Abstract
Campylobacter jejuni is a leading food-borne pathogen causing gastroenteritis in humans. Although OxyR is a widespread oxidative stress regulator in many Gram-negative bacteria, C. jejuni lacks OxyR and instead possesses the metalloregulator PerR. Despite the important role played by PerR in oxidative stress defense, little is known about the factors influencing perR expression in C. jejuni. In this study, a perR promoter-lacZ fusion assay demonstrated that iron significantly reduced the level of perR transcription, whereas other metal ions, such as copper, cobalt, manganese, and zinc, did not affect perR transcription. Notably, a perR mutation substantially increased the level of perR transcription and in trans complementation restored the transcriptional changes, suggesting perR is transcriptionally autoregulated in C. jejuni. In the perR mutant, iron did not repress perR transcription, indicating the iron dependence of perR expression results from perR autoregulation. Electrophoretic mobility shift assays showed that PerR binds to the perR promoter, and DNase I footprinting assays identified a PerR binding site overlapping the −35 region of the two perR promoters, further supporting perR autoregulation at the transcriptional level. Alignment of the PerR binding sequence in the perR promoter with the regulatory region of other PerR regulon genes of C. jejuni revealed a 16-bp consensus PerR binding sequence, which shares high similarities to the Bacillus subtilis PerR box. The results of this study demonstrated that PerR directly interacts with the perR promoter and regulates perR transcription and that perR autoregulation is responsible for the repression of perR transcription by iron in C. jejuni.
INTRODUCTION
Defense against reactive oxygen species is crucial to the survival of bacteria possessing aerobic metabolisms. Reactive oxygen species, such as the superoxide anion, hydrogen peroxide, and hydroxyl radical, are inevitably generated as by-products of aerobiosis, causing damage to biological molecules (16). In the generation of reactive oxygen species, intracellular free iron also plays a central role through the Fenton reaction (17). To achieve prompt protection from the damaging effects of reactive oxygen species, bacteria produce various oxidative stress defense proteins, and their expressions are controlled by elaborate regulatory mechanisms. In Escherichia coli and Salmonella, for example, SoxRS and OxyR are well-known regulators to facilitate bacterial resistance to superoxide and hydrogen peroxide stresses, respectively (16). Particularly, OxyR is a hydrogen peroxide-sensing regulator activating a number of genes highly induced by hydrogen peroxide (39). Although OxyR is a widespread oxidative stress regulator, some bacterial species, including Bacillus subtilis, Staphylococcus aureus, Streptococcus pyogenes, and Campylobacter jejuni, lack OxyR and instead harbor PerR to regulate oxidative stress genes (24).
Campylobacter is a Gram-negative food-borne pathogen, causing estimated 400 to 500 million infections in humans worldwide per year (30, 38). Among Campylobacter species, Campylobacter jejuni is most frequently implicated in human campylobacteriosis, causing fever, diarrhea, and in some cases Guillain-Barré syndrome as a postinfection complication (1). As a microaerophile, C. jejuni requires 5 to 10% oxygen for its optimal growth and is sensitive to atmospheric oxygen levels. Due to the oxygen requirement for growth, C. jejuni is subject to oxidative stress and produces various oxidative resistance proteins, such as superoxide dismutase (SodB), catalase (KatA), and alkyl hydroperoxide reductase (AhpC) (2). While SodB detoxifies the superoxide anion, KatA and AhpC are involved in the peroxide stress resistance of C. jejuni (3, 11, 28). C. jejuni PerR regulates peroxide stress genes, including katA and ahpC, although PerR and PerR-like proteins have usually been reported in Gram-positive bacteria (34). In fact, C. jejuni is the first Gram-negative bacterium where a non-OxyR-dependent regulation system is reported to control peroxide stress genes (34). Since PerR represses peroxide resistance genes, a perR mutation renders C. jejuni hyper-resistant to hydrogen peroxide (34). In addition to the role of PerR in oxidative stress resistance, recent studies have demonstrated that perR expression is upregulated by acid shock (29), and a perR mutation significantly impairs C. jejuni's ability to colonize the intestines of chickens (25), suggesting that PerR mediates various pathobiological functions of C. jejuni. Although a couple of studies have thus far investigated the PerR regulon of C. jejuni, little is known about factors influencing perR expression in this bacterium despite its importance.
In the present study, we investigated the role of various metal ions in the expression of the metalloregulator PerR and presented the PerR binding sequence, the first described in Gram-negative bacteria. In addition, we demonstrate that perR is transcriptionally autoregulated and that perR autoregulation is responsible for the iron-responsive repression of perR in C. jejuni.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
C. jejuni NCTC11168 was used (27). C. jejuni NCTC11168 and its derivatives were grown at 42°C in Mueller-Hinton (MH) media (Oxoid) or MEMα (minimum essential medium alpha; Gibco, catalog no. 41061) under microaerobic conditions generated by Anoxomat (Mart Microbiology B.V, Netherlands). Culture media were occasionally supplemented with kanamycin (50 μg ml−1) or chloramphenicol (10 μg ml−1) where required. Broth cultures were microaerobically grown with shaking at 180 rpm. All of the strains and plasmids used in the present study are listed in Table 1.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F′ φ80dlacZΔM15 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 Δ(lacZYA-argF)U169 deoR λ− | Invitrogen |
| BL21(DE3) | F′ dcm ompT hsdS(rB− mB−) galDE3(lacI lacUV5-T7 gene 1 ind-1 sam-7 nin-5) | Novagen |
| C. jejuni | ||
| NCTC11168 | Wild type, a human isolate | 27 |
| FMB3001 | pMW10::PperR/NCTC11168 | This study |
| FMB3003 | perR mutant (perR::cat) | This study |
| FMB3007 | pMW10::PperR/FMB3003 | This study |
| FMB3008 | perR complementation | This study |
| Plasmids | ||
| pUC19 | Cloning and suicide vector; Ampr | New England Biolabs |
| pUC19-perR | pUC19 carrying perR | This study |
| pUC19-perR::cat | pUC19 carrying perR::cat | This study |
| pET15b | Ampr; N-terminal His tag, T7 promoter, pBR322 ori | Novagen |
| pET15b-perR | pET15b carrying perR | This study |
| pMW10 | E. coli-C. jejuni shuttle plasmid; lacZ mob repB; Kanr | 36 |
| pMW10::PperR | pMW10 carrying perR promoter | This study |
| pMW10::PperR-perR | pMW10 carrying perR promoter and perR | This study |
| pRY112 | E. coli-C. jejuni shuttle plasmid; Cmr | 37 |
Cmr, chloramphenicol resistance; Ampr, ampicillin resistance; Kanr, kanamycin resistance.
Construction of a perR mutant and a complementation strain.
A DNA region containing perR and its flanking area was amplified by PCR with perR_F_EcoRI and perR_R_PstI primers (Table 2). The PCR product was ligated into pUC19 after digestion with EcoRI and PstI. A chloramphenicol resistance cassette (cat) was PCR amplified from pRY112 (37) with Vent DNA polymerase (New England Biolabs) using the catF(SmaI) and catR(SmaI) primers and inserted into an EcoRV site of the pUC19::perR plasmid. The orientation of the cat cassette was confirmed by sequencing. The constructed plasmid (pUC19-perR::cat) was electroporated into C. jejuni, and a perR mutant was screened by growing on MH agar plates supplemented with chloramphenicol (10 μg ml−1). The allelic exchange in the perR mutant was confirmed by PCR (data not shown). A perR flanking region was PCR amplified with c_perR_F(SmaI) and c_perR_R(SmaI) primers and cloned into a noncoding region of the E. coli-C. jejuni shuttle plasmid pMW10, which had been digested with SmiI. The pMW10 harboring perR was introduced into the perR mutant by conjugation, as described previously (23).
Table 2.
Primers used in this study
| Primer | DNA sequence (5′-3′) |
|---|---|
| catF(SmaI) | GTGTTCCTTTCCCGGGTAATTGCG |
| catR(SmaI) | CATAAAAACCCGGGGGAACTAAAGGG |
| perR_F_EcoRI | TCTAGAAAAAGAATTCAATAGTTTGTTG |
| perR_R_PstI | ACACTATGATCTGCAGTTATTTGAG |
| c_perR_F(SmaI) | AATAGTTTGTTGTCTAGAGTTTTAAAAT |
| c_perR_R(SmaI) | ATTTCATAAACCTCTAGAATAACCC |
| perR_F(BamHI) | ATGATTTGGATCCAAAAGTTGTTAGCTTTG |
| perR_R(BamHI) | CGCTAAAGGGATCCAAGGGTATTC |
| perR_his_F(NdeI) | AAGCAAGGAATAAATCATATGGAATTAC |
| perR_his_R(BamHI) | CATTAAAAGGATCCTTAAAATATATGGG |
| perR_F | GATTTGCATGTAAAAGTTGTTAGCTTTGAATATGAA |
| perR_R | CGTCGCTAAAGAGATTGAAGGGTATTCTT |
| perR_in_F | AAAAGACATGAGCATCCTAATATTGATG |
| perR_in_R | GACACTTTTTGCAATTATCAACATAAGC |
| ahpC_F | ATCATCAACGATAGCATTCACTGGACAC |
| ahpC_R | AATACTACCGCTCCTTTTGGACCTATG |
| ahpC_in_F | ACTCCAGTAAATCAAGGTGGTATTGGTC |
| ahpC_in_R | TTTTTGCCAAGATATTCAGCCACGCC |
| perR_FP_F | AGCCTTGCAAGAAATGAATAATAATGC |
| perR_FP_R | ATTCATCAATATTAGGATGCTCATGTC |
| perR_PE_F | AGCCTTGCAAGAAATGAATAATAATGC |
| perR_PE_R | GCCTATCTTTTTTTCTAAATGCTCTTG |
| PE_R | ATTCATCAATATTAGGATGCTCATGTC |
Determination of the perR transcriptional level with reporter gene assays.
The promoter and partial coding region of perR were amplified with perR_F(BamHI) and perR_R(BamHI) primers and cloned into a BamHI site of pMW10 that contains the promoterless lacZ gene. The plasmid was mobilized to C. jejuni strains by conjugation. β-Galactosidase assays were performed with C. jejuni strains harboring the perR promoter (PperR)-lacZ transcriptional fusion construct, as described previously (36). To examine the effect of metal ions on perR transcription, the defined culture medium MEMα was used in the assay after supplementation with CoCl2, CuCl2,FeSO4, Fe2(SO4)3, MnCl2, or ZnCl2.
Purification of rPerR.
Recombinant PerR (rPerR) was expressed in E. coli BL21(DE3) using the pET15b vector (Novagen). The perR gene was PCR amplified with perR_his_F(NdeI) and perR_his_R(BamHI) primers. After digestion with NdeI and BamHI, the PCR products were cloned into pET15b that had been digested with the same enzymes. The constructed plasmids were transformed into E. coli BL21(DE3). E. coli harboring pET15b::perR was grown to an optical density at 600 nm of 1.0 at 37°C, and the expression of rPerR was induced by 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 6 h at 20°C. Bacterial cells were harvested by centrifugation and resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 300 mM NaCl). Cells were lysed by sonication and the cell extracts were purified under native conditions using Ni2+ affinity chromatography (Qiagen). The His tag of rPerR was removed with a thrombin cleavage capture kit (Novagen). Briefly, rPerR was treated with biotinylated thrombin, and His tag-free rPerR was separated with streptavidin agarose and spin filters. Purified rPerR protein was visualized by SDS-PAGE.
EMSA.
An electrophoretic mobility shift assay (EMSA) was performed as described elsewhere, with some modifications (19). The promoter regions of perR and ahpC and the internal coding regions of each gene for competition were amplified by PCR with the primer pairs of perR_F and perR_R (product size, 373 bp), ahpC_F and ahpC_R (403 bp), perR_in_F and perR_in_R (331 bp), and ahpC_in_F and ahpC_in_R (323 bp), respectively (Table 2). The PCR products were purified from an agarose gel using Wizard SV Gel and the PCR Clean-Up System (Promega) and labeled with [γ-32P]ATP (GE Healthcare). Unincorporated radioisotope was removed with a MicroSpinG-25 column (GE Healthcare). The 0.2 nM 32P-labeled DNA probe was incubated with rPerR at different concentrations at 37°C for 15 min in 10 μl of the gel shift assay buffer [20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 5 mM dithiothreitol (DTT), 0.2% Tween 20, 30 mM KCl, and 0.1 μg of poly(dI-dC)]. The reaction mixtures were resolved in a 6% polyacrylamide gel, and the radioactivity of the DNA probes was visualized with the BAS2500 system (Fuji Film).
Primer extension assays.
Primer extension assays were performed as described previously (31). Briefly, C. jejuni was grown to the mid-exponential phase for approximately 8 h in MH broth with shaking and harvested by centrifugation at 10,000 × g for 5 min. The total RNA was purified with TRIzol (Invitrogen) according to the manufacturer's instructions. Purified RNA was resuspended in sterile distilled RNase-free water, and the RNA concentration was determined by measuring the optical density of the solution at 260 and 280 nm using NanoVue (GE Healthcare). A portion (10 pmol) of the PE_R primer was labeled with 32P at the 5′ end by 10 U of T4 polynucleotide kinase (Invitrogen) and 80 μCi of [γ-32P]dATP for 30 min at 37°C. The labeling mixture was heated at 70°C for 10 min and purified with MicroSpin G-25 columns (GE Healthcare). The γ-32P-end-labeled primer (0.5 pmol) was coprecipitated with 15 μg of total RNA by the addition of sodium acetate and absolute ethanol. The pellet was washed with 75% ethanol, dried at room temperature, and resuspended in 20 μl of 250 mM KCl, 2 mM Tris (pH 7.9), and 0.2 mM EDTA. The mixture was heated to 65°C and then was allowed to cool to room temperature for 1 h. After annealing, 50 μl of reaction solution containing 5 μg of actinomycin D, 700 μM deoxynucleoside triphosphates, 10 mM MgCl2, 5 mM DTT, 20 mM Tris (pH 7.6), 30 U of RNasin (Promega), and 150 U of Superscript III reverse transcriptase (Invitrogen) was added. The mixture was incubated at 42°C for 70 min and treated with 100 U of RNase T1 (Invitrogen) at 37°C for 15 min. The sample was ethanol precipitated after addition of 1.4 μl of 5 M NaCl with 2.5 volumes of absolute ethanol and then washed with 75% ethanol. Each sample was resuspended with 6 μl of formamide dye and 4 μl of Tris-EDTA (pH 8.0) buffer and then denatured at 90°C for 3 min. The samples were resolved on 6% polyacrylamide-8 M urea gels, and the reverse transcription signals were analyzed by using BAS 2500 (Fuji Film). The PE_R primer (Table 2) was used for sequencing the perR promoter region with a SequiTherm EXCELII DNA sequencing system (Epicentre).
DNase I footprinting assays.
DNase I footprinting assays were performed as reported elsewhere (7). A DNA region containing the perR promoter was PCR amplified with a 32P-labeled primer perR_FP_R and a nonradiolabeled primer perR_FP_F, and the PCR product was purified from an agarose gel with a Wizard SV Gel and PCR Clean-Up System (Promega). rPerR was incubated with the 32P-labeled perR promoter at 37°C for 15 min in 40 μl of the DNA-binding buffer [20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 5 mM DTT, 0.2% Tween 20, 30 mM KCl, 10 mM MgCl2, and 0.1 μg of poly(dI-dC)], and the reaction mixture was treated with 0.1 U of DNase I (Takara). The reactions were stopped by adding 200 μl of ice-cold stop solution (0.4 M sodium acetate, 2.5 mM EDTA), and the DNA products were purified by phenol extraction and ethanol precipitation. The digested DNA fragments were separated by electrophoresis on 6% polyacrylamide-8 M urea gels alongside sequencing ladders that had been generated with the same 32P-labeled PerR_FP_R primer used to amplify DNA fragments for the DNase I digestion.
RESULTS
Amino acid sequence analysis of C. jejuni PerR.
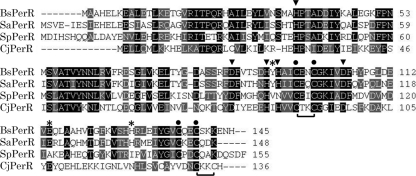
Comparison of amino acid sequences revealed that C. jejuni PerR shares 32, 31, and 31% identity to the PerR proteins from the Gram-positive B. subtilis, S. aureus, and S. pyogenes, respectively (Fig. 1). In B. subtilis, Mn2+ and Fe2+ are metal ions involved in the regulatory function of PerR, and five amino acid residues (H37, D85, H91, H93, and D104) form a square pyramid to mediate the binding of PerR either to Mn2+ or Fe2+ (18, 21). These five amino acid residues are also highly conserved in C. jejuni PerR, suggesting that C. jejuni PerR may require Mn2+ or Fe2+ for gene expression regulation as does B. subtilis PerR. The metal ion Zn2+ has a structural role in B. subtilis PerR (20), and four cysteine residues (C96-XX-C99 and C136-XX-C139 of B. subtilis PerR; “X” represents any amino acid residue) are associated with Zn2+ binding in a tetrahedral fashion (18, 21, 33). The cysteine pair C96-XX-C99 of B. subtilis PerR is highly conserved among the PerR proteins, including C. jejuni PerR (C89-XX-C92 of C. jejuni PerR), and another cysteine pair (C132-XX-C135) is also present in C. jejuni PerR near the conserved B. subtilis C136-XX-C139 motif (Fig. 1). Amino acid residues of the less conserved metal binding site, such as Y92, E114, and H128 of B. subtilis PerR, are known to influence metal ion selectivity between Mn2+ and Fe2+ (22). Although these three amino acid residues are relatively well conserved in Gram-positive bacteria, they are not conserved in C. jejuni except E114 (E107 of C. jejuni PerR) (Fig. 1). These results suggest that C. jejuni PerR may bind to Zn2+ and either Mn2+ or Fe2+ as does B. subtilis PerR; however, poor conservation of the amino acid residues facilitating metal-ion selectivity suggests that C. jejuni PerR may have different metal-ion selectivity from B. subtilis PerR.
Fig. 1.
Sequence alignment of the PerR proteins. The amino acid residues required for binding to a regulatory metal ion (Mn2+ or Fe2+) in PerR (21, 33) are indicated with an arrowhead. Two zinc-binding Cys-XX-Cys motifs reported in B. subtilis (18, 21, 33) are marked with a dot, and Cys-XX-Cys motifs in C. jejuni are indicated with a bracket. Amino acid residues involved in metal ion selectivity (22) are indicated with an asterisk. The sequences for B. subtilis PerR (BsPerR; GenBank accession number NP_388753.1), S. aureus PerR (SaPerR; NP_646618), S. pyogenes (SpPerR; YP_595895.1), and C. jejuni PerR (CjPerR; YP_002343760) are shown.
Effect of metal ions on perR expression.
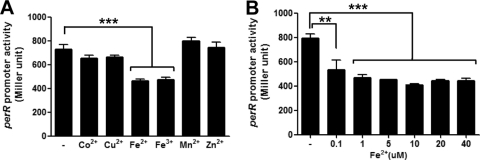
To investigate the role of metal ions in perR expression, the level of perR transcription was measured with β-galactosidase assays after the addition of various metal ions, such as Co2+, Cu2+, Fe2+, Fe3+, Mn2+, and Zn2+, to the defined medium MEMα, which does not contain any of these metal ions. The PperR-lacZ fusion assay showed that perR is constitutively expressed independent of the growth phase of C. jejuni (date not shown). The tested metal ions exhibited differential effects on perR transcription. The presence of iron, either ferrous (Fe2+) or ferric (Fe3+), significantly repressed perR transcription (Fig. 2A), whereas Co2+, Cu2+, Mn2+, and Zn2+ did not alter the level of perR transcription (Fig. 2A). When different concentrations of iron were used in the assay, as low as 0.1 μM iron significantly decreased perR expression, and the reduction of perR transcription by iron was saturated at concentrations greater than 5 μM (Fig. 2B). These findings demonstrate that, in C. jejuni, perR expression is primarily regulated in response specifically to iron even at low concentrations. To address whether hydrogen peroxide influences the responsiveness of perR transcription to metal ions, the promoter fusion assay was conducted in the presence of hydrogen peroxide; however, hydrogen peroxide did not make any significant changes (see Fig. S1 in the supplemental material). Regardless of the presence or absence of hydrogen peroxide, iron was the major metal ion affecting perR transcription.
Fig. 2.
Effects of metal ions on perR transcription. (A) PperR-lacZ fusion assays in the presence of 20 μM CoCl2, 20 μM CuCl2,40 μM FeSO4, 40 μM Fe2(SO4)3, 40 μM MnCl2, or 10 μM ZnCl2. The “-” indicates the negative control without any metal ions tested. (B) Concentration-dependent reduction of perR transcription by iron. The “-” indicates the negative control without iron. Metal ions were added to MEMα, and C. jejuni cultures were grown for 8 h before carrying out the assay. The results show the means and standard deviations of three independent experiments. **: P < 0.01; ***, P < 0.001. The significance of the results was statistically analyzed by one-way analysis of variance (ANOVA) with Dunnett's post tests at a 95% confidence interval using Prism software (version 5.01; GraphPad Software, Inc.).
Derepression of perR transcription by a perR mutation.
A sequence resembling the B. subtilis PerR box was found upstream of perR in C. jejuni (35); however, it has been unknown whether PerR is autoregulated in C. jejuni. To address whether PerR regulates perR expression in C. jejuni, a PperR-lacZ fusion assay was performed with a perR mutant. Due to the effect of iron on perR transcription (Fig. 2), the assay was conducted with the defined medium MEMα in the presence or absence of iron. Interestingly, the PperR-lacZ fusion assay demonstrated that perR expression was significantly increased in the perR mutant compared to the wild type (Fig. 3), suggesting that perR is autoregulated in C. jejuni. Compared to the wild type, in trans complementation of perR slightly reduced the level of perR transcription (Fig. 3), presumably because PerR produced from the perR gene in the multicopy shuttle plasmid further repressed perR expression. Interestingly, the perR mutation eliminated iron-dependent repression of perR (Fig. 3), indicating that the perR repression by iron is mediated by perR autoregulation. These results clearly demonstrated that perR is autoregulated at the transcriptional level in C. jejuni and that perR autoregulation accounts for the repression of perR transcription by iron.
Fig. 3.
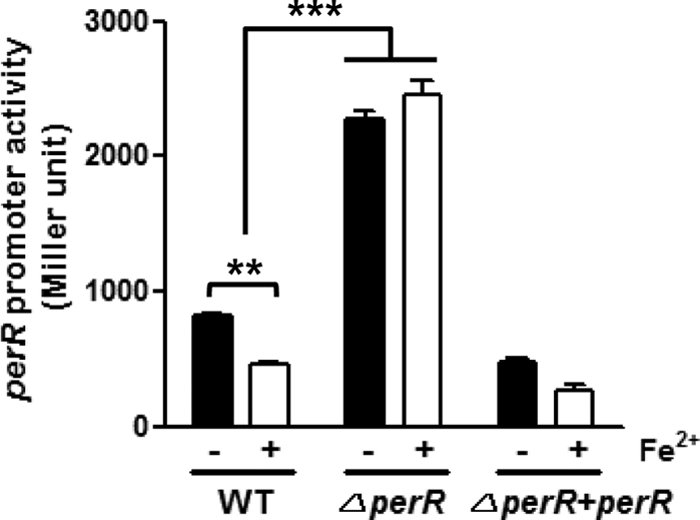
Derepression of perR expression by a perR mutation. The results of PperR-lacZ fusion assays show the level of perR transcription in the wild type (WT), the perR mutant (ΔperR), and the complementation strain (ΔperR+perR) in the presence (+) or absence (−) of iron. The results show the means and standard deviations of three independent experiments. **, P < 0.01; ***, P < 0.001. The significance of results was statistically analyzed by two-way ANOVA of variance with Bonferroni's post tests at a 95% confidence interval using Prism software (version 5.01; GraphPad Software, Inc.).
Interaction of PerR with the perR promoter.
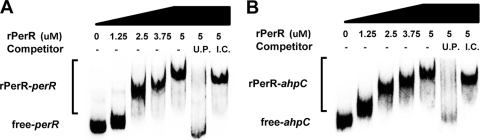
Because the PperR-lacZ fusion assay showed that perR is transcriptionally autoregulated (Fig. 3), an EMSA was performed to investigate whether PerR directly binds to the perR promoter. The rPerR protein was expressed in and purified from E. coli and was used in EMSA. The results of the EMSA demonstrated that PerR bound to the perR promoter, whereas DNA fragments amplified from the perR coding region did not compete with the DNA probe of the perR promoter region, confirming the specificity of PerR binding to the perR regulatory region (Fig. 4A). To confirm the binding of PerR to a known PerR-regulated gene, the ahpC promoter was included as a positive control in EMSA (Fig. 4B), because PerR is known to regulate ahpC in C. jejuni (34). These findings show that PerR binds to the perR promoter, strongly indicating PerR directly regulates its own transcription.
Fig. 4.
Binding of PerR to the promoter regions of perR (A) and ahpC (B). The ahpC promoter was used as a positive control of EMSA. The concentrations of rPerR are indicated above the panel. “U.P.” and “I.C.” stand for “unlabeled probe” and “internal coding region” used as competitors in the assay.
Determination of the PerR binding site in the perR promoter.
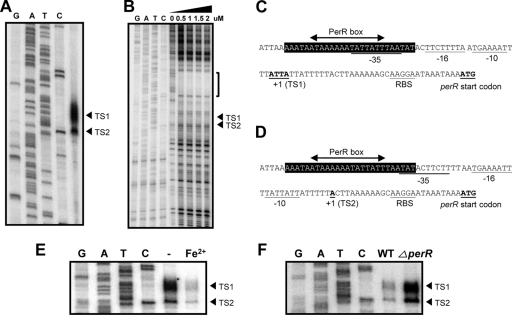
DNase I footprinting assays were performed to determine the PerR binding site in the perR promoter. Prior to DNase I footprinting analysis, primer extension assays were conducted to define the perR promoter. Two transcriptional start sites were found in the promoter region of perR (Fig. 5A). The transcriptional start site (TS2; Transcriptional Start site 2) located at 26-bp upstream of the perR start codon was sharp, whereas the other transcriptional start site (TS1; Transcriptional Start site 1) was broad; presumably, the repeated A and T around TS1 made the control of transcription initiation at TS1 loose. DNA sequences similar to the consensus RpoD promoter of C. jejuni were found upstream of each transcriptional start site (Fig. 5C and D). DNase I footprinting assay exhibited that a 26-bp AT-rich region from −36 to −61 was protected from DNase I cleavage (Fig. 5B). The protected region contained a sequence resembling the canonical PerR box of B. subtilis, and the PerR binding site determined by DNase I footprinting overlaps with the putative −35 region of the two perR promoters (Fig. 5C and D); thus, binding of PerR would hinder access of RNA polymerase to the perR promoter and represses perR transcription. Consistent with the effects of iron and the perR mutation on perR transcription (Fig. 2 and 3), primer extension assays showed that iron significantly reduced the level of perR transcripts from the two transcriptional start sites (Fig. 5E), and the perR mutation notably increased the signal intensity of the two start sites (Fig. 5F), indicating that both iron and PerR affects the initiation of perR transcription at the two transcriptional start sites. The results demonstrated that PerR recognizes the PerR box present in the perR promoter, and its binding to the perR regulatory region may interfere with perR transcription.
Fig. 5.
Characterization of the perR transcriptional start site and identification of the PerR binding site. (A) Determination of the transcriptional start site of perR by a primer extension assay. Two transcriptional start sites of perR are designated TS1 (Transcriptional Start site 1) and TS2, which are indicated with an arrowhead on the right. (B) Determination of the PerR binding site in the perR promoter by DNase I footprinting. The PerR binding region is indicated with a bracket and the transcriptional start sites are marked with an arrowhead. The perR promoters initiating transcription at TS1 (C) and TS2 (D). The −10, −16, and −35 elements of the perR promoter are underlined. The region protected from DNase I cleavage is indicated with black background, and a sequence similar to the B. subtilis PerR box is marked with an arrowed line. (E) Repression of perR transcription by iron at the transcriptional start site TS1 and TS2. Primer extension assay was conducted with 10 μg of total RNA isolated from C. jejuni cultures grown in MEMα in the absence (-) or presence of 40 μM FeSO4. (F) Derepression of perR transcription by the perR mutation. Primer extension assay was performed with 14 μg of total RNA from the wild type (WT) and the perR mutant (ΔperR) grown in MEMα in the presence of 40 μM FeSO4.
Consensus PerR binding sequence in C. jejuni.
Based on the PerR binding sequence present in the perR promoter, putative PerR binding sequences were identified in the promoter region of PerR regulon genes, such as ahpC, katA, and dps (25, 34). Sequence alignment revealed the PerR binding sequence of C. jejuni (ATaAtAATwATTaTwA; “w” means A or T, capital letters are identical residues, and lowercase letters indicate less-conserved residues) (Fig. 6A), which is quite similar to the canonical PerR boxes reported in other Gram-positive bacteria (Fig. 6B). The conserved PerR binding sequence determined by aligning the PerR binding sequences of several bacterial species is identical to the B. subtilis PerR box (Fig. 6B). These findings demonstrate that the C. jejuni PerR binding site shares high sequence similarities with the PerR boxes of Gram-positive bacteria.
Fig. 6.
Determination of the perR binding sequence in C. jejuni. (A) Multiple alignment of the PerR box present in the perR promoter and putative PerR binding sequences of the PerR-regulated genes of C. jejuni. The black background indicates identical or highly conserved residues. Sequence logo was generated with the WebLogo version 2.8.2 (5). (B) Comparison of consensus PerR binding sequences of C. jejuni, B. subtilis (9), S. aureus (15), and S. pyogenes (4). Identical residues are written in boldface capital letters.
DISCUSSION
As a metalloregulatory protein, PerR has exhibited differential metal ion dependency in multiple bacterial species. The B. subtilis PerR protein contains two metal ions; Zn2+ and either Fe2+ or Mn2+, where the former is required for protein stability and the latter participates in regulation (13, 20). While Mn-bound PerR prevents full derepression of the PerR regulon genes by hydrogen peroxide, Fe-bound PerR demonstrates complete derepression of the PerR regulon genes by hydrogen peroxide (13). Although both Fe2+ and Mn2+ can bind to PerR and modulate the expression of the PerR regulon genes, only Mn2+ is involved in the regulation of perR expression in B. subtilis in a way that Mn2+ represses perR expression (10). Similarly, in S. aureus, perR expression is repressed by Mn2+, and Fe2+ rather increases the level of perR transcription (15). However, the CatR regulator, a PerR homolog in Streptomyces coelicolor, does not appear to be responsive to metal ions (12). Previous proteomic and transcriptomic analyses have reported that the expression level of perR and the PerR regulon genes, such as katA and ahpC, are reduced by iron in C. jejuni (14, 26); however, details of the effect of iron on perR expression and the roles of other metal ions in perR expression have not been understood. In the present study, perR expression was significantly repressed by iron even at low (0.1 μM) concentrations (Fig. 2). Unlike B. subtilis and S. aureus, Mn2+ did not affect perR expression in C. jejuni (Fig. 2A), and other metal ions, including Co2+, Cu2+, and Zn2+, did not change the level of perR transcription (Fig. 2A). Both Fe2+ and Fe3+ similarly repressed perR transcription (Fig. 2A), probably due to intracellular conversion between Fe2+ and Fe3+.
Notably, a perR mutation eliminated iron-mediated repression of perR transcription (Fig. 3), suggesting the repression of perR by iron results from perR autoregulation. In B. subtilis and S. aureus, perR is autoregulated; however, in these bacteria, perR is repressed by Mn2+, but not by Fe2+ (10, 15). Structural analysis of the B. subtilis PerR protein revealed that five amino acid residues (H37, D85, H91, H93, and D104) of the regulatory metal binding site form a square pyramidal environment to capture either Mn2+ or Fe2+ (21, 33). Amino acid sequence analysis of PerR proteins showed that these five amino acid residues are well conserved in most PerR proteins except H93 of S. pyogenes PerR, which is replaced with an asparagine (N101; Fig. 1). Recently, Ma et al. reported that amino acid residues in the less-conserved metal binding site of B. subtilis PerR, including Y92, E114, and H128, significantly affected the metal ion selectivity between Mn2+ and Fe2+, possibly by influencing the coordination geometry of the PerR protein (22). The three amino acid residues are highly conserved in S. aureus PerR, whereas only E114 (E107 of C. jejuni PerR) is conserved in C. jejuni PerR (Fig. 1). Consistently, perR transcription is repressed by Mn2+ in B. subtilis and S. aureus (10, 15); however, in C. jejuni, perR expression was insensitive to Mn2+ but exhibited substantial responsiveness to Fe2+ (Fig. 2). Based on this report, the poor conservation of the amino acid residues mediating the metal ion selectivity may result in different metal ion dependence of perR expression in C. jejuni compared to B. subtilis and S. aureus.
It has been reported that PerR regulates the oxidative stress defense genes in C. jejuni (25, 34); however, interaction of PerR with the promoter of PerR-regulated genes has not yet been empirically proven. In the present study, we first demonstrated PerR binding to the promoter region of the perR-regulated genes in C. jejuni (Fig. 4). PerR bound to the regulatory regions of perR and ahpC (Fig. 4); ahpC was used as a positive control in EMSA, because ahpC is known to be regulated by PerR in C. jejuni (25, 34). Two adjacent transcriptional start sites of perR were identified by primer extension analysis (Fig. 5A). The same results were obtained in our multiple trials of primer extension analysis by using several different primers and RNA samples from different batches (data not shown). Two perR promoters resembling the consensus RpoD promoter sequence of C. jejuni were identified upstream of the transcriptional start sites (Fig. 5C and D). DNase I footprinting assays identified a 26-bp region overlapping with the −35 region of two perR promoters (Fig. 5B, C, and D), suggesting that PerR binding may interfere with perR transcription driven by the two perR promoters. In order to assess which promoter is primarily affected by iron and PerR, primer extension analysis was performed in the presence or absence of iron and with the perR mutant. The results demonstrated that the transcript levels of both TS1 and TS2 were affected by iron and the perR mutation (Fig. 5E and F). This is consistent with the finding that the PerR binding site overlaps with the −35 region of two perR promoters (Fig. 5C and D). Although both of the perR promoters were regulated by iron in the present study, it is highly possible that these two promoters can be differentially regulated under certain conditions. It would be an interesting future study to investigate the regulation of the two perR promoters in C. jejuni.
In B. subtilis and S. aureus, the PerR binding sequences are known to be an AT-rich inverted repeat (9, 15). Particularly, in B. subtilis, the PerR target site consists of 15 bp with a core 7-1-7 inverted repeat (9). Similarly, the PerR binding site of C. jejuni is an inverted repeat consisting of only A and T (Fig. 6A). To the best of our knowledge, this is the first presentation of the PerR binding sequence in Gram-negative bacteria. Alignment of the PerR binding sequences of a few bacterial species revealed a consensus PerR binding sequence which is identical to that of B. subtilis (Fig. 6B). This suggests that the PerR binding sequences are highly conserved in both Gram-positive bacteria and the Gram-negative C. jejuni. In B. subtilis, two PerR boxes are present in the perR promoter, and one of the two PerR boxes overlaps with the −10 region of the perR promoter (10). A PerR box was identified by bioinformatics analysis in the downstream region of the perR transcriptional start site in S. aureus (15). Consistent with the presence of PerR box in the perR promoter regions of B. subtilis and S. aureus, perR is autoregulated in these bacteria, since a perR mutation derepresses perR transcription based on promoter fusion assays (10, 15). In contrast to B. subtilis and S. aureus, no PerR box was observed in the perR promoter of S. pyogenes (4), indicating that perR may not be autoregulated in this catalase-negative bacterium. A previous in silico analysis of the C. jejuni genome revealed the presence of a region in the perR promoter, whose DNA sequence is similar to the B. subtilis PerR box (35). In fact, the PerR binding region determined by DNase I footprinting in this study overlapped with the PerR box predicted by the in silico analysis due to its high sequence similarity to the PerR box of B. subtilis.
In C. jejuni, PerR represses katA and ahpC, encoding the major enzymes conferring resistance to peroxide stress (25, 34). As the sole catalase present in C. jejuni, KatA significantly contributes to the peroxide stress resistance and intracellular survival of C. jejuni (6, 11). The alkyl hydroperoxide reductase AhpC scavenges low levels of hydrogen peroxide in E. coli (32) and is involved in the defense against organic peroxide (e.g., cumene hydroperoxide) in C. jejuni (3). Reduced expression of perR will lead to derepression of katA and ahpC, and subsequently derepressed expression of katA and ahpC will render C. jejuni resistant to peroxide stress, whereas increased expression of perR will make C. jejuni susceptible to peroxide stress due to enhanced repression of katA and ahpC by PerR. By perR autoregulation, C. jejuni can tightly maintain the intracellular level of PerR and controls the expression of peroxide resistance genes. Therefore, perR autoregulation is important in the regulation of the oxidative stress defense of C. jejuni. In addition, iron is the metal ion involved in the generation of reactive oxygen species, particularly hydroxyl radicals, via the Fenton reaction. In fact, there is no enzyme capable of detoxifying hydroxyl radicals, although hydroxyl radicals can give damages to most biomolecules (16). Therefore, the iron dependence of perR autoregulation may enable C. jejuni to sense the level of intracellular free iron available for the Fenton reaction.
Multiple studies reported that both perR and perR regulon genes, including katA and ahpC, are repressed by iron (3, 14, 26), although the iron-dependent repression of ahpC and katA is mediated primarily by PerR (34). This cannot be simply explained by the role of PerR as a repressor of katA and ahpC, because reduced perR expression by iron should derepress katA and ahpC. It can be postulated that high concentrations of iron reduce perR expression but rather increase the level of iron-bound PerR, which in turn may further repress katA and ahpC at high iron concentrations. In order to prove this, however, we still need to understand the interaction of PerR with iron in C. jejuni and the contribution of iron to the DNA-binding activity of PerR. According to a report by van Vliet et al. (34), although the perR mutation substantially increased the KatA activity in C. jejuni, the KatA activity was still affected by iron in the perR mutant; the KatA activity was 2-fold higher in the low-iron condition than in the high-iron condition (34). However, the effect of iron on the KatA activity disappeared in the perR and fur double mutant, whereas a fur single mutation alone did not eliminate the iron-dependent regulation of the KatA activity (34). This suggests that the PerR regulon is perhaps regulated by the interplay between PerR and Fur in association with iron. In addition, we still cannot exclude the involvement of an unknown regulator, which may overcome the derepression of katA and ahpC by PerR and still can repress the PerR regulon genes in the presence of iron.
A transcriptomic analysis of the perR regulon performed by Palyada et al. showed that the microarray expression ratio of perR is not different between the perR mutant and the wild-type C. jejuni regardless of the presence of iron (25). This discrepancy can be attributed to the intrinsic differences between microarray analysis and promoter-fusion assay under different experimental settings. DNA microarray is best designed to analyze transcriptomic changes, but the results can be unavoidably affected by multiple factors, such as the high or low expression levels of transcripts (8). The perR transcription level was higher in the perR mutant than in the wild type by approximately 2- and 3-fold in the absence and presence of iron, respectively, as determined by the PperR-lacZ fusion assay (Fig. 3). Such fold changes could not be fully reflected in the microarray analysis. In addition, the experimental conditions were also different. To assess the effect of excess iron, Palyada et al. treated C. jejuni with iron for short time (15 min) prior to RNA extraction, whereas iron was added at the beginning of bacterial culture in the present study; microarray requires RNA samples that are produced right after condition changes, whereas the lacZ promoter fusion assay needs sufficient time to synthesize the β-galactosidase enzyme, whose expression is driven by the fused promoter.
In summary, we clearly demonstrated here that perR transcription is reduced by iron in C. jejuni and that perR autoregulation mediates the iron-responsive repression of perR. Based on the important roles played by PerR and iron in oxidative stress resistance, these findings provide new insights into the regulation of oxidative stress resistance in C. jejuni. In addition, we first reported the PerR box in Gram-negative bacteria, showing that the PerR box of C. jejuni is highly homologous to that of other Gram-positive bacteria, such as B. subtilis and S. aureus. Future studies will investigate how PerR senses peroxide stress and how PerR interplays with other regulatory elements in C. jejuni.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the startup funding from UPEI to B.J. and the Korean Health Technology R&D Project (A09058009010000100) to S.R. from the Ministry for Health, Welfare and Family Affairs, Republic of Korea. M.K. and S.H. are recipients of a graduate study fellowship provided by the Ministry of Education through the Brain Korea 21 Project.
Footnotes
Supplemental material for this article may be found at hhttp://jb.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Allos B. M. 2001. Campylobacter jejuni Infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201–1206 [DOI] [PubMed] [Google Scholar]
- 2. Atack J. M., Kelly D. J. 2009. Oxidative stress in Campylobacter jejuni: responses, resistance, and regulation. Future Microbiol. 4:677–690 [DOI] [PubMed] [Google Scholar]
- 3. Baillon M. L., van Vliet A. H., Ketley J. M., Constantinidou C., Penn C. W. 1999. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 181:4798–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brenot A., King K. Y., Caparon M. G. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55:221–234 [DOI] [PubMed] [Google Scholar]
- 5. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Day W. A., Jr., Sajecki J. L., Pitts T. M., Joens L. A. 2000. Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68:6337–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delany I., Spohn G., Rappuoli R., Scarlato V. 2002. Growth phase-dependent regulation of target gene promoters for binding of the essential orphan response regulator HP1043 of Helicobacter pylori. J. Bacteriol. 184:4800–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Draghici S., Khatri P., Eklund A. C., Szallasi Z. 2006. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 22:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fuangthong M., Helmann J. D. 2003. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J. Bacteriol. 185:6348–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fuangthong M., Herbig A. F., Bsat N., Helmann J. D. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grant K. A., Park S. F. 1995. Molecular characterization of katA from Campylobacter jejuni and generation of a catalase-deficient mutant of Campylobacter coli by interspecific allelic exchange. Microbiology 141:1369–1376 [DOI] [PubMed] [Google Scholar]
- 12. Hahn J. S., Oh S. Y., Chater K. F., Cho Y. H., Roe J. H. 2000. H2O2-sensitive Fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 275:38254–38260 [DOI] [PubMed] [Google Scholar]
- 13. Herbig A. F., Helmann J. D. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41:849–859 [DOI] [PubMed] [Google Scholar]
- 14. Holmes K., et al. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151:243–257 [DOI] [PubMed] [Google Scholar]
- 15. Horsburgh M. J., Clements M. O., Crossley H., Ingham E., Foster S. J. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imlay J. A. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imlay J. A. 2006. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59:1073–1082 [DOI] [PubMed] [Google Scholar]
- 18. Jacquamet L., et al. 2009. Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol. Microbiol. 73:20–31 [DOI] [PubMed] [Google Scholar]
- 19. Jeon B., Zhang Q. 2007. Cj0011c, a periplasmic single- and double-stranded DNA-binding protein, contributes to natural transformation in Campylobacter jejuni. J. Bacteriol. 189:7399–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee J. W., Helmann J. D. 2006. Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J. Biol. Chem. 281:23567–23578 [DOI] [PubMed] [Google Scholar]
- 21. Lee J. W., Helmann J. D. 2006. The PerR transcription factor senses H2O2 by metal-catalyzed histidine oxidation. Nature 440:363–367 [DOI] [PubMed] [Google Scholar]
- 22. Ma Z., Lee J. W., Helmann J. D. 2011. Identification of altered function alleles that affect Bacillus subtilis PerR metal ion selectivity. Nucleic Acids Res. 39:5036–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller W. G., et al. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mongkolsuk S., Helmann J. D. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9–15 [DOI] [PubMed] [Google Scholar]
- 25. Palyada K., et al. 2009. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Palyada K., Threadgill D., Stintzi A. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parkhill J., et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed] [Google Scholar]
- 28. Pesci E. C., Cottle D. L., Pickett C. L. 1994. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect. Immun. 62:2687–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reid A. N., Pandey R., Palyada K., Naikare H., Stintzi A. 2008. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl. Environ. Microbiol. 74:1583–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruiz-Palacios G. M. 2007. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44:701–703 [DOI] [PubMed] [Google Scholar]
- 31. Ryu S., Garges S. 1994. Promoter switch in the Escherichia coli pts operon. J. Biol. Chem. 269:4767–4772 [PubMed] [Google Scholar]
- 32. Seaver L. C., Imlay J. A. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Traore D. A., et al. 2006. Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol. Microbiol. 61:1211–1219 [DOI] [PubMed] [Google Scholar]
- 34. van Vliet A. H., Baillon M. L., Penn C. W., Ketley J. M. 1999. Campylobacter jejuni contains two Fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181:6371–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Vliet A. H., Ketley J. M., Park S. F., Penn C. W. 2002. The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26:173–186 [DOI] [PubMed] [Google Scholar]
- 36. Wösten M. M., Boeve M., Koot M. G., van Nuenen A. C., A. van der Zeijst B. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yao R., Alm R. A., Trust T. J., Guerry P. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127–130 [DOI] [PubMed] [Google Scholar]
- 38. Young K. T., Davis L. M., Dirita V. J. 2007. Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5:665–679 [DOI] [PubMed] [Google Scholar]
- 39. Zheng M., et al. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.