Abstract
Prophylactic probiotic therapy has shown beneficial effects in an experimental rat model for acute pancreatitis on the health status of the animals. Mechanisms by which probiotic therapy interferes with severity of acute pancreatitis and associated sepsis, however, are poorly understood. The aims of this study were to identify the probiotic-induced changes in the gut microbiota and to correlate these changes to disease outcome. Duodenum and ileum samples were obtained from healthy and diseased rats subjected to pancreatitis for 7 days and prophylactically treated with either a multispecies probiotic mixture or a placebo. Intestinal microbiota was characterized by terminal-restriction fragment length polymorphism (T-RFLP) analyses of PCR-amplified 16S rRNA gene fragments. These analyses showed that during acute pancreatitis the host-specific ileal microbiota was replaced by an “acute pancreatitis-associated microbiota.” This replacement was not reversed by administration of the probiotic mixture. An increase, however, was observed in the relative abundance of a novel bacterial phylotype most closely related to Clostridium lituseburense and referred to as commensal rat ileum bacterium (CRIB). Specific primers targeting the CRIB 16S rRNA gene sequence were developed to detect this phylotype by quantitative PCR. An ileal abundance of CRIB 16S rRNA genes of more than 7.5% of the total bacterial 16S rRNA gene pool was correlated with reduced duodenal bacterial overgrowth, reduced bacterial translocation to remote organs, improved pancreas pathology, and reduced proinflammatory cytokine levels in plasma. Our current findings and future studies involving this uncharacterized bacterial phylotype will contribute to unraveling one of the potential mechanisms of probiotic therapy.
INTRODUCTION
Systemic inflammatory response syndrome (SIRS), sepsis, and multiple organ dysfunction syndrome (MODS) are major causes of mortality in severely ill patients. The role of the intestinal microbiota in the initiation and propagation of critical illness has been increasingly acknowledged during the past decades. In several disease models it has been demonstrated that intestinal barrier dysfunction allows gut-derived bacteria to translocate to extraintestinal organs and cause sepsis and secondary infectious complications resulting in high mortality rates (8).
Severe acute pancreatitis is an example of a critical illness that is characterized by intestinal barrier dysfunction. Acute pancreatitis usually has a mild and self-limiting clinical course; however, in about 20 to 30% of the patients it develops into a serious disease with SIRS and MODS. In these patients, infection of necrotic pancreatic tissue with gut-derived bacteria occurs in 30 to 70% of the cases (5). These infectious complications are frequently the cause of mortality in patients with acute pancreatitis, with a mortality rate up to 50% (1, 4, 20).
We have previously shown in an experimental rat model for acute pancreatitis that prophylactic administration of a mixture of probiotic bacterial species resulted in reduced bacterial translocation to the pancreas and other extraintestinal organs, improved disease outcome, and reduced late mortality of acute pancreatitis (26). Although the effects of the probiotic therapy were highly significant, not all treated animals were protected from the adverse consequences of experimental acute pancreatitis. Furthermore, the underlying mechanism(s) by which probiotics ameliorate the severity of acute pancreatitis is incompletely understood. In order to get a better insight in the role of gut microbiota in the process of bacterial translocation during experimental pancreatitis, a detailed analysis of the bacterial communities present in the various parts of the intestine was made. The aims of this study were to identify the probiotic-induced changes in the gut microbiota and to correlate these changes to disease outcome. As described here, molecular analysis of intestinal samples of rats treated with probiotics has led to a new hypothesis on how probiotics can improve the clinical course of acute pancreatitis.
MATERIALS AND METHODS
Animals.
Male specific-pathogen-free Sprague-Dawley rats, 250 to 350 g (Harlan, Horst, The Netherlands), were kept under stable housing conditions with a 12-hour light/dark cycle and free access to water and food (RMH 1110; Hope Farms, Woerden, The Netherlands) throughout the experiment. All animals were allowed to adjust to these conditions for 1 week prior to surgery. Rats were randomized between two experimental groups: 17 rats were included in the group prophylactically treated with probiotics, and 21 rats were assigned to the placebo group. Both groups were subjected to the surgical procedures described below. In addition, six rats which did not receive any surgical intervention or treatment were used as healthy controls. All animal procedures were performed in accordance with institutional guidelines and with approval from the institutional animal care committee of the University Medical Center, Utrecht, The Netherlands.
Probiotics and placebo.
The multispecies probiotic mixture consisted of equal amounts of six different viable, freeze-dried probiotic strains, Bifidobacterium bifidum (W23), Bifidobacterium animalis subsp. lactis (W52), Lactobacillus acidophilus (W70), Lactobacillus casei (W56), Lactobacillus salivarius (W24), and Lactococcus lactis (W58), blended in a carrier material consisting of maize starch, maltodextrins, and a mineral mix (Ecologic 641; Winclove Bio Industries, Amsterdam, The Netherlands). The placebo product consisted of carrier material only. Directly before administration, both the freeze-dried placebo product and the probiotic mixture were reconstituted in sterile water for 15 min at 37°C. A single probiotic dose of 1.0 ml contained a total of about 5 × 109 CFU. Probiotics or placebo were administered intragastrically through a permanent gastric cannula once daily, starting 5 days prior to induction of acute pancreatitis, and twice daily for 6 days after induction of acute pancreatitis (see Fig. S1 in the supplemental material).
Surgical procedures.
Surgical procedures were performed as described previously (26). Briefly, all procedures were performed under general anesthesia using a combination of 2% isoflurane gas and 0.3 ml 10% buprenorphine intramuscularly administered (Temgesic; Reckitt Benckiser Healthcare Ltd., Hull, United Kingdom). At the start of the experiment, a permanent gastric cannula was fitted by tunneling a silicone cannula subcutaneously from the abdominal wall to the back of the animal. The gastric end of the cannula was inserted into the stomach through a puncture within a purse-string suture on the greater curvature. Animals were allowed to recover for 3 days prior to the start of daily probiotics or placebo administration. Five days after starting daily administration of probiotics or placebo, acute pancreatitis was induced as described previously, with minor adaptations (21). The common bile duct was clamped, and 0.5 ml sterilized glycodeoxycholic acid in 10 mM glycylglycine-NaOH-buffered solution (pH 8.0, 37°C; chemicals obtained from Sigma-Aldrich Chemie B.V., Zwijndrecht, The Netherlands) was infused, after which hepato-duodenal bile flow was restored. Next, the right jugular vein was cannulated for continuous intravenous infusion of cerulein (5 μg/kg per hour, for 6 h; chemicals obtained from Sigma-Aldrich Chemie B.V.).
Tissue and fluid samples.
Seven days after induction of pancreatitis, surviving rats were anesthetized to allow aseptic removal of tissue samples and sampling of peritoneal fluid and blood. After sample collection, rats were euthanized by blood loss. Mesenteric lymph nodes (MLNs), liver, spleen, pancreas, and duodenum were removed for microbiological analysis. After carefully removing all pancreatic and mesenteric tissue from the proximal duodenum and distal part of the terminal ileum, sections of approximately 2 cm were excised at both locations. Both segments were then transferred to a sterile vial, snap-frozen in liquid nitrogen, and stored at −80°C. A portion of pancreatic tissue was fixed in 4% formalin and analyzed histopathologically, using standard hematoxylin and eosin (H&E) staining. The histopathological severity of acute pancreatitis was assessed based on a scoring system modified from Schmidt et al. (21) as previously described (26). Several aspects of (peri)pancreatic histopathology were assessed (peritonitis, edema, ductal pathology, inflammatory infiltrate, acinar cells, acinar dilatation, and hemorrhagic changes). These aspects were scored on a scale, with maxima varying from 2 to 5 points per aspect.
All tissue samples were weighed and processed immediately for quantitative and qualitative culturing of aerobic and anaerobic microorganisms in appropriate media as described previously (26). Tissue samples were homogenized in cysteine broth and cultured in 10-fold dilution series. The samples were cultured aerobically on blood agar, MacConkey agar (Gram-negative bacteria), and Columbia colistin nalidixic acid agar (staphylococci and streptococci), microaerobically on Man-Rogosa-Sharpe agar (lactobacilli), and anaerobically on Schaedler agar. Bacterial counts are presented in CFU/g tissue. Threshold detection level of bacterial growth was >102 CFU/g.
Multiplex cytokine assays.
Plasma cytokine levels of interleukin-1α (IL-1α), IL-6, IL-12p70, IL-18, interferon-γ (IFN-γ), CXCL1 (growth-related oncogene; GRO/KC), and CCL2 (monocyte chemoattractant protein-1 [MCP-1]), all from Linco Research, Inc. (St. Louis, MO), and IL-1β, IL-2, IL-10, and tumor necrosis factor alpha (TNF-α), all from Bio-Rad Laboratories (Hercules, CA), were analyzed using rat cytokine multiplex assays according to the manufacturer's instructions.
DNA extraction.
Duodenum and ileum samples were taken from the freezer (−20°C), and a 1-cm section from the center of the sample was excised on a sterile field. A longitudinal incision over the full 1-cm section was made, and the sample was left to thaw on a piece of sterile aluminum foil placed directly on a cooled metal plate at 4°C. After thawing, the intestinal content, including the mucosa, was scraped off with the back of a sterile surgical blade and collected in a 2-ml screw-cap tube containing a buffer solution of 6 M guanidine thiocyanate, 0.6% Tween 20 (vol/vol), 10 mM EDTA, 50 mM Tris-HCl (pH 6.5), and 2 g of zirconia beads (<0.1 mM; Biospec, Bartlesville, OK). For DNA extraction from pure bacterial cultures and the complete probiotic mixture, approximately 108 CFU was used. Cells were physically disrupted by shaking for 4 min in a MiniBeadbeater-96 (Biospec), followed by heating for 5 min at 90°C. Samples were centrifuged for 1 min at 10,000 × g, and 200 μl of the supernatant was subsequently used for DNA isolation. DNA isolation was performed as described previously (7), except for omitting the final washing step with acetone.
T-RFLP analysis.
Terminal-restriction fragment length polymorphism (T-RFLP) analysis was performed as described previously (15). For PCR amplification of 16S rRNA gene fragments, the universal primers 8F and 926R (Table 1), with fluorescent dyes 6-FAM and NED, respectively, at the 5′ end, were used. Reaction mixtures (15 μl) contained PCR buffer (Applied Biosystems, Foster City, CA), 62.5 μM each deoxynucleoside triphosphate (dNTP; Applied Biosystems), 1.5 mM MgCl2, a 0.5 μM concentration of each primer, 0.5 U of Taq DNA polymerase (Promega, Madison, WI), and 1 μl of DNA sample. DNA amplification was performed with a 9700 thermal cycler (Perkin-Elmer, Norwalk, CT) using the following program: 94°C for 5 min, followed by 35 cycles of 30 s at 94°C, 45 s at 56°C, and 2 min at 72°C, and a final extension for 5 min at 72°C. PCR products were digested with MspI and HinP1I, with both enzymes in the same reaction (New England BioLabs, Ipswich, MA). The resulting fragments were size separated using an ABI 3100 genetic analyzer (Applied Biosystems) equipped with 36-cm capillaries using POP4 gel matrix. A custom size standard with ROX-labeled fragments, MapMarker 30–1,000 (Bioventures, Murfreesboro, TN), was added to each sample prior to electrophoresis for accurate sizing of terminal-restriction fragments (T-RFs). After electrophoresis, sizes of fluorescently labeled T-RFs were determined using GeneScan software (Applied Biosystems).
Table 1.
Primers and their targets used in this study
| Primer | Primer sequence (5′–3′) | Target | Reference |
|---|---|---|---|
| 8F | AGA GTT TGA TCC TGG CTC (AG) | Universal 16S rRNA gene | 14 |
| 926R | CCG TCA ATT CCT TTR AGT TT | Universal 16S rRNA gene | 17 |
| T7 | TAA TAC GAC TCA CTA TAG G | pGEM-T vector specific | Promega |
| SP6 | GAT TTA GGT GAC ACT ATA G | pGEM-T vector specific | Promega |
| 519R | GWA TTA CCG CGG CKG CTG | Universal 16S rRNA gene | 14 |
| 533F | GTG CCA GCA GCC GCG GTA A | Universal 16S rRNA gene | 27 |
| Bact1369F | CGG TGA ATA CGT TCY CGG | Universal 16S rRNA gene | 24 |
| Prok1492R | GGW TAC CTT GTT ACG ACT T | Universal 16S rRNA gene | 24 |
| CRIB-61F | GTC GAG CGA TTT ACT TCG GTA | CRIB-specific 16S rRNA gene | This study |
| CRIB-235R | GGG TCC ATC CTG TAC CGC AAA | CRIB-specific 16S rRNA gene | This study |
Observed T-RFs were assigned to microbial taxa using the MCPC database (Dr. Van Haeringen Laboratorium B.V., Wageningen, The Netherlands), which was built using in silico T-RF predictions from data that were extracted from prokaryote sections of the EMBL sequence database (release 82, March 2005) using Patscan (9). For each of the extracted sequences the T-RF size using the aforementioned combination of primers and restriction enzymes was calculated for both primers, and calculated values were validated for a number of pure cultures, including Escherichia coli, Clostridium perfringens, and Staphylococcus aureus (data not shown). The relative peak intensity of each T-RF (referred to as relative abundance) was defined as the height of a specific peak as a percentage of the total sum of all peak heights for a given sample. Relative abundance was calculated for all T-RFs between 50 and 600 bp long and with peak heights of >50 fluorescence units (FU), which was well above the background noise level (10 to 20 FU) of nontemplate control reactions. This procedure was followed for both primers; the data obtained with primer 8F were used in the figures.
Identification of the 16S rRNA gene sequence of CRIB.
PCR mixtures (50 μl) were prepared using Taq DNA polymerase kit from Invitrogen (Gaithersburg, MD) and contained 0.5 μl of Taq DNA polymerase (1.25 U), 20 mM Tris-HCl (pH 8.5), 50 mM KCl, 3.0 mM MgCl2, 5 pmol of the primers 8F and 926R (Table 1), 200 μM each dNTP, and 1 μl of template DNA. Amplification was performed with a T1 thermocycler (Whatman Biometra, Göttingen, Germany) using the following program: 94°C for 5 min, followed by 35 cycles of 30 s at 94°C, 20 s at 56°C, and 40 s at 68°C, and a final extension for 7 min at 68°C. Aliquots (5 μl) were analyzed by agarose electrophoresis to check for product size and quantity.
Products were purified with the QIAquick PCR purification kit (Westburg, Leusden, The Netherlands) according to the manufacturer's instructions, and cloned into the pGEM-T easy vector system (Promega) using competent E. coli JM109 as a host. Colonies of ampicillin-resistant transformants were transferred with a sterile toothpick to 15 μl of Tris-EDTA buffer and boiled for 15 min at 95°C. PCR was performed using vector-specific primers T7 and SP6 (Table 1) to check the sizes of the inserts using the cell lysate as a template. PCR products of ∼0.95 kb were purified as described above and digested at 37°C for 90 min with CfoI, which is an isoschizomer of HinP1I, and MspI (Boehringer, Mannheim, Germany). One clone, containing an insert yielding a fragment of approximately 450 bp after restriction analysis, was grown in Luria broth liquid medium (5 ml) with ampicillin (100 μg/ml) and used for further 16S rRNA gene sequence analysis. Plasmid DNA was isolated using the Wizard Plus purification system (Promega) and sequenced by using the Sequenase (T7) sequencing kit (Amersham Life Sciences, Slough, United Kingdom) according to the manufacturer's specifications using primers T7, SP6, 519R, and 533F (Table 1) labeled with IRD-800. Sequences were automatically analyzed on a LiCor DNA Sequencer 4000L (LiCor, Lincoln, NE) and manually assembled.
Phylogenetic analysis and primer design.
The CRIB 16S rRNA gene sequence was aligned with published 16S rRNA sequences (see Fig. S2 in the supplemental material) using the ARB software package and release 90 of the ARB-SILVA reference database (16, 18). Different primer sets were developed using the ProbeDesign and ProbeMatch functions in ARB and were tested for specificity by conventional PCR. The primer set CRIB-61F/CRIB-235R (Table 1) was further tested by qPCR. For phylogenetic analyses as presented here, the sequence was realigned using the SINA WebAligner (http://www.arb-silva.de) and compared to published sequences using ARB-Silva release 102.
qPCR analysis.
Quantitative PCR (qPCR) was performed using the iQ5 Real-Time PCR detection system (Bio-Rad). Reaction mixtures (25 μl) contained 12.5 μl iQ SYBR green supermix (Bio-Rad), 0.2 μM each primer, and 5 μl of template DNA. All reactions were performed in triplicate. Standard curves were generated from a dilution series of 16S rRNA gene fragments amplified from the target sequence (108 to 100 copies/μl). The universal primer set Bact1369/Prok1492 (Table 1) was used for quantification of total bacterial 16S rRNA gene copies using the following program: 3 min at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 56°C, and 30 s at 72°C. Primers CRIB-61F/CRIB-235R were used to determine the relative abundance of CRIB 16S rRNA gene copies in the samples. Amplification conditions included 3 min at 95°C, followed by 40 cycles of 20 s at 95°C and 1 min at 68°C. Gradient PCR was initially used to determine the optimal annealing temperature, with amplification efficiency, range of linearity, and lowest detectable concentration as criteria. The predicted specificity of the CRIB-specific qPCR primer set was confirmed using nearly full-length 16S rRNA gene amplicons that were obtained, using primer pair 8F/Prok1492 (Table 1), from genomic DNA of closely related bacterial species: Clostridium lituseburense (DSM 797T), Clostridium irregulare (DSM 2635T), Clostridium hiranonis (DSM 13275T), and Clostridium bartlettii (DSM 16795T).
Relative abundance of CRIB was calculated by dividing CRIB 16S rRNA gene copies amplified using the primer set CRIB-61F/CRIB-235R by total 16S rRNA gene copies amplified using the primer set Bact1369/Prok1492 per μl of isolated DNA.
Statistical analysis.
T-RFLP profiles were compared by visual inspection and principal component analysis (PCA) using VHL analysis software (Dr. Van Haeringen Laboratorium B.V.) that was custom built in collaboration with Dalicon B.V. (Wageningen, The Netherlands) using IDL (RSI, Boulder, CO). The number of T-RFs is presented as mean ± standard error of the mean (SEM).
The nonparametric Kruskal-Wallis test, followed by post hoc Mann-Whitney U tests with Bonferroni correction, was used to test for statistical differences in the relative abundance of CRIB between the different independent treatment groups. The intraclass correlation coefficient (ICC) was calculated to compare the T-RFLP to qPCR analysis. The correlation between two variables was described with Spearman's rank correlation coefficients and corresponding P values. Differences in bacterial counts, histopathological scores, and plasma cytokine levels between different groups of animals were analyzed using the Mann-Whitney U test. Differences with P values of <0.05 were considered statistically significant.
Nucleotide sequence accession number.
The partial 16S rRNA gene sequence of CRIB has been submitted to the GenBank/EMBL/DBBJ databases under accession number HQ224563.
RESULTS
Acute pancreatitis induces changes in duodenal and ileal microbiota.
To study the changes in duodenal and ileal microbiota as result of acute pancreatitis, duodenal and ileal samples were studied by T-RFLP analysis of PCR-amplified 16S rRNA gene fragments.
Previously we have demonstrated that there was a small increase in the total bacterial counts in the duodenum and a significant increase in culturable opportunistic pathogens 7 days after the induction of acute pancreatitis (26). In the present study, T-RFLP analysis of duodenal microbiota demonstrated that the number of T-RFs almost doubled in the diseased animals (7 days after induction of acute pancreatitis) compared to healthy control animals (25 ± 3.5 versus 41 ± 8.4, healthy control versus placebo, respectively). T-RFLP analysis of the bacterial communities in the terminal ileum, however, showed that induction of acute pancreatitis resulted in only a modest increase in the number of T-RFs in diseased animals compared to healthy controls (60.3 ± 7.2 versus 69.0 ± 7.5, healthy control versus placebo, respectively).
The composition of the ileal microbiota, as evaluated by PCA, was altered in the diseased animals compared to the healthy controls. Each healthy rat displayed a unique microbiota fingerprint, visible as a characteristic scattering of the samples in the two-dimensional space of the first principal components (Fig. 1). In contrast, the samples of the diseased animals, with one exception, show a strong clustering, demonstrating that the diseased animals apparently acquired a rather uniform “acute pancreatitis-associated microbiota” independent of which treatment was administered.
Fig. 1.
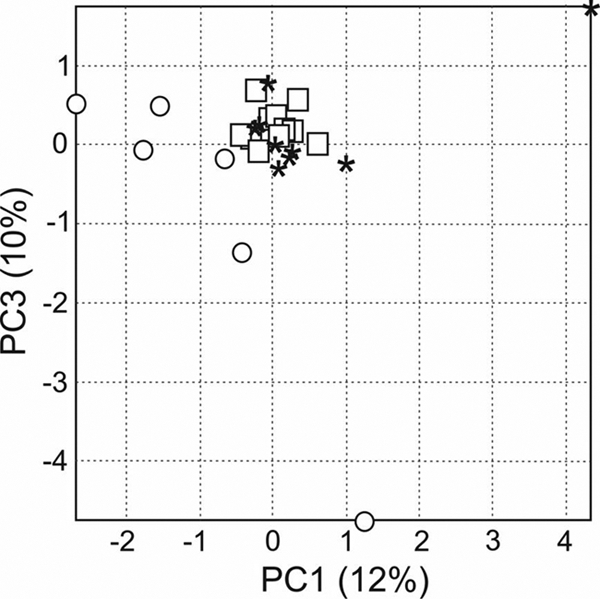
Correlations between the T-RFLP profiles obtained from the ileal samples mapped onto the first and third principal components of PCA. Healthy rats are indicated with circles. Diseased rats are indicated with asterisks (placebo-treated animals) or squares (probiotic-treated animals).
Probiotics stimulate a not previously described bacterium in the terminal ileum.
T-RFLP analysis demonstrated that T-RFs of the length predicted for those of the probiotic bacteria could also be detected in placebo-treated animals, indicating that the endogenous microbiota shares similar T-RFs with the probiotic bacteria (see Fig. S3 and S4 in the supplemental material).
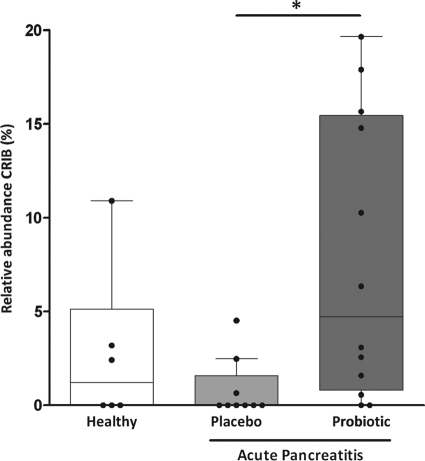
Probiotic-induced changes in the duodenal microbiota could not be detected based on T-RFLP analysis (Fig. S3). In contrast, in the ileal samples a significant increase of an unknown bacterial phylotype (T-RF 457 bp, primer 8F, MspI and HinP1I digested) was observed in the probiotic-treated animals compared to the placebo-treated animals (Fig. 2 and Fig. S4). This T-RF could also be detected in a number of healthy control animals. It could be excluded that this bacterial phylotype was administered inadvertently, because it was absent in both the multispecies probiotic mixture and the carrier material (placebo) (data not shown).
Fig. 2.
Effect of pancreatitis and treatment with either placebo or the probiotic mixture on the relative ileal abundance of T-RF 457, corresponding to the as yet uncharacterized bacterial phylotype referred to as CRIB. Indicated are healthy control (white bar) and diseased animals, treated either with placebo (light gray bar) or the probiotic mixture (dark gray bar), respectively. Box-and-whisker diagrams represent median with interquartile range (boxes) and minimum and maximum nonoutlier values (whiskers). An asterisk indicates a significant difference (P < 0.05).
In order to further characterize this phylotype, a 16S rRNA gene clone library was generated from ileal DNA samples showing the 457-bp T-RF. Clones were screened by RF analysis using enzymes recognizing the same restriction sites as those applied for T-RFLP analysis, and a representative clone containing an insert leading to a fragment of approximately 450 bp was selected for further sequence analysis. The cloned 16S rRNA gene fragment, yielding a predicted T-RF of 457 bp, showed less than 97% sequence identity to the 16S rRNA gene sequences of bacterial isolates (Table 2), suggesting that it might represent a not-previously described bacterial species, which in the following is referred to as “commensal rat ileum bacterium” (CRIB). Higher sequence identities were observed with environmental sequences mostly retrieved from mammalian intestinal samples, including mouse and human (Fig. 3).
Table 2.
16S rRNA gene sequence identities of the CRIB clone with published sequences
| GenBank accession no. | Description | % identitya |
|---|---|---|
| EU089965 | Clostridium maritimum strain G12 | 96% |
| EU089964 | Clostridium ruminantium strain LA1 | 96% |
| M59107 | Clostridium lituseburense (DSM 797) | 96% |
| AB538434 | Clostridium bifermentans (JCM 7832) | 95% |
| AB550230 | Clostridium sordellii (JCM 3814) | 94% |
| AF320283 | Clostridium bifermentans (DSM 13560) | 94% |
| AB075771 | Clostridium sordellii (ATCC 9714) | 94% |
| NR_027573 | Clostridium bartlettii (DSM 16795) | 94% |
| X73451 | Clostridium ghonii (NCIMB 10636) | 94% |
| X73450 | Clostridium difficile (DSM 11209) | 94% |
| NR_029249 | Clostridium irregulare strain 6VI (DSM 2635) | 93% |
| NR_028611 | Clostridium hiranonis strain TO-931 (DSM 13275) | 92% |
The percentage of identity was determined by comparison of the partial 16S rRNA gene sequence of CRIB with sequences present in the database using the BLAST tool from NCBI.
Fig. 3.
Neighbor-joining tree based on the 16S rRNA gene sequence of CRIB and other related clostridia. E. coli was included as an outgroup. Alignment and phylogenetic analysis were performed with the ARB software (16). The tree was calculated for E. coli positions 55 to 926. The reference bar indicates 10% sequence divergence. GenBank accession numbers are given in parentheses.
Based on the 16S rRNA gene sequence of the CRIB clone, a primer set was designed for the specific quantitative detection of CRIB in environmental samples by qPCR. In order to demonstrate that these primers indeed specifically detect the same 16S rRNA gene sequences that give rise to the 457-bp T-RF, qPCR was performed on the ileal DNA samples and the results were compared to the T-RFLP results. An overall good agreement between the two techniques for calculation of the relative abundance of CRIB was demonstrated by the highly significant correlation between the two data sets (ICC = 0.893, P < 0.001; see Fig. S5 in the supplemental material).
Relative ileal abundance of CRIB is associated with decreased disease severity.
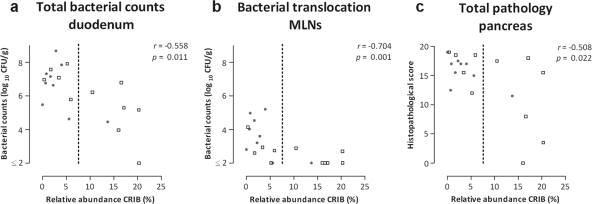
Different measures of disease outcome were analyzed in order to assess disease severity, including duodenal bacterial overgrowth, bacterial translocation to remote organs, and pancreas pathology. The relative abundance of CRIB in the ileum of the diseased rats was inversely correlated with the degree of bacterial overgrowth in the duodenum (Fig. 4a). Total bacterial counts were lower in the duodenum of animals with high relative ileal abundance of CRIB. In addition, bacterial infection of the MLNs (Fig. 4b), as well as the liver, spleen, and pancreas (see Fig. S6 in the supplemental material), was also inversely correlated with the relative ileal abundance of CRIB. Furthermore, histopathological evaluation of pancreatic tissue obtained 7 days after the induction of acute pancreatitis demonstrated that pancreas pathology was significantly and inversely correlated with the relative ileal abundance of CRIB (Fig. 4c). Acinar cell pathology and inflammatory cell infiltrate were the aspects which contributed most to this correlation (see Fig. S6 in the supplemental material). It should be noted that the relative ileal numbers of the administered probiotic strains, or endogenous strains with identical T-RFs, did not correlate with the clinical and histological severity of the pancreatitis.
Fig. 4.
Associations between different measures of disease outcome and the relative ileal abundance of CRIB in diseased rats after 7 days of acute pancreatitis. Animals were treated with either placebo (gray dots) or probiotics (squares). Indicated are total bacterial counts in the duodenum (a), bacterial translocation to the MLNs (b), and total pathology of the pancreas (c). The relative abundance of CRIB was determined by qPCR analysis. Bacterial counts are expressed in log10 CFU per gram of sample. Pearson's correlation coefficients are provided, with corresponding P values. The dotted lines indicate the divisions between the samples with a low (<7.5%) or high (>7.5%) relative ileal abundance of CRIB (>7.5%).
The data presented above suggest that the clinical effects of intervention with probiotics are mediated by CRIB. In order to substantiate this association, the diseased animals were divided into two groups with either a low (<7.5%) or a high (>7.5%) relative ileal abundance of CRIB, based on their average relative ileal abundance of CRIB as determined by qPCR analysis. A significantly lower total number of duodenal bacteria was detected in the animals with >7.5% CRIB compared to the animals with <7.5% CRIB (P < 0.05). In addition, a relative abundance of CRIB above 7.5% was also significantly correlated (P < 0.05) with less bacterial infection of the MLNs and lower total histopathological scores of the pancreas.
Relative ileal abundance of CRIB is associated with altered cytokine levels during acute pancreatitis.
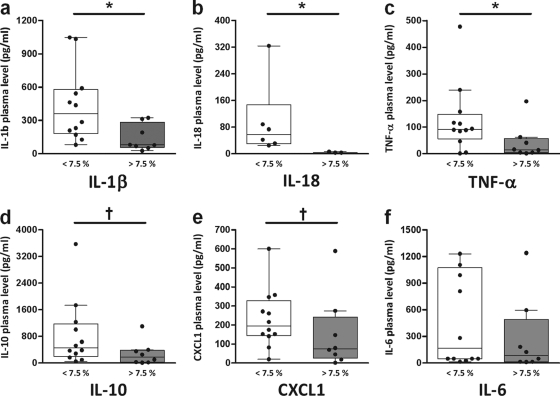
To determine the possible association between high relative abundance of CRIB in the ileum and cytokine plasma levels, a cytokine multiplex assay was performed on the plasma samples obtained 7 days after induction of acute pancreatitis. In a previous study, we identified high plasma levels of IL-1β, IL-6, IL-10, CXCL1, and TNF-α as strong predictors of mortality and bacteremia during the course of acute pancreatitis in rats, and it was found that treatment with a multispecies probiotic mixture causes a mild reduction of these cytokines (data not shown). In line with these observations, the animals with high relative ileal abundance of CRIB (>7.5%) showed significantly lower plasma levels of IL-1β and TNF-α and a tendency to reduced levels of IL-10 and CXCL1 (Fig. 5). In addition, elevated plasma levels of IL-18, a proinflammatory cytokine, which is significantly increased in acute pancreatitis patients with local or systemic complications (19), were detected only in animals having a low relative ileal abundance of CRIB (<7.5%; Fig. 5b).
Fig. 5.
Effect of low (<7.5%; white bars) or high (>7.5%; gray bars) relative ileal abundance of CRIB in diseased rats on plasma cytokine levels of IL-1β (a), IL-18 (b), TNF-α (c), IL-10 (d), CXCL1 (e), and IL-6 (f). The relative abundance of CRIB was determined by qPCR analysis. Box-and-whisker diagrams represent median with interquartile range (boxes) and minimum and maximum nonoutlier values (whiskers). Significant differences: *, P < 0.05; †, P < 0.10.
DISCUSSION
A not previously described bacterial phylotype (CRIB) was identified in this study to be associated with reduced severity of pancreatitis and associated sepsis. A higher than average relative ileal abundance of CRIB (i.e., >7.5%) was significantly correlated with decreased duodenal bacterial overgrowth, reduced bacterial translocation to remote organs, reduced infection of pancreatic necrosis, and improved pancreas histology. In addition, high relative abundance of this bacterial phylotype was associated with less severe immune responses during acute pancreatitis as demonstrated by lower plasma levels of proinflammatory cytokines. Moreover, we demonstrated that there is an association between the presence of CRIB in the ileum of rats and the administration of a multispecies probiotic mixture. Together, these results suggest that effects of this multispecies probiotic mixture (Ecologic 641) are mediated by stimulation of a not previously described gut commensal bacterium (CRIB), which protects the host from severe sepsis. The bacterial species most closely related to this not previously described bacterial phylotype is Clostridium lituseburense. For the detection of this bacterial phylotype, a specific and quantitative PCR (qPCR) assay was developed using 16S rRNA gene-targeted primers. This qPCR can be used in future studies to detect this phylotype in clinically relevant samples.
Dysbiosis of the small intestinal microbiota is often seen in critically ill patients and can be a cause of the development of sepsis. For example, a low level of intestinal bacterial overgrowth can be detected in patients with hepatic cirrhosis and is associated with systemic endotoxemia (3, 28). In the case of acute pancreatitis, the occurrence of small intestinal bacterial overgrowth is a pathological event during the course of acute pancreatitis due to impaired intestinal motility and is quantitatively and qualitatively correlated with bacterial translocation and infection of pancreatic necrosis (25, 26). In addition, it has recently been demonstrated in a rat model that during acute pancreatitis bacterial translocation occurs from the small intestine rather than from the colon (10). The duodenal bacterial counts found in this study confirm our previous findings that acute pancreatitis leads to duodenal bacterial overgrowth. Despite the occurrence of bacterial overgrowth in the duodenum, there was no change in composition of the duodenal microbiota. In contrast, the composition of the ileal microbiota was considerably changed upon induction of acute pancreatitis. PCA revealed that each healthy rat displayed a unique ileal microbiota fingerprint, as previously observed for ileal microbiota of healthy humans (6). In contrast, increased similarity of the T-RFLP profiles of diseased animals indicated that these animals acquired an “acute pancreatitis-associated microbiota” independent of which treatment was deployed. Apparently, pancreatitis induces such a strong modification of the ileal microbiota that the unique individual microbial inhabitants are replaced by other, potentially pathogenic bacteria.
Previous studies have demonstrated that although the effects of this specific probiotic mixture were highly significant, not all probiotic-treated animals were protected from the adverse effects of experimental acute pancreatitis (26). Here we show that one bacterial phylotype (referred to as CRIB) was significantly more abundant in the ileum of animals prophylactically treated with the probiotic mixture than in that of the placebo-treated animals. However, both T-RFLP and qPCR analysis showed high interindividual variation in the relative abundance of CRIB in the terminal ileum, which suggests that this bacterium is not an equally dominant member of the normal microbiota in every animal. This might explain why the administration of the multispecies probiotic mixture could not prevent pancreatitis-associated infectious complications in every animal. In fact, in animals that reacted poorly to probiotic treatment (e.g., the animals with high pancreatic bacterial counts), CRIB was present only in relatively low numbers (<7.5%). The chosen threshold value was set at 7.5%, based on the average relative abundance of CRIB in the total group of diseased animals. The relatively small sample size prevented a more accurate estimate of the threshold by ROC analysis. Still, the data point toward an important role of CRIB in the effectiveness of probiotic treatment. As the present study had not been designed to provide mechanistic insight in this association, it is currently unknown if and how probiotic administration can lead to an increase in relative abundance of CRIB, as well as a decrease in disease severity. The causality of the observed association between CRIB and protection against severe sepsis will be addressed in future experiments.
The terminal ileum is the main site for interaction of commensal microbiota with the host immune system. This is reflected by the fact that the Peyer's patches, organized lymphoid tissue important for immune surveillance and initiating of immune responses in the gut, are mainly located in the ileum (13). In our study we demonstrated that CRIB can be a dominant member of the ileal microbiota in rats, reaching up to 20% of the total microbiota in some animals. The mechanisms by which CRIB confers protective effects may include modulation of the mucosal immune system. This is suggested by the observation that the relative abundance of CRIB was correlated with altered plasma cytokine levels during acute pancreatitis.
Recently, important roles in counterbalancing dysbiosis and regulation of immune responses have been suggested for several specific members of the gut microbiota. Faecalibacterium prausnitzii has been described to have anti-inflammatory properties in both in vitro and in vivo models (22), and underrepresentation of this bacterium in the gut microbiota was suggested to be involved in IBD pathogenesis in both animals and humans (23). Furthermore, segmented filamentous bacteria (SFB) were shown to have a crucial role in maturation of T cell responses in the gut (11, 12). Faecalibacterium prausnitzii and SFB are members of the same taxonomic class as CRIB, the Clostridia; however, they belong to different subgroups (Fig. 3). In addition, a recent study has demonstrated a significant induction of colonic regulatory T cells as result of the colonization by a specific mix of indigenous Clostridium species in a mouse experiment (2). In that study, early inoculation of these Clostridium species resulted in the amelioration of the effects of experimental colitis and a lower IgE production level. Altogether, these studies demonstrate that specific members of the Clostridia can have health-promoting effects.
We anticipate that the findings reported here will be of key importance in unraveling one of the potential mechanisms of probiotic action and fostering a further understanding of the relation between gut microbiota and the host. Future experiments, including efforts toward the isolation of CRIB, will evaluate whether CRIB also plays a role in human disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. Akkermans-van Vliet (Wageningen University), W. Harmsen, W. de Jager, F. Lutgendorff, A. Verheem, and M. R. Visser (all from UMC Utrecht) for expert technical assistance and advice.
Part of this study was supported by Senter-Novem, an agency of the Dutch Ministry of Economic Affairs (grant number TSGE-3109). Susana Fuentes was supported by a fellowship of the Fundación Alfonso Martín Escudero (FAME).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Ammori B. J. 2003. Role of the gut in the course of severe acute pancreatitis. Pancreas 26:122–129 [DOI] [PubMed] [Google Scholar]
- 2. Atarashi K., et al. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bauer T. M., et al. 2002. Small intestinal bacterial overgrowth in human cirrhosis is associated with systemic endotoxemia. Am. J. Gastroenterol. 97:2364–2370 [DOI] [PubMed] [Google Scholar]
- 4. Beger H. G., Rau B., Isenmann R. 2003. Natural history of necrotizing pancreatitis. Pancreatology 3:93–101 [DOI] [PubMed] [Google Scholar]
- 5. Beger H. G., Rau B. M. 2007. Severe acute pancreatitis: clinical course and management. World J. Gastroenterol. 13:5043–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Booijink C. C., et al. 2010. High temporal and inter-individual variation detected in the human ileal microbiota. Environ. Microbiol. 12:3213–3227 [DOI] [PubMed] [Google Scholar]
- 7. Carter M. J., Milton I. D. 1993. An inexpensive and simple method for DNA purifications on silica particles. Nucleic Acids Res. 21:1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clark J. A., Coopersmith C. M. 2007. Intestinal crosstalk: a new paradigm for understanding the gut as the “motor” of critical illness. Shock 28:384–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dsouza M., Larsen N., Overbeek R. 1997. Searching for patterns in genomic data. Trends Genet. 13:497–498 [DOI] [PubMed] [Google Scholar]
- 10. Fritz S., et al. 2010. Bacterial translocation and infected pancreatic necrosis in acute necrotizing pancreatitis derives from small bowel rather than from colon. Am. J. Surg. 200:111–117 [DOI] [PubMed] [Google Scholar]
- 11. Gaboriau-Routhiau V., et al. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31:677–689 [DOI] [PubMed] [Google Scholar]
- 12. Ivanov I. I., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iwasaki A. 2007. Mucosal dendritic cells. Annu. Rev. Immunol. 25:381–418 [DOI] [PubMed] [Google Scholar]
- 14. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Inc., New York, NY [Google Scholar]
- 15. Liu W. T., Marsh T. L., Cheng H., Forney L. J. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ludwig W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muyzer G., Teske A., Wirsen C. O., Jannasch H. W. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165–172 [DOI] [PubMed] [Google Scholar]
- 18. Pruesse E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rau B., Baumgart K., Paszkowski A. S., Mayer J. M., Beger H. G. 2001. Clinical relevance of caspase-1 activated cytokines in acute pancreatitis: high correlation of serum interleukin-18 with pancreatic necrosis and systemic complications. Crit. Care Med. 29:1556–1562 [DOI] [PubMed] [Google Scholar]
- 20. Schmid S. W., Uhl W., Friess H., Malfertheiner P., Büchler M. W. 1999. The role of infection in acute pancreatitis. Gut 45:311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt J., et al. 1992. A better model of acute pancreatitis for evaluating therapy. Ann. Surg. 215:44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sokol H., et al. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105:16731–16736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sokol H., et al. 2009. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 15:1183–1189 [DOI] [PubMed] [Google Scholar]
- 24. Suzuki M. T., Taylor L. T., DeLong E. F. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Felius I. D., et al. 2003. Interdigestive small bowel motility and duodenal bacterial overgrowth in experimental acute pancreatitis. Neurogastroenterol. Motil. 15:267–276 [DOI] [PubMed] [Google Scholar]
- 26. van Minnen L. P., et al. 2007. Modification of intestinal flora with multispecies probiotics reduces bacterial translocation and improves clinical course in a rat model of acute pancreatitis. Surgery 141:470–480 [DOI] [PubMed] [Google Scholar]
- 27. Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang C. Y., Chang C. S., Chen G. H. 1998. Small-intestinal bacterial overgrowth in patients with liver cirrhosis, diagnosed with glucose H2 or CH4 breath tests. Scand. J. Gastroenterol. 33:867–871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.