet and oral hypoglycemic medications, were without history of neuropathic ulcerations, and had healed wounds at the 6-month follow-up.
Table 1.
Patient informationc
| Patient no. | Wound location | Age (yr) | Gender | Wound area (cm2) | Wound durationa (mo) | Time to healingb (mo) | Antibiotics and antiseptics |
|---|---|---|---|---|---|---|---|
| 1 | Right medial leg | 70 | M | 4.71 | 8.0 | Not healed | TAS: yes (PHB); TAB: no; SAB: no |
| 2 | Left lateral leg | 61 | M | 51.04 | >120 | Not healed | TAS: yes (PHB and cadexomer iodine); TAB: no; SAB: no |
| 3 | Right medial leg | 46 | F | 34.56 | >120 | Not healed | TAS: yes (SSD); TAB: no; SAB: yes (ciprofloxacin discontinued 3 wk prior) |
| 4 | Right medial leg | 82 | M | 12.92 | 68.6 | Not healed | TAS: no; TAB: yes (mupirocin); SAB: no |
| 5 | Right medial leg | 84 | M | 18.02 | 1.9 | 2 | TAS: yes (SSD, mupirocin); TAB: no; SAB: yes (cephalexin discontinued 2 wk prior) |
| 6 | Right medial leg | 88 | M | 2.2 | 11.2 | 4 | TAS: yes (SSD and silver dressing); TAB: no; SAB: no |
| 7 | Right medial leg | 80 | M | 1.27 | 1.0 | 3.25 | TAS: yes (SSD); TAB: no; SAB: no |
| 8 | Right anterior shin | 45 | M | 1.79 | 7.0 | Not healed | TAS: no; TAB: no; SAB: yes (TMP-SMX) |
| 9 | Left lateral leg | 78 | F | 1.0 | 13.8 | 4.75 | TAS: yes (silver dressing); TAB: no; SAB: no |
| 10 | Left medial leg | 42 | F | 6.6 | 1.7 | 2.4 | TAS: no; TAB: no; SAB: no |
Wound duration at time of first sample per patients' report.
Time to healing after first sample, with follow-up of at least 6 months.
Abbreviations: TAS, topical antiseptic; TAB, topical antibiotic; SAB, systemic antibiotic; PHB, polyhexanide biguanide; SSD, silver sulfadiazine; TMP-SMX, trimethoprim sulfamethoxazole; F, female; M, male.
Table 2 summarizes the number of different taxa reported by each analytic method. Pyrosequencing identified significantly more members at each taxonomic level, with Ibis showing improved performance compared to that of culture. Sequence lengths from pyrosequencing averaged 400 bp, with a total of 154,376 sequences after quality reductions were analyzed across 14 samples studied (averaging 4,828 per sample). A total of 143,190 sequences were identified with >95% identity to known bacteria (genus-level identity); 133,412 sequences were able to be resolved at >97% identity and 83,478 at >99% identity.
Table 2.
Number of taxa identified by each analytic method
| Taxon | No. identified by: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pyrosequencing |
Ibis T5000 |
Culture |
|||||||
| Total | Range | Mean (SE) | Total | Range | Mean (SE) | Total | Range | Mean (SE) | |
| Phylum | 6 | 2–5 | 3.43 (0.25) | 4 | 1–3 | 1.78 (0.19) | 2 | 1–2 | 1.07 (0.18) |
| Class | 11 | 3–7 | 4.64 (0.37) | 8 | 1–5 | 2.29 (0.30) | 3 | 1–2 | 1.07 (0.18) |
| Order | 15 | 3–8 | 5.71 (0.51) | 13 | 1–5 | 2.86 (0.36) | 5 | 1–2 | 1.07 (0.18) |
| Family | 27 | 3–12 | 7.86 (0.78) | 15 | 1–5 | 2.93 (0.37) | 7 | 1–2 | 1 (0.20) |
| Genus | 43 | 3–17 | 9.64 (1.04) | 20 | 1–8 | 3.50 (0.52) | 7 | 1–2 | 1 (0.20) |
| Species | 55 | 4–15 | 8.78 (0.87) | 29 | 1–7 | 3.29 (0.50) | 8 | 1–2 | 1 (0.20) |
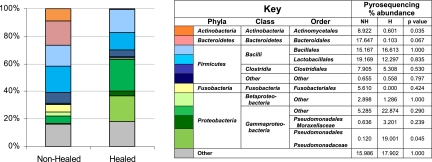
The more comprehensive data obtained from pyrosequencing showed significant differences when the data from baseline samples were grouped as nonhealed or healed based on whether the patient was healed at 6 months (Fig. 1). Figure 1 illustrates the more diverse microbial populations in nonhealed wounds identified by pyrosequencing. In comparisons of the percent abundance of taxa from the nonhealed to the healed samples according to pyrosequencing and using the Wilcoxon signed-rank test, there were significantly more Actinomycetales and a trend toward more Bacteroidales in nonhealed wounds. Members of the class Gammaproteobacteria, and specifically the family Pseudomonadaceae, were elevated in healed wounds.
Fig. 1.
Taxa identified by pyrosequencing. The stacked graph illustrates the relative abundance of each taxon identified by pyrosequencing from the data from baseline samples grouped as nonhealed (NH) or healed (H) based on whether the patient was healed at 6 months (color coded according to the key). The table shown in the key details the total percent abundance of each taxon in the nonhealed versus healed wounds, showing significantly more Actinomycetales in nonhealed wounds and Pseudomonadaceae in healed wounds.
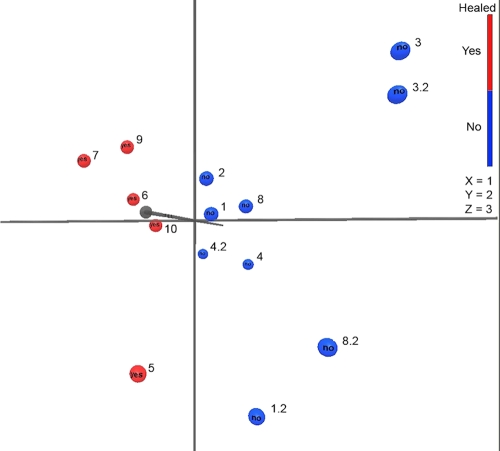
The increase in bacterial diversity in wounds not healed by 6 months was substantiated by microbial diversity analyses of the pyrosequencing data, performed as previously described (35). Table 3 shows the microbial diversity estimates obtained with the parametric and nonparametric modeling of rarefaction, Shannon diversity index, and abundance-based coverage estimator (ACE) for the initial samples. Divergence of 3 and 5% is indicative of sequences differing at the species and genus levels, respectively. Table 4 summarizes the data from Table 3, with averages of patients grouped as nonhealed or healed based on their status at 6 months and showing statistically significantly higher abundance and diversity levels in nonhealed wounds. No statistically significant differences were found when bacterial abundance and diversity estimates were grouped according to baseline wound area (<15 cm2 versus >15 cm2 or <10 cm2 versus >10 cm2) or wound duration (<24 months versus >24 months or <12 months versus >12 months) (data not shown). Baseline wound surface area, duration, or analysis by Ibis or conventional culture did not reveal significant differences between wounds that healed by 6 months and those that did not. However, as shown in Table 4, both baseline wound surface area and duration trended toward significance; thus, inadequate sample size could explain the lack of significance for these measures. An unweighted principle component analysis (PCA) was performed as previously described (15, 35) on the pyrosequencing data from all of the samples. Figure 2 shows the PCA plot, illustrating clear and significant (P < 0.05) separation of wound samples from patients not healed (blue) and healed (red) at 6 months.
Table 3.
Diversity and richness data for wound samplesa
| Patient no. | Wound healedb | OTU 3% | OTU 5% | Shannon 3% | Shannon 5% | ACE 3% | ACE 5% | Cleaned 3% | Assemble 3% | Clusters 3% |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | No | 136 | 55 | 4.15 | 3.22 | 163 | 59 | 10.18 | 12 | 14 |
| 2 | No | 469 | 181 | 5.47 | 4.45 | 565 | 204 | 11.2 | 12 | 10 |
| 3 | No | 652 | 321 | 5.84 | 5.00 | 816 | 384 | 14 | 13 | 14 |
| 4 | No | 326 | 148 | 4.60 | 3.66 | 397 | 180 | 11 | 10 | 13 |
| 8 | No | 814 | 391 | 5.98 | 5.04 | 1077 | 478 | 12.9 | 13 | 13 |
| 5 | Yes | 65 | 34 | 3.36 | 2.80 | 72 | 37 | 8.3 | 6 | 7 |
| 6 | Yes | 89 | 36 | 3.78 | 2.97 | 117 | 37 | 8.6 | 10 | 8 |
| 7 | Yes | 38 | 20 | 2.88 | 2.31 | 63 | 26 | 6.2 | 5 | 6 |
| 9 | Yes | 202 | 80 | 4.74 | 3.89 | 242 | 87 | 8.2 | 8 | 7 |
| 10 | Yes | 201 | 92 | 4.35 | 3.56 | 229 | 105 | 8.2 | 7 | 7 |
OTU, operational taxonomic unit; ACE, abundance-based coverage estimator; Shannon, Shannon diversity index; 3%, index derived from sequences with <3% divergence (>97% alignment with database sequence); 5%, index derived from sequences with <5% divergence (>95% alignment with database sequence).
Six months after baseline sample.
Table 4.
Comparison of averages of nonhealed and healed woundsa
| Wound status | OTU 3% | OTU 5% | Shannon 3% | Shannon 5% | ACE 3% | ACE 5% | Cleaned 3% | Assemble 3% | Clusters 3% | Wound area (cm2) | Wound durationb (mo) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nonhealed | 479 | 219 | 5.21 | 4.28 | 603 | 261 | 11.86 | 12.0 | 12.8 | 21.0 | 64.7 |
| Healed | 119 | 52 | 3.82 | 3.11 | 145 | 58 | 7.90 | 7.2 | 7.0 | 5.8 | 5.92 |
| P value | 0.036 | 0.049 | 0.022 | 0.035 | 0.043 | 0.053 | 0.002 | 0.002 | 0.001 | 0.189 | 0.079 |
OTU, operational taxonomic unit; ACE, abundance-based coverage estimator; Shannon, Shannon diversity index; 3%, index derived from sequences with <3% divergence (>97% alignment with database sequence); 5%, index derived from sequences with <5% divergence (>95% alignment with database sequence).
Wound duration at the time of the first sample per patients' reports.
Fig. 2.
Principle component analysis for wound samples. Principle component analysis plot showing separation of wound samples from patients nonhealed (blue) and healed (red) at 6 months. All samples were included in this analysis, with second samples as x.2, where x is the patient number.
Agreement between culture, Ibis, and pyrosequencing was estimated for genera identified by each method, as shown in Tables 5 and 6. The kappa statistic (κ) was used to estimate the agreement between tests and can take on values between 1 (indicating positive agreement between tests) and −1 (indicating negative agreement between tests), with 0 indicating that agreement between tests is equal to chance alone. Table 5 shows no evidence of agreement between culture and pyrosequencing and culture and Ibis, except for the genus Enterobacter, which had total agreement between culture and both Ibis and pyrosequencing. Table 6 shows evidence of agreement between Ibis and pyrosequencing for Streptococcus and Bacteroides, as well as total agreement between Enterobacter, Neisseria, Serratia, and Moraxella.
Table 5.
Agreement between culture, Ibis T5000, and pyrosequencinga
| Test result |
Staphylococcus |
Streptococcus |
Enterobacter |
Proteus |
Pseudomonas |
Alcaligenes |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cx− | Cx+ | κ | M | Cx− | Cx+ | κ | M | Cx− | Cx+ | κ | M | Cx− | Cx+ | κ | M | Cx− | Cx+ | κ | M | Cx− | Cx+ | κ | M | |
| Ibis− | 1 | 0 | 0.143 (−0.124, 0.410) | 6 (0.0143) | 8 | 0 | 0.364 (−0.043, 0.770) | 4 (0.0455) | 12 | 0 | 1 (1, 1) | 0 (1.00) | 12 | 1 | −0.0769 (−0.183, 0.029) | 0 (1.00) | 11 | 1 | 0.417 (−0.252, 1.085) | 0 (1.00) | 13 | 1 | NA | NA |
| Ibis+ | 6 | 7 | 4 | 20 | 2 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | ||||||||||||
| Pyro− | 3 | 2 | 0.143 (−0.354, 0.640) | 0.667 (0.414) | 5 | 0 | 0.169 (−0.073, 0.412) | 7 (0.0082) | 12 | 0 | 1 (1, 1) | 0 (1.00) | 10 | 0 | 0.323 (−0.177, 0.822) | 3 (0.0833) | 8 | 0 | 0.364 (−0.043, 0.770) | 4 (0.0455) | 11 | 0 | 0.44 (−0.155, 1.035) | 2 (0.1573) |
| Pyro+ | 4 | 5 | 7 | 2 | 0 | 2 | 3 | 1 | 4 | 2 | 2 | 1 | ||||||||||||
Cx−/Cx+, number of culture samples negative or positive, respectively, for Cx; Ibis−/Ibis+, number of samples negative or positive, respectively, for Ibis; Pyro−/Pyro+, number of samples negative or positive, respectively, for pyrosequencing; κ, kappa statistic (95% confidence interval); M, result for McNemar's test (P).
Table 6.
Agreement between Ibis T5000 and pyrosequencinga
| Genus and parameter | Pyro− | Pyro+ |
|---|---|---|
| Staphylococcus | ||
| Ibis− | 1 | 0 |
| Ibis+ | 4 | 9 |
| κ | 0.243 (−0.166, 0.653) | |
| M | 4 (0.046) | |
| Streptococcus | ||
| Ibis− | 5 | 3 |
| Ibis+ | 0 | 6 |
| κ | 0.588 (0.212, 0.965) | |
| M | 3 (0.083) | |
| Enterobacter | ||
| Ibis− | 12 | 0 |
| Ibis+ | 0 | 2 |
| κ | 1 (1, 1) | |
| M | 0 (1.0) | |
| Proteus | ||
| Ibis− | 10 | 3 |
| Ibis+ | 0 | 1 |
| κ | 0.323 (−0.178, 0.822) | |
| M | 3 (0.083) | |
| Pseudomonas | ||
| Ibis− | 8 | 4 |
| Ibis+ | 0 | 2 |
| κ | 0.364 (−0.043, 0.770) | |
| M | 4 (0.046) | |
| Bacteroides | ||
| Ibis− | 9 | 3 |
| Ibis+ | 0 | 2 |
| κ | 0.462 (0.0064, 0.917) | |
| M | 3 (0.083) | |
| Clostridium | ||
| Ibis− | 12 | 1 |
| Ibis+ | 0 | 1 |
| κ | 0.632 (−0.0153, 1.278) | |
| M | 1 (0.317) | |
| Finegoldia | ||
| Ibis− | 10 | 3 |
| Ibis+ | 0 | 1 |
| κ | 0.323 (−0.178, 0.822) | |
| M | 3 (0.083) | |
| Peptostreptococcus | ||
| Ibis− | 12 | 1 |
| Ibis+ | 1 | 0 |
| κ | −0.077 (−0.183, 0.029) | |
| M | 0 (1.0) | |
| Neisseria | ||
| Ibis− | 12 | 0 |
| Ibis+ | 0 | 2 |
| κ | 1 (1, 1) | |
| M | 0 (1.0) | |
| Escherichia | ||
| Ibis− | 9 | 3 |
| Ibis+ | 2 | 0 |
| κ | −0.207 (−0.403, −0.001) | |
| M | 0.2 (0.655) | |
| Serratia | ||
| Ibis− | 13 | 0 |
| Ibis+ | 0 | 1 |
| κ | 1 (1, 1) | |
| M | 0 (1.0) | |
| Moraxella | ||
| Ibis− | 13 | 0 |
| Ibis+ | 0 | 1 |
| κ | 1 (1, 1) | |
| M | 0 (1.0) | |
Cx−/Cx+ number of culture samples negative or positive, respectively, for Cx; Ibis−/Ibis+, number of samples negative or positive, respectively, for Ibis; Pyro−/Pyro+, number of samples negative or positive, respectively, for pyrosequencing; κ, kappa statistic (95% confidence interval); M, result for McNemar's test (P).
Alternatively, McNemar's test was used to compare the proportion of discordant detection of the genera identified by each method. Results indicated evidence of a difference in the proportion of Staphylococcus detection by Ibis compared to those detected by culture and pyrosequencing, as well as Pseudomonas detection by pyrosequencing compared to those by culture and Ibis. Streptococcus also was disproportionately detected by Ibis and pyrosequencing compared to culture. Although these statistical analyses are limited by low sample numbers, in general our data show that discordance between culture and pyrosequencing was due to culture-negative/pyrosequencing-positive discordance in all genera except Staphylococcus, making culture-positive/pyrosequencing-negative discordance rare. Discordance between culture and Ibis occurred in more genera, including Proteus, Pseudomonas, and Alcaligenes. Ibis-positive/pyrosequencing-negative discordance occurred in three genera, including Staphylococcus, Peptostreptococcus, and Escherichia, as opposed to eight genera with pyrosequencing-positive/Ibis-negative discordance. This suggests that even for the genera that were detected by both Ibis and pyrosequencing, pyrosequencing detected them more frequently. Genera that were reported by pyrosequencing but not Ibis T5000 in these samples include the following: Actinomyces, Brevibacterium, Corynebacterium, Enterococcus, Peptoniphilus, Parvimonas, Anaerococcus, Helcococcus, Alcaligenes, and Acinetobacter. Of note, Brevibacterium and Corynebacterium were detected by Ibis in some samples but were not reported based on quality control standards.
DISCUSSION
Modern molecular tools provide powerful means to define chronic wound microbial communities. To our knowledge, this is the first study to use longitudinal healing parameters to contrast the baseline bacterial composition of wounds revealed by pyrosequencing. We found that wounds that did not heal by 6 months had increased bacterial abundance and diversity compared to that of wounds that healed within 6 months, with significant increases in members of Actinomycetales in nonhealed wounds and Pseudomonadaceae in wounds that healed. No significant differences were found when bacterial abundance and diversity were compared to other baseline measurements of wound surface area and patient-reported wound duration. Additionally, baseline area, wound duration, or analysis by Ibis or conventional culture did not reveal significant differences between wounds that healed by 6 months and those that did not. However, wound area and duration measures did trend toward significance in this study and have been shown in other studies to be reasonable prognostic indicators for healing (21); thus, the lack of significance in these measures may be related to small sample size.
Although the relationship between community diversity and stability has been widely debated, there is emerging consensus that diversity increases stability on the level of communities (15, 23, 28, 29). A recent comparison of bacterial composition in diabetic ulcers and contralateral intact skin showed decreased diversity and increased microbial abundance in diabetic ulcers compared to those of contralateral intact skin (15). Gontcharova et al. imply that the more diverse flora of the intact skin may contribute to its robust environment, protecting the ecosystem from the spread of infection or accumulation of opportunistic and pathogenic populations. Gontcharova et al. also found intact skin and wounds to have distinctly different microbial communities with little effect on one another (15). Our study, showing less diversity in wounds that heal, suggests that it is unlikely that the microbial communities of wounds become more similar to intact skin throughout the healing process but rather shift to become less diverse with healing. The further study of larger patient populations monitored longitudinally is needed to more fully understand the role of microbial diversity and biofilm stability throughout the healing process. The role of host innate immunity in this increase in microbial diversity in chronic wounds also will be an important consideration in future studies.
Other chronic diseases in which the microbiome has been longitudinally studied have shown a similar increased diversity associated with an aberrant microbiota and infection, including bacterial vaginosis (BV) and celiac disease (18, 31, 32). In BV, the bacterial diversity decreases and the lactobacillus abundance increases with recovery, coinciding with a decrease in members of the phyla Actinobacteria and Bacteroides (18). In the current study, wounds that healed within 6 months had fewer members of Actinobacteria and Bacteroides than wounds that did not heal, but further longitudinal study is needed to understand whether increases in the abundance of other microbes are associated with healing. We found an increase in the family Pseudomonadaceae in wounds that healed within 6 months compared to nonhealed wounds, although the significance of this is unclear as Pseudomonas species generally are regarded as pathogenic in wounds (10, 41); however, we have previously observed very high levels of Pseudomonas species in the normal nasopharynx, suggesting that this species is important in the normal flora (24). In a study by Price et al., increased Pseudomonadaceae colonization was shown to be associated with antibiotic use in wounds, although no trends in bacterial taxa diversity values were evident among the patients treated with antibiotics and those left untreated, and longitudinal healing trends were not evaluated (27). As we did not control for antibiotic administration, we are unable to evaluate the contribution of antibacterial use to Pseudomonas colonization or other aspects of our data. A speculative explanation for the relationship between increased Pseudomonadaceae and decreased diversity observed in our study is the production of potent bacteriocins by Pseudomonas species that can kill other microbes, as has been observed in lungs of cystic fibrosis (CF) patients (1). However, increased Pseudomonas species and decreased overall bacterial diversity in CF patients portends a poor prognosis rather than clinical improvement, as was found for the wounds that healed in our study (40). Differences in pathophysiology between CF and wounds make further comparison difficult. More study is needed to determine whether the shift toward Pseudomonadaceae with healing and antibiotic treatment is an important factor in wound healing, constituting a shift to a more benign microbiome.
In the present study, the microbes that were increased in nonhealed wounds, particularly of the phyla Actinobacteria and Bacteroidetes, consisted mostly of anaerobes and facultative anaerobes, supporting previous reports of anaerobes as an important population in chronic wound biofilms (36, 38). Although the superficial surface of wounds is exposed to air, anaerobes may survive by symbiotically existing with aerobic bacteria or existing within internal, oxygen-depleted regions of biofilms (3, 30). Anaerobes likely contribute to delayed reepithelialization and defective extracellular matrix reorganization and angiogenesis, as shown in vitro (36). In spite of the likely importance of anaerobic pathogens in wounds and the use of conventional anaerobic culture methods for every sample in this study, no anaerobic species were found using conventional culture methods. Many organisms, such as anaerobic and other fastidious bacteria, typically will not be diagnosed with routine clinical diagnostic culture methods, as they require very specialized collection and handling, as well as specialized growth media and long incubation periods (often >14 days) that are outside the standard operating procedures of most diagnostic laboratories (37). On average, the suggested incubation time for many culture methods is 3 to 5 days to maintain the cost-effectiveness of culture based diagnostic (12, 25). We found that even the detection of culturable bacteria such as Staphylococcus and Streptococcus was consistently greater by the molecular diagnostic methods than by conventional culture. At least some of the differences between the culture and molecular methods in this study could be due to the inability to differentiate species on the culture plates. The differences also may be attributable to a fraction of the bacteria being dead or in a viable but unculturable state (such as a more dormant state within the biofilm) or from the recent use of topical or systemic antimicrobials. While the potential bias exists for the detection of nonviable bacteria with molecular methods, our data suggest that underestimation by conventional culture-based methods is more likely.
Comparison of the data from Ibis T5000 and pyrosequencing revealed that, for genera detected by both methods, pyrosequencing-positive/Ibis-negative discordance was roughly three times more common than Ibis-positive/pyrosequencing-negative discordance. This difference most likely is due to the increased depth of coverage provided by pyrosequencing. Ibis cannot reliably identify species that make up less than 3% of the microbiome, whereas pyrosequencing can detect species that are several orders of magnitude less numerous, as it provides for the direct sequencing of each 16S molecule. Ibis thus is more sensitive to factors that influence the sample DNA levels, including cell lysis methods for DNA extraction and the efficiency of PCR amplification, making studies that use different extraction and PCR methods difficult to compare. In this study, the increased depth of coverage provided by pyrosequencing led to the identification of differences between nonhealed and healed wounds, whereas no differences between the two populations could be detected by Ibis. Thus, the use of the more comprehensive sequencing technologies in studies of larger, longitudinally sampled patient populations likely will be important in further defining the significant differences between the microbiomes of nonhealing and healing wounds. More information on the microbiomes that lead to delayed healing could enable the more targeted use of the less comprehensive technologies, such as different primer targets for Ibis (using standardized DNA extraction and PCR amplification techniques) or the isolation of the key microbes in culture, leading to the more cost-effective and meaningful use of these clinically applicable technologies.
ACKNOWLEDGMENTS
M.S.T. is supported by the National Psoriasis Foundation Medical Dermatology Fellowship and the Dermatology Foundation Dermatologist Investigator Research Fellowship. M.A.G. is supported by funds from the National Institutes of Health (NIH) (BRS ACURE-Q0600136) to the Oral HIV/AIDS Research Alliance (OHARA), NIH/NIAID (RO1 AI035097), and NIH/NIDCR (R01 DE017486-01A1 and R01DE 13932-4). P.M. is supported by funds from the NIH/NIAID (1R21AI074077-01A2). G.D.E. is supported by grants from the NIH (R01 AI080935-01, R01 DC02148, U19AI084024, and U01 DK082316) and is a consultant to Abbott Molecular. F.Z.H. is supported by a grant from Abbott Molecular.
S.E.D. is a coowner and CEO of Pathogenius Diagnostics, a clinical diagnostic facility specializing in wound diagnostics, and is a coowner and CEO of Research and Testing Laboratory, which develops molecular diagnostic methods for licensing purposes and performs fee-for-service and collaborative research in molecular biology and bioinformatics.
Footnotes
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Bakkal S., Robinson S. M., Ordonez C. L., Waltz D. A., Riley M. A. 2010. Role of bacteriocins in mediating interactions of bacterial isolates taken from cystic fibrosis patients. Microbiology 156:2058–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borriello G., et al. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 48:2659–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bradshaw D. J., Marsh P. D., Allison C., Schilling K. M. 1996. Effect of oxygen, inoculum composition and flow rate on development of mixed-culture oral biofilms. Microbiology 142:623–629 [DOI] [PubMed] [Google Scholar]
- 4. Dowd S. E., et al. 2011. Survey of fungi and yeast in polymicrobial infections in chronic wounds. J. Wound Care 20:40–47 [DOI] [PubMed] [Google Scholar]
- 5. Dowd S. E., et al. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dowd S. E., et al. 2008. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 3:e3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ecker D. J., et al. 2009. Molecular genotyping of microbes by multilocus PCR and mass spectrometry: a new tool for hospital infection control and public health surveillance. Methods Mol. Biol. 551:71–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ecker D. J., et al. 2008. Ibis T5000: a universal biosensor approach for microbiology. Nat. Rev. Microbiol. 6:553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ehrlich G. D., et al. 2010. The distributed genome hypothesis as a rubric for understanding evolution in situ during chronic bacterial biofilm infectious processes. FEMS Immunol. Med. Microbiol. 59:269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fazli M., et al. 2009. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 47:4084–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fonder M. A., et al. 2008. Treating the chronic wound: a practical approach to the care of nonhealing wounds and wound care dressings. J. Am. Acad. Dermatol. 58:185–206 [DOI] [PubMed] [Google Scholar]
- 12. Forward K. R. 2006. An evaluation of extended incubation time with blind subculture of blood cultures in patients with suspected endocarditis. Can. J. Infect. Dis. Med. Microbiol. 17:186–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frankel Y. M., et al. 2009. Defining wound microbial flora: molecular microbiology opening new horizons. Arch. Dermatol. 145:1193–1195 [DOI] [PubMed] [Google Scholar]
- 14. Gallo P. H., et al. Demonstration of Bacillus cereus in peri-implant infection using a multi-primer PCR-mass spectrometric assay: report of two cases. J. Bone Joint Surg. Am. 93(15):e851–e856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gontcharova V., Youn E., Sun Y., Wolcott R. D., Dowd S. E. 2010. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol. J. 4:8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gontcharova V., et al. 2010. Black Box Chimera Check (B2C2): a Windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol. J. 4:47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu F. Z., Ehrlich G. D. 2008. Population-level virulence factors amongst pathogenic bacteria: relation to infection outcome. Future Microbiol. 3:31–42 [DOI] [PubMed] [Google Scholar]
- 18. Hummelen R., et al. 2010. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One 5:e12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kantor J., Margolis D. J. 1998. Efficacy and prognostic value of simple wound measurements. Arch. Dermatol. 134:1571–1574 [DOI] [PubMed] [Google Scholar]
- 20. Lozupone C., Lladser M. E., Knights D., Stombaugh J., Knight R. 2010. UniFrac: an effective distance metric for microbial community comparison. ISME J. 5:169–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Margolis D. J., Berlin J. A., Strom B. L. 2000. Which venous leg ulcers will heal with limb compression bandages? Am. J. Med. 109:15–19 [DOI] [PubMed] [Google Scholar]
- 22. Martin J. M., Zenilman J. M., Lazarus G. S. 2010. Molecular microbiology: new dimensions for cutaneous biology and wound healing. J. Investig. Dermatol. 130:38–48 [DOI] [PubMed] [Google Scholar]
- 23. McCann K. S. 2000. The diversity-stability debate. Nature 405:228–233 [DOI] [PubMed] [Google Scholar]
- 24. Nistico L., et al. 2011. Pathogenic biofilms in adenoids: a reservoir for persistent bacteria. J. Clin. Microbiol. 49:1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petti C. A., et al. 2006. Utility of extended blood culture incubation for isolation of Haemophilus, Actinobacillus, Cardiobacterium, Eikenella, and Kingella organisms: a retrospective multicenter evaluation. J. Clin. Microbiol. 44:257–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pitta D. W., et al. 2010. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb. Ecol. 59:511–522 [DOI] [PubMed] [Google Scholar]
- 27. Price L. B., et al. 2009. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One 4:e6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Proulx R., et al. 2010. Diversity promotes temporal stability across levels of ecosystem organization in experimental grasslands. PLoS One 5:e13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ptacnik R., et al. 2008. Diversity predicts stability and resource use efficiency in natural phytoplankton communities. Proc. Natl. Acad. Sci. U. S. A. 105:5134–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rasmussen K., Lewandowski Z. 1998. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol. Bioeng. 59:302–309 [DOI] [PubMed] [Google Scholar]
- 31. Reid G., et al. 2011. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9:27–38 [DOI] [PubMed] [Google Scholar]
- 32. Schippa S., et al. 2010. A distinctive ‘microbial signature’ in celiac pediatric patients. BMC Microbiol. 10:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schloss P. D., et al. 2009. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sen C. K., et al. 2009. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 17:763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith D. M., et al. 2010. Evaluation of the bacterial diversity of pressure ulcers using bTEFAP pyrosequencing. BMC Med. Genomics 3:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stephens P., et al. 2003. Anaerobic cocci populating the deep tissues of chronic wounds impair cellular wound healing responses in vitro. Br. J. Dermatol. 148:456–466 [DOI] [PubMed] [Google Scholar]
- 37. Stott M. B., et al. 2008. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ. Microbiol. 10:2030–2041 [DOI] [PubMed] [Google Scholar]
- 38. Thomsen T. R., et al. The bacteriology of chronic venous leg ulcer examined by culture-independent molecular methods. Wound Repair Regen. 18:38–49 [DOI] [PubMed] [Google Scholar]
- 39. Wolcott R. D., Gontcharova V., Sun Y., Dowd S. E. 2009. Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded FLX and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiol. 9:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zemanick E. T., Sagel S. D., Harris J. K. 2011. The airway microbiome in cystic fibrosis and implications for treatment. Curr. Opin. Pediatr. 23:319–324 [DOI] [PubMed] [Google Scholar]
- 41. Zhao G., et al. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: a model for the study of chronic wounds. Wound Repair Regen. 18:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]