Abstract
In this study, whole-genome microarrays were used to gain insights into the global molecular response of Lactococcus lactis subsp. lactis IL1403 at an early stage of infection with the lytic phage c2. The bacterium differentially regulated the expression of 61 genes belonging to 14 functional categories, including cell envelope processes (12 genes), regulatory functions (11 genes), and carbohydrate metabolism (7 genes). The nature of these genes suggests a complex response involving four main mechanisms: (i) induction of membrane stress proteins, (ii) d-alanylation of cell wall lipoteichoic acids (LTAs), (iii) maintenance of the proton motive force (PMF), and (iv) energy conservation. The phage presence is sensed as a membrane stress in L. lactis subsp. lactis IL1403, which activated a cell wall-targeted response probably orchestrated by the concerted action of membrane phage shock protein C-like homologues, the global regulator SpxB, and the two-component system CesSR. The bacterium upregulated genes (ddl and dltABCD) responsible for incorporation of d-alanine esters into LTAs, an event associated with increased resistance to phage attack in Gram-positive bacteria. The expression of genes (yshC, citE, citF) affecting both PMF components was also regulated to restore the physiological PMF, which was disrupted following phage infection. While mobilizing the response to the phage-mediated stress, the bacterium activated an energy-saving program by repressing growth-related functions and switching to anaerobic respiration, probably to sustain the PMF and the overall cell response to phage. To our knowledge, this represents the first detailed description in L. lactis of the molecular mechanisms involved in the host response to the membrane perturbations mediated by phage infection.
INTRODUCTION
Virulent bacteriophages (phages) are ubiquitous in the dairy industry environment, where they can largely affect the technological efficiency of Lactococcus lactis strains used as starters in the production of fermented foods. Researchers have been able to minimize phage impact and extend the life span of lactococcal cultures by creating isogenic derivatives with enhanced phage resistance and by devising rotation regimes of strains carrying various protection mechanisms (16). However, the phages' evolutionary ability to circumvent these mechanisms challenges the long-term effectiveness of such strategies, and thus, alternative and more sophisticated antiphage solutions are constantly required. These can be generated from a better understanding of the molecular mechanisms underlying phage-host interaction, which can be achieved by using high-throughput transcriptomic technologies such as microarrays and quantitative real time-PCR (qRT-PCR). These techniques allow a genome-wide transcriptional profiling analysis and have been successfully used to follow changes in gene expression of the bacterial host during infection with lytic phages (50, 52). In L. lactis, the genes responsive to the early stages of a phage infection could allow the identification of potential targets (i.e., gene networks, promoters, and regulatory elements) for the rational design of new antiphage strategies for the improvement of starter cultures.
Phage c2 and L. lactis subsp. lactis IL1403 (L. lactis IL1403) provide an excellent model system for such a study, as their complete genome sequences are known (5, 39). Twelve groups have been defined for lactococcal phages, but only three main species of the Siphoviridae, namely, c2, 936, and P335, are commonly found in dairy plants (23). Phage c2 is a member of the c2 group, which represents almost one-quarter of the industrial phage isolates and is composed of virulent phages with isometric and moderately elongated (prolate) heads sharing extensive DNA homology and similar structural protein profiles. Phage c2 is also one of the few lactococcal phages requiring a two-step mechanism to successfully infect the bacterial host. The phage initially adsorbs reversibly to a primary receptor, suggested to be composed of rhamnose moieties (42). Subsequently, phage c2 binds irreversibly to the membrane-anchored phage infecting protein, Pip (42, 64). The expression of phage c2 genes is temporally but not tightly regulated and is heavily dependent on the host machinery. While no middle expression period is observed, transcription of 22 early genes is driven by five highly active early promoters, and transcription of the 17 late genes is controlled from a single late promoter (38).
An efficient bacterial host for phage c2 is L. lactis IL1403, one of the best-characterized lactic acid bacteria. Its 2.3-Mbp genome contains 2,310 protein-encoding genes, which include six prophages, 43 insertion sequence (IS) elements, and gene sets for DNA transformation and possible fermentation pathways and aerobic respiration (5). It also encodes a type I restriction-modification (R/M) system able to restrict by 2 log units the efficiency of plaquing (EOP) of the 936-type phage bIL67 (55).
In this study, we have used whole-genome microarrays to obtain a holistic view of the L. lactis subsp. lactis IL1403 response at the onset of infection by the lytic phage c2. Our objective was to gain insights into whether any defense strategy is activated by this phage-sensitive strain in response to phage infection. Our results suggest that the host activates a sophisticated response involving induction of membrane stress proteins, d-alanylation of the cell wall, maintenance of the proton motive force (PMF), and energy conservation. The host response is strongly targeted to the cell wall, thus suggesting that the phage presence is sensed as an extracytoplasmic stress affecting membrane integrity. The regulation and implications of this response on the chances of cell survival to phage infection are discussed.
MATERIALS AND METHODS
Bacterial strains.
L. lactis subsp. lactis IL1403 was used for whole-genome expression profiling of the host response to phage infection. L. lactis subsp. cremoris MG1363 (L. lactis MG1363) served as the sensitive host for propagation of phage c2 at high titers. Lactococcal strains were routinely propagated at 30°C in M17 medium (Oxoid, Hampshire, England) supplemented with 0.5% (wt/vol) glucose (GM17). Solid media contained 1.0% (wt/vol) bacteriological agar (Oxoid, Hampshire, England). Bacterial strains were stocked in M17 containing 40% glycerol at −80°C. Working cultures were stored at 4°C and transferred periodically.
Preparation of high-titer stock solutions of bacteriophage c2.
Phage c2 was propagated to high titers on L. lactis MG1363 as previously described (23). Concentration and purification of the phage lysate by polyethylene glycol (PEG) precipitation followed by cesium chloride (CsCl) gradient centrifugation were performed as follows. Briefly, cell debris was removed from the phage lysate by centrifugation at 11,000 × g for 30 min at 4°C. The phage-containing supernatant was recovered, and the phage was concentrated by addition of 15% PEG 8000 and 1 M NaCl, followed by slow agitation for 24 h at 4°C. Phage was precipitated by centrifugation at 16,700 × g for 60 min at 4°C, and the phage pellet was resuspended in 4 ml of TMN buffer (10 mM Tris-HCl, 10 mM MgSO4, 500 mM NaCl; pH 7.5). Concentrated phage was purified by centrifugation at 400,000 × g for 3 h at 4°C on a discontinuous-step CsCl gradient (1.3, 1.5, and 1.7 g/ml). The phage preparation was desalted by dialysis against 10 mM Na3PO4 buffer (pH 7.0) for 24 h at 4°C, recovered in 1 ml TMN buffer, and titrated at 30°C according to standard procedures (59). This protocol consistently yielded phage c2 stock solutions with titers in the range of 1013 PFU/ml. Phage stock solutions were stored at 4°C.
Determination of MOI value for phage-host challenge.
The multiplicity of infection (MOI) of phage c2 necessary to completely lyse a L. lactis IL1403 population within 40 min was determined by infecting cells in mid-log phase (optical density at 600 nm [OD600] = 0.4) with increasing phage titers. Clearing of the bacterial population was evaluated by measuring the cell density at 600 nm.
Microarray design.
The whole-genome sequences of L. lactis IL1403 (2,310 genes) and phage c2 (39 genes) were used to design 50-mer oligonucleotide probes for each gene which were subsequently spotted onto glass slides in duplicate. Control elements on each array included negative-control probes from unrelated Arabidopsis thaliana genes. Oligonucleotide probe design and microarray production were carried out by Ocimum Biosolutions Ltd. (Hyderabad, India).
Total RNA isolation and mRNA enrichment.
Total RNA was isolated as follows. An overnight culture of L. lactis IL1403 was used to prepare a fresh 0.5% inoculum in 40 ml of GM17 broth, which was propagated at 30°C to mid-log phase (OD600 = 0.4). The cell culture was split into two 20-ml aliquots (experimental and reference) to which were added 10 mM CaCl2 and phage c2 (MOI = 800) (experimental) or TMN buffer (reference). Following incubation at 30°C for 10 min to allow phage adsorption on cells, samples were immersed in a −80°C ethanol bath for 5 min to prevent any changes in gene expression profiles. After centrifugation at 11,000 × g for 5 min, cell pellets were resuspended in 200 μl of Tris-EDTA buffer (pH 8.0) containing 6 mg/ml lysozyme and incubated at 37°C for 10 min. Total RNA was extracted using a RiboPure-Bacteria kit (Ambion) and contaminating DNA was removed with a DNA-free kit (Ambion), according to the manufacturer's recommendations. The amount and purity of recovered RNA were determined by reading the absorbance at 260 nm for RNA concentration and the absorbances at 230 and 280 nm for solvent and protein contamination, respectively, by means of a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). RNA integrity was verified on glyoxal-agarose gels after ethidium bromide staining. Total RNA samples (50 μg) were enriched for mRNA by using an mRNA-ONLY prokaryotic mRNA isolation kit (Epicentre Biotechnologies).
cDNA preparation and labeling.
Reverse transcription of enriched mRNA into single-stranded cDNA was initiated by adding 9 μg of random hexamers (Invitrogen) and 2 μl of a 50× aminoallyl-deoxynucleoside triphosphate stock mixture (25 mM dATP, dGTP, and dCTP, 10 mM dTTP, and 15 mM aminoallyl-dUTP). The volume was adjusted to 34 μl with RNase-free water before the mixture was incubated at 70°C for 10 min, followed by 10 min at 25°C to allow primer annealing. After addition of 12 μl of 5× First Strand buffer (Invitrogen), 3 μl of 0.1 M dithiothreitol (Invitrogen), 3 μl of SUPERase-In (20 U/μl; Ambion), and 6 μl of SuperScript III reverse transcriptase (200 U/μl; Invitrogen), the mixture was first incubated at 25°C for 10 min and then at 42°C for 24 h. cDNA synthesis was stopped by heating at 95°C for 10 min, and the RNA template was removed by alkaline treatment with 20 μl of 0.5 M NaOH and 10 μl of 0.5 M EDTA at 65°C for 30 min. Ten microliters of 1 M HCl was added to neutralize the cDNA mixture, which was then purified using a QIAquick PCR purification kit (Qiagen, Germany), according to the manufacturer's instructions, except that the free amine-containing wash and elution buffers were replaced with an ethanol-based phosphate wash buffer (80% ethanol, 5 mM KPO4, pH 8.5) and a phosphate elution buffer (4 mM KPO4, pH 8.5), respectively. Purified cDNA was dried in a Speed Vac apparatus, resuspended in 60 μl of 0.1 M sodium bicarbonate (pH 9.0), and labeled by incubation with Cy3 (reference) or Cy5 (experimental) mono-N-hydroxysuccinimide ester (GE Healthcare, United Kingdom) at room temperature for 90 min in the dark. The labeling reaction was quenched by adding 5 μl of 3 M sodium acetate (pH 5.2) and incubating at room temperature for 15 min in the dark. Uncoupled dye was removed using the QIAquick PCR purification kit (Qiagen, Germany) and the buffers provided with the kit, according to the manufacturer's instructions. The efficiency of labeling was determined by reading the absorbance at 260 nm for cDNA concentration, the absorbance at 550 nm for Cy3 incorporation, and the absorbance at 650 nm for Cy5 incorporation.
Microarray hybridization, scanning, and data analysis.
The two differentially labeled cDNA probes were pooled, dried in a Speed Vac apparatus, and resuspended in 120 μl of a preheated salt-based hybridization buffer (Ocimum Biosolutions, India) at 42°C. Microarray slides were prehybridized in a solution containing 5× saline-sodium citrate (SSC; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer, 0.1% sodium dodecyl sulfate (SDS), and 1% bovine serum albumin (BSA) at 42°C for 60 min. After two washes with room temperature double-distilled water for 10 min each, arrays were dried by centrifugation at 1,000 rpm for 5 min. The cDNA probe was denatured by heating at 95°C for 3 min and cooled on ice for 1 min. After addition of 40 μl of the background blocker KREAblock (Kreatech Biotechnology, The Netherlands), the hybridization mixture was heated at 42°C before application to a microarray slide. The probe was kept in place with a coverslip, placed in a metal chamber, and incubated in a water bath at 42°C for 24 h. Following overnight hybridization, slides were washed under gentle agitation for 1 min in preheated 5× SSC, 0.1% SDS buffer at 42°C, followed by washes at room temperature for 2 min in 2× SSC, 2 min in 1× SSC, and 1 min in 0.2× SSC. Slides were quickly rinsed in room temperature double-distilled water and dried in 50-ml conical-bottom tubes by centrifugation at 1,000 rpm for 5 min at room temperature. Microarrays were scanned with an Affymetrix 428 array scanner (Affymetrix) by using laser lights at wavelengths of 532 nm and 635 nm to excite the Cy3 and Cy5 dyes, respectively. Images were captured as 16-bit TIFF files by using an Affymetrix array reader (version 1.1) and analyzed using ImaGene software (version 7.5; BioDiscovery Inc.). This software uses a patented automatic segmentation algorithm to differentiate between signal and background values and a complex tool including seven different criteria for a spot fail/pass test to detect and flag low-quality spots. The presence of additional poor or empty spots not detected automatically was checked manually, and flagged spots were excluded from quantification analysis. Background correction was applied globally by using the median of the signal values of all negative-control spots. A locally weighted regression scatter plot smoothing (LOWESS)-based procedure (67) was applied globally, using all spots, to normalize background-corrected microarray data. The experiments were biologically replicated twice. Considering that each biological replicate also contained two technical duplicates on the array, the two hybridizations generated eight data points from both channels for each gene. The statistical significance of differential gene expression was calculated with the Significance Analysis of Microarray (SAM) software (62) by using a minimal 2-fold change in expression ratio and a maximal false discovery rate (FDR) of 1%.
qRT-PCR.
Enriched mRNA (1 μg) was reverse transcribed using 6 ng of random hexamers (Invitrogen), 5 mM deoxynucleoside triphosphate mix, 1× First Strand buffer (Invitrogen), 5 mM dithiothreitol (Invitrogen), 40 U of SUPERase-In (Ambion), and 400 U of SuperScript III reverse transcriptase at 55°C for 1 h. Primers for the qRT-PCR (see Table S1 in the supplemental material) were designed by using the PrimerSelect program (version 8.0.2) from the Lasergene suite (DNAStar Inc.). The qRT-PCR mixture was set up by using SYBR green I master mix (Roche), 200 nM each forward and reverse primers, and 10 ng of cDNA template. The alaS housekeeping gene was used as an internal control and for normalization of results. The reactions were performed in triplicate by using an ABI Prism 7000 detection system (Applied Biosystems). Cycling conditions for all amplifications were one cycle of 95°C for 10 min and 45 cycles of 95°C for 15 s, 55°C for 5 s, and 72°C for 15 s. From the qRT-PCR data, an average cycle threshold (CT) value was calculated from the triplicate reactions. A mathematical model based on PCR efficiency and crossing-point deviation (49) was used to determine the relative amounts of target and reference gene transcripts and, consequently, the average relative expression ratio between phage-treated and untreated samples.
Computational prediction of protein SCL and function.
Predictions of subcellular localization (SCL) of L. lactis IL1403 proteins were retrieved from the PSORTdb (version 2.0) database. This database includes information from a manually curated data set of ∼2,000 proteins of experimentally verified localization as well as SCL computational predictions of 2,165 proteins from 140 completely sequenced microbial genomes (96 Gram-negative and 44 Gram-positive organisms) generated by using the PSORTb program. This predictive tool uses the data set of proteins of known SCL to train multiple SCL predictors and relies on a series of analytical modules to identify typical sequence features known to correlate with specific localizations. In the case of Gram-positive bacteria, four single (cytoplasm, cytoplasmic membrane, cell wall, extracellular) and two multiple (cytoplasm/cytoplasmic membrane, cytoplasmic membrane/cell wall) localization sites are predicted by PSORTb and included in PSORTdb (53). Prediction of topology in putative membrane proteins, including a specification of the membrane-spanning segments and the in/out orientation relative to the membrane, was obtained by using the servers TMHMM (version 2.0; www.cbs.dtu.dk/services/TMHMM) and TOPCONS (http://topcons.cbr.su.se/index.php). The latter tool uses the prediction from five different topology prediction algorithms (SCAMPI-seq, single-sequence mode; SCAMPI-msa, multiple-sequence mode; PRODIV-TMHMM; PRO-TMHMM; and OCTOPUS) as input to the TOPCONS hidden Markov model (HMM), which gives a consensus prediction for the protein, together with a reliability score based on the agreement of the included methods across the sequence (4).
Prediction of function in proteins annotated as hypothetical in the genome of L. lactis IL1403 was achieved by using the PFAM database (http://pfam.sanger.ac.uk) (17), the Conserved Domain Database (CDD) (40), and the InterProScan service (www.ebi.ac.uk/Tools/InterProScan). The last tool predicts the occurrence of functional domains and motifs/signatures in a protein by combining 12 different databases and their relative protein signature recognition methods (69).
Fluorometric determination of ΔΨ in L. lactis IL1403 following phage adsorption.
Changes in the membrane potential (ΔΨ) of L. lactis IL1403 cells following phage adsorption were estimated by using flow cytometry and the lipophilic cationic dye rhodamine 123 (Sigma, St. Louis, MO). This fluorochrome tends to accumulate on the inner side of energized membranes (i.e., cells maintaining an active ΔΨ), and consequently, its intracellular fluorescence intensity decreases following membrane depolarization. Rhodamine 123 was prepared as a stock solution of 1 mg/ml in methanol. The uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) was used as a positive control for lack of activity of the electron transport system. CCCP is a lipophilic weak acid that binds protons on the acidic side of the microbial membrane, diffuses through, and releases them inside the cell. As a consequence, the formation of a proton gradient is blocked, ΔΨ is reduced to zero, and ATP is not synthesized (46). Cells were grown at 30°C to mid-log phase (OD600 = 0.4), 2 μg/ml rhodamine 123 was added, and the cells were incubated for an additional 15 min to allow cell uptake of the dye. At this time point, a 1-ml aliquot was withdrawn for flow cytometric analyses, whereas the remaining sample was infected with phage c2 (MOI = 800) in the presence of 10 mM CaCl2 and further incubated at 30°C. One-milliliter aliquots were withdrawn after 10, 15, and 25 min and processed for flow cytometric analyses. Aliquots were centrifuged at 20,800 × g for 2 min to remove the growth media. Cell pellets were washed twice in filtered (pore size, 0.22 μm) 50 mM sodium phosphate buffer (pH 7) containing 0.01% Tween 20 and resuspended in the same buffer to an approximate concentration of 5 × 106 to 107 cells per ml. Fluorescence measurements were made with a FACSCanto II flow cytometer (BD Biosciences, San Jose, CA) equipped with two air-cooled lasers, a 20-mW solid-state laser (emission, 488 nm) and a 17-mW HeNe laser (emission, 633 nm), and five sensors for the detection of forward (FSC) and sideward (SSC) light scatter and green (FL1, 525 nm), yellow (FL2, 575 nm), and far red (FL3, 695 nm) fluorescence. Cell samples were delivered at a low flow rate (300 to 500 cells per s) until a total number of 10,000 events was acquired by using the FACSDiva software (version 5.0.2; BD Biosciences, San Jose, CA). Fluorescence signals were recorded by using the following detector settings: FSC, 300; SSC, 300; and FL1, 400 with logarithmic amplifications. A threshold was set at FSC and SSC signals of 200 to reduce background noise deriving from cellular debris.
Microarray data accession number.
The microarray data have been submitted to the Gene Expression Omnibus (GEO) database at the website http://www.ncbi.nlm.nih.gov/geo/ under accession number GSE26042.
RESULTS
Determination of conditions for synchronous c2 infection.
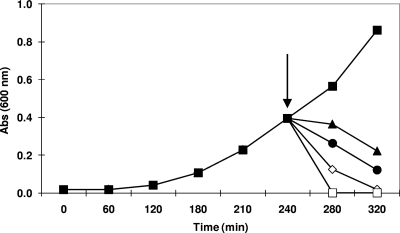
The biology of phage c2 infection has been studied so far using L. lactis MG1363, a sensitive host that lyses in 30 min (38, 39). Less well characterized is phage c2 infection of L. lactis IL1403, but our data show that the lytic cycle is extended to an average of 40 min, probably as a result of the activity of a host-encoded restriction/modification (R/M) system (54). To obtain a homogeneous gene expression profile of the L. lactis IL1403 response to phage c2 attack, we deemed it necessary to use infection conditions ensuring synchronous infection and, consequently, complete lysis of the cell population within this time frame. A number of phage-host challenge tests were performed, and an MOI of 800 was determined to be required to achieve complete lysis within 40 min (Fig. 1). Production of phage progeny was verified by the titer of infection.
Fig. 1.
Phage-host challenges to determine the MOI necessary to lyse completely a bacterial population within 40 min. Effects of the addition of phage c2 (indicated by an arrow) at MOIs of 0 (▪), 200 (▴), 400 (•), 600 (⋄), and 800 (□) on the viability of L. lactis IL1403 cells growing in log phase.
Transcriptional profiling of L. lactis IL1403 response to phage c2 infection.
The early step of the infection process, involving phage adsorption and DNA injection, is crucial to understand the host response to phage interaction at the molecular level. As regards phage c2, viral DNA appears in L. lactis MG1363 5 min after phage addition and replication starts at between 15 and 20 min (18, 19, 47). Consequently, 10 min after phage addition was chosen as a representative time point of an early stage of infection, prior to DNA replication, to determine the response of L. lactis IL1403 to phage c2 attack. Analysis of the microarray data of two independent biological replicates revealed that L. lactis IL1403 differentially regulated the expression of 61 genes (Table 1 ), with an even distribution between up- and downregulated genes. This represents ca. 3% of the overall host genetic makeup and suggests that no significant changes in host metabolism are caused by phage infection. Similarly, low levels (4 to 9%) of Escherichia coli genes were found to respond to infection by lytic phages (50, 52). A third of the regulated genes (19 out of 61) and most of the positively induced genes (17 out of 32) encoded products annotated as hypothetical proteins. The 61 regulated genes belonged to 14 functional categories, with components of the cell envelope (12 genes), regulatory functions (11 genes), and carbohydrate metabolism (7 genes) accounting for half of the regulated genes.
Table 1.
Transcriptional response of L. lactis IL1403 following infection by phage c2
| Functional category/pathwaya | Gene | Expression ratio (fold) | Gene product/putative functionb | Subcellular localizationc |
|---|---|---|---|---|
| Carbohydrate metabolism | ||||
| Citrate (TCAd) cycle | citE | −2.25 | Citrate lyase beta chain (EC 4.1.3.6) | Cytoplasmic |
| Citrate (TCA) cycle | citF | −2.28 | Citrate lyase alpha chain (EC 4.1.3.6) | Cytoplasmic |
| Glycolysis/gluconeogenesis | enoB | −2.26 | Phosphopyruvate hydratase (EC 4.2.1.11) | Cytoplasmic |
| Glycolysis/gluconeogenesis | adhE | −2.35 | Acetaldehyde-CoA-/alcohol dehydrogenase (EC 1.2.1.10) | Cytoplasmic |
| Starch and sucrose metabolism | malQ | −2.18 | 4-Alpha-glucanotransferase (EC 2.4.1.25) | Cytoplasmic |
| Amino sugar and nucleotide sugar metabolism | pmi | −2.05 | Mannose-6-phosphate isomerase (EC 5.3.1.8) | Cytoplasmic |
| Starch and sucrose metabolism | pgmB | 2.08 | Beta-phosphoglucomutase (EC 5.4.2.6) | Cytoplasmic |
| Cell envelope | ||||
| Surface (lipo-)polysaccharides and antigens | yndF | 3.16 | Hypothetical protein | Cell wall |
| Surface (lipo-)polysaccharides and antigens | floL | −2.72 | Flotillin-like protein | Cytoplasmic membrane |
| Surface (lipo-)polysaccharides and antigens | ywaF | −2.34 | Glycosyltransferase | Cytoplasmic |
| Peptidoglycan biosynthesis | dltA | 2.11 | d-Alanine–poly(phosphoribitol) ligase (EC 6.1.1.13) | Cytoplasmic |
| Peptidoglycan biosynthesis | dltD | 2.11 | d-Alanine transfer protein | Unknown |
| Peptidoglycan biosynthesis | ddl | 2.18 | d-Alanine-d-alanine ligase (EC 6.3.2.4) | Cytoplasmic |
| ABC transporters | optS | −2.13 | Oligopeptide ABC transporter substrate binding protein | Cell wall |
| Unknown | yfbG | 2.09 | Hypothetical protein | Cytoplasmic membrane |
| Unknown | ybfC | 2.08 | Hypothetical protein | Cytoplasmic membrane |
| Unknown | yghD | 2.54 | Hypothetical protein | Cytoplasmic membrane |
| Unknown | ythB | 2.46 | Putative stress response protein, PspC-like | Cytoplasmic membrane |
| Unknown | ythC | 2.83 | Hypothetical protein | Cytoplasmic |
| Energy metabolism | ||||
| Oxidative phosphorylation | yshC | −2.51 | Putative Mn-dependent inorganic pyrophosphatase (EC 3.6.1.1) | Cytoplasmic |
| Electron transport | ndrH | 2.18 | Glutaredoxin-like protein NrdH | Cytoplasmic |
| Lipid metabolism | ||||
| Fatty acid biosynthesis | fabG1 | −2.23 | 3-Oxoacyl-acyl carrier protein reductase (EC 1.1.1.100) | Cytoplasmic |
| Fatty acid biosynthesis | acpD | 2.04 | Acyl carrier protein phosphodiesterase (EC 3.1.4.14) | Unknown |
| Pyruvate metabolism | pflA | −2.29 | Pyruvate-formate lyase activating enzyme (EC 1.97.1.4) | Cytoplasmic |
| Terpenoid backbone biosynthesis | yebB | −2.00 | Isopentenyl-diphosphate delta-isomerase (EC 5.3.3.2) | Cytoplasmic |
| Metabolism of cofactors and vitamins | ||||
| Pantothenate and CoA biosynthesis | coaA | −2.55 | Pantothenate kinase (EC 2.7.1.33) | Cytoplasmic |
| Riboflavin metabolism | ribC | 2.29 | Riboflavin kinase (EC 2.7.1.26) | Cytoplasmic |
| Protein and amino acid metabolism | ||||
| Amino acid transport and metabolism | yteB | −2.54 | Putative d-amino acid oxidase protein | Cytoplasmic |
| Amino acid transport and metabolism | yeiG | −2.67 | Aminotransferase (EC 2.6.1.–) | Cytoplasmic |
| Arginine and proline metabolism | argC | 2.27 | N-Acetyl-γ-glutamyl-phosphate reductase (EC 1.2.1.38) | Unknown |
| Arginine and proline metabolism | argJ | 2.06 | Ornithine acetyltransferase (EC 2.3.1.35) | Cytoplasmic |
| Arginine and proline metabolism | argD | 2.22 | Acetylornithine aminotransferase (EC 2.6.1.11) | Cytoplasmic |
| Degradation of proteins and (glyco-)peptides | pepXP | −2.10 | X-prolyl dipeptidyl aminopeptidase (EC 3.4.14.11) | Cytoplasmic |
| Purines, pyrimidines, nucleosides and nucleotides | ||||
| Purine metabolism, pyrimidine metabolism | nrdE | −2.29 | Ribonucleoside diphosphate reductase alpha chain (EC 1.17.4.1) | Unknown |
| Purine metabolism, pyrimidine metabolism | nrdD | 2.65 | Anaerobic ribonucleoside triphosphate reductase (EC 1.17.4.2) | Cytoplasmic |
| Purine metabolism, pyrimidine metabolism | nrdG | 2.87 | Anaerobic ribonucleoside triphosphate reductase activating protein (EC 1.97.1.4) | Cytoplasmic |
| Regulatory functions | ||||
| Transcription factors | rliA | −2.85 | Transcription regulator, LacI family | Cytoplasmic |
| Transcription factors | yddC | 2.23 | Putative transcription regulator | Cytoplasmic |
| Transcription factors | yveF | 2.24 | Putative PadR family transcriptional regulator | Cytoplasmic |
| GTP-binding proteins | ysxL | −2.17 | GTPase EngB | Cytoplasmic |
| GTP-binding proteins | yphL | −2.04 | GTP-binding protein EngA | Cytoplasmic membrane |
| GTP-binding proteins | yyaL | 2.10 | GTP-dependent nucleic acid-binding protein | Cytoplasmic |
| General | yeeG | −2.28 | Transcription regulator | Cytoplasmic membrane |
| General | ahrC | −2.04 | Transcription regulator of arginine metabolism | Cytoplasmic |
| General | yjaI | 2.27 | Hypothetical protein | Unknown |
| Unknown | yneH | 2.72 | Putative transcription regulator | Unknown |
| Two-component systems | kinD | 2.11 | Sensor protein kinase KinD | Cytoplasmic membrane |
| Replication and repair | ||||
| DNA replication, nucleotide excision repair | ligA | −2.45 | NAD-dependent DNA ligase (EC 6.5.1.2) | Cytoplasmic |
| DNA replication | dnaC | −2.05 | Replicative DNA helicase (EC 3.6.1.–) | Cytoplasmic |
| DNA repair | ywaC | 2.81 | Hypothetical protein | Cytoplasmic |
| Homologous recombination | ruvB | −2.26 | DNA helicase RuvB (EC 3.1.22.4) | Cytoplasmic |
| Transcription | ||||
| RNA processing | rheB | −2.62 | ATP-dependent RNA helicase | Cytoplasmic |
| RNA synthesis/modification, DNA transcription | nusG | −2.32 | Transcription antitermination protein | Cytoplasmic |
| Aminoacyl-tRNA biosynthesis | gatA | −2.11 | Aspartyl/glutamyl-tRNA amidotransferase subunit A (EC 6.3.5.6, EC 6.3.5.7) | Cytoplasmic |
| Aminoacyl-tRNA biosynthesis | argS | 2.04 | Arginyl-tRNA synthetase (EC 6.1.1.19) | Cytoplasmic |
| Unknown | ||||
| Unknown | yhfA | 2.02 | Hypothetical protein | Cytoplasmic |
| Unknown | yveE | 2.10 | Hypothetical protein | Cytoplasmic |
| Unknown | ywaH | 2.11 | Hypothetical protein | Cytoplasmic |
| Unknown | yccL | 2.34 | Hypothetical protein | Cytoplasmic |
| Unknown | yhbF | 2.34 | Hypothetical protein | Cytoplasmic |
| Unknown | yhbE | 2.46 | Hypothetical protein | Cytoplasmic |
Pathway assignment is based on the KEGG GENES database (www.genome.jp/kegg/genes.html).
Putative function is based on homology to conserved domains and motifs/signatures by using the InterProScan tool (69).
Subcellular localization is based on predictions retrieved from the PSORTdb (version 2.0) database (53).
TCA, tricarboxylic acid.
L. lactis IL1403 increases expression of genes involved in d-alanylation of cell wall LTAs or TAs in response to phage c2 infection.
Half of the genes regulated by L. lactis IL1403 encode proteins whose putative functions are involved to different extents in biogenesis, modification, and regulatory processes of the cell envelope. Among these, the bacterium upregulated genes (ddl, dltA, and dltD) playing a role in the peptidoglycan biosynthesis pathway. ddl encodes a d-alanine–d-alanine ligase that converts d-alanine to its dimer in a reversible reaction important to both d-alanine metabolism and cell wall biosynthesis. The d-alanine monomers are then activated and incorporated as d-alanine esters (d-alanylation) into teichoic acids (TAs), either cell wall teichoic acids (WTAs) or LTAs, by the Dlt proteins. dltA and dltD form part of the four-gene operon dltABCD, where they overlap with the dltB and dltC genes, respectively. This is indicative of transcriptional coupling, and, indeed, RT-PCR confirmed that dltB and dltC were also upregulated at levels similar to those for dltA and dltD (Table 2). Induction of the dlt operon should result in increased d-alanylation of TAs, an event known to exert pleiotropic effects on the cell physiology of Gram-positive bacteria (28, 45).
Table 2.
qRT-PCR validation of microarray data for selected genes differentially expressed in L. lactis IL1403 following infection by phage c2
| Gene | Expression ratio (fold) |
|
|---|---|---|
| qRT-PCR | Microarrays | |
| ddl | 2.42 | 2.18 |
| dltA | 2.80 | 2.11 |
| dltB | 2.50 | NSDa |
| dltC | 3.78 | NSD |
| dltD | 2.94 | 2.11 |
| nrdG | 15.88 | 2.87 |
| yhbF | 5.18 | 2.34 |
| yndF | 3.26 | 3.16 |
| yneH | 7.50 | 2.72 |
| ythB | 7.12 | 2.46 |
| ythC | 4.10 | 2.83 |
| kinD | 4.82 | 2.11 |
| llrD | 5.96 | NSD |
| yhbE | 11.02 | 2.46 |
| ywaF | −2.90 | −2.34 |
| yveF | 2.40 | 2.24 |
NSD, not significantly different.
A bacterial activity directed toward strengthening and modifying membrane TAs is also suggested by the downregulation of the amylomaltase gene malQ and upregulation of the β-phosphoglucomutase gene pgmB. This increases the intracellular pool of d-glucose-1-phosphate required for the synthesis of UDP-glucose, which is a precursor of TAs (66) (36). In addition, the strongest upregulated gene (yndF) in L. lactis IL1403 encodes a protein (653 amino acids [aa]) likely to interact with or to be regulated by the dlt genes. Indeed, YndF contains the highly conserved LPXTGE motif (Protein Family PF00746), common to many Gram-positive surface proteins. These include the M protein of Streptococcus pyogenes, whose expression has recently been shown to be modulated by the d-alanylation pathway (13). L. lactis IL1403 also highly upregulated a number of genes encoding hypothetical proteins with predicted localization on the cytoplasmic membrane. These include genes (yeeG and yjaI) encoding LytR-like transcriptional attenuators (Protein Family PF03816) that, in Staphylococcus aureus, control peptidoglycan hydrolase activity (9) and are part of the cell wall stress stimulon induced by antibiotics (63).
L. lactis IL1403 restores the physiological PMF altered by phage infection.
Phage c2 infection triggered in L. lactis IL1403 the downregulation of genes actively involved in the primary (yshC) and secondary (citEF) mechanisms of generation of the PMF, which is a major store of free energy in any bacterial cell (37). yshC encodes a manganese-dependent inorganic pyrophosphatase that plays a key role in the oxidative phosphorylation pathway and has been shown to be essential to E. coli growth (10). YshC activity provides the inorganic phosphate necessary for the F0F1-ATPase to generate ATP while driving an influx of protons through the cell membrane. Downregulation of YshC activity would move the equilibrium of this reaction toward ATP hydrolysis. Consequently, the energy released would be used to pump protons out of the cell against their thermodynamic gradient. The citEF genes encode the beta and alpha chains, respectively, of the three-subunit enzyme citrate lyase, which catalyzes the first step of the citrolactic fermentation pathway leading to the intracellular conversion of citrate to lactate. In L. lactis, this pathway generates metabolic energy (ΔΨ and ΔpH) by a secondary mechanism, which results in a PMF sufficiently high to drive ATP synthesis via F0F1-ATPase (22). Consequently, the downregulation of yshC and citEF genes by L. lactis IL1403 is suggestive of a PMF collapse and, in particular, of a hyperpolarization of the membrane. This suggestion finds support in the downregulation of ahrC and upregulation of the biosynthetic genes argCJD, which would switch arginine catabolism off to favor accumulation of the amino acid. The former is usually the active pathway in the cell, which leads to ammonia production and is proposed to maintain pH homeostasis in L. lactis (32, 33). However, the F0F1-ATPase-mediated export of protons would leave a more alkaline intracellular pH, thus generating no need for ammonia neutralization.
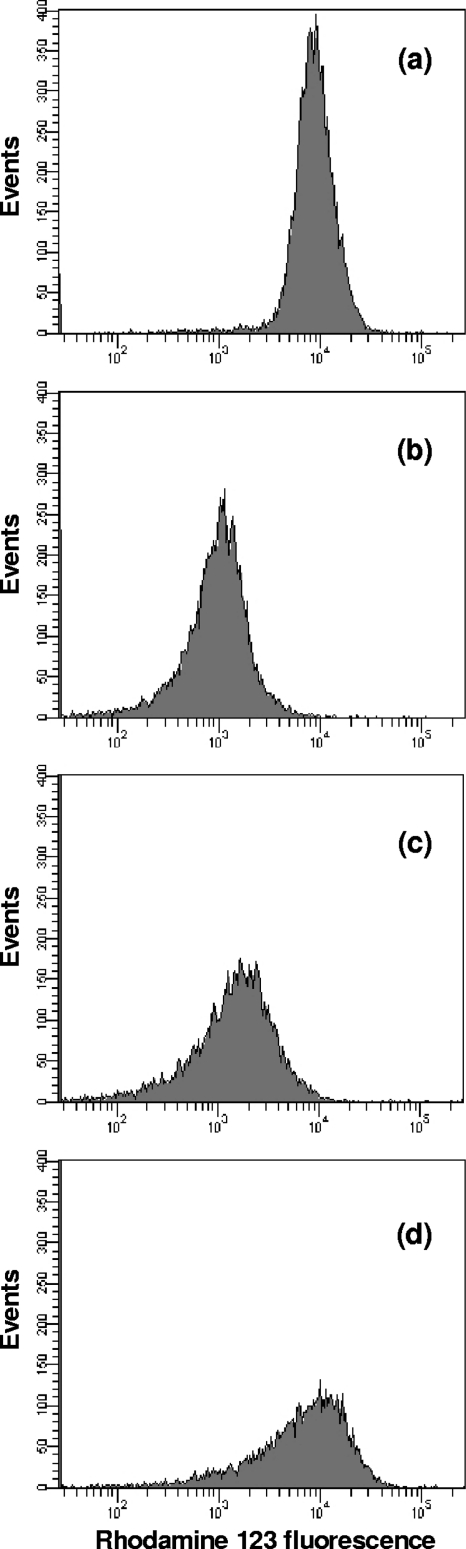
In E. coli, adsorption of phages T4 and T5 induces a partial and transient depolarization of the host membrane, to which the infected bacteria respond by a repolarization that leads to a new steady state of reduced ΔΨ (27, 31). The membrane hyperpolarization predicted in L. lactis IL1403 would be consistent with the repolarization phenomenon observed in E. coli. Consequently, putative changes in ΔΨ of L. lactis IL1403 during phage c2 infection were investigated by using flow cytometry and the ΔΨ-sensitive dye rhodamine 123. Exponentially growing cells actively accumulated rhodamine 123 at fluorescence levels averaging around 104 arbitrary units (Fig. 2 a). Ten minutes after addition of phage c2, the average intracellular fluorescence decreased by 1 log unit, which was indicative of a membrane depolarization and consequent efflux of the dye out of the cells (Fig. 2b). A similar ΔΨ collapse was observed when rhodamine 123-stained cells were incubated with the uncoupler CCCP for 10 min (data not shown). A gradual reuptake of fluorochrome up to preinfection levels was observed at between 15 and 25 min within the infection process (Fig. 2c and d), suggesting that bacteria had regained an energized membrane. These data indicate that phage c2 infection causes an initial dissipation of the ΔΨ in L. lactis IL1403, followed by a gradual restoration of its physiological value, as previously reported in E. coli (27, 31).
Fig. 2.
Flow cytometric analysis of membrane potential in rhodamine 123-stained cells of L. lactis IL1403 before (a) and 10 min (b), 15 min (c), and 25 min (d) after addition of phage c2.
L. lactis IL1403 represses energy-consuming functions and switches to anaerobiosis in response to phage c2 infection.
L. lactis IL1403 also downregulated genes involved in metabolism of lipids (pflA, fabG, yebB), carbohydrates (enoB and adhE), cofactors (coaA), and proteins and amino acids (pepXP, yeiG, yteB) and in DNA replication (ruvB, dnaC, ligA), transcription (rheB, nusG), and translation (gatA) (Table 1). This suggests a global reduction of growth- and development-promoting activities to which the bacterium generally commits considerable amounts of energy, such as the biogenesis of the lipid membrane (pflA, fabG, yebB) and of the cell wall (ywaF, pmi).
Repression of pflA exerts a rate-limiting action on lipid biogenesis as it prevents the formation of acetyl coenzyme A (acetyl-CoA), which is a central intermediate of the fatty acid biosynthesis pathway (11). fabG encodes a reductase enzyme participating in the first step of each cycle of chain elongation, and its repression in E. coli blocks both fatty acid biosynthesis and cell growth (70). YebB is a key enzyme in the biosynthesis of isoprenoids, which serve numerous essential functions in prokaryotic bacteria, including structural components of membranes (21). ywaF encodes a glycosyltransferase (Protein Family PF00534) transferring activated sugars to an acceptor molecule localized at the outer surface of the cell membrane and was found to be regulated in L. lactis following treatment with lactococcin 972 (Lcn972) (41). Finally, the mannose-6-phosphate isomerase Pmi catalyzes the first step in the synthesis of mannose-containing sugar chains and seems to play a central regulatory role in both cell wall synthesis and energy production (34). Downregulation of genes (ysxL and yphL) encoding GTPases is also in tune with the overall repression of growth-promoting functions, as the activity of these DNA-binding enzymes has been demonstrated to link various aspects of the cell cycle and metabolism with translation by playing key roles in the assembly of bacterial ribosomes (8).
While repressing energy-consuming functions, L. lactis IL1403 also upregulated the nrdDG genes and concomitantly downregulated nrdE. The operons nrdEF and nrdDG encode ribonucleotide reductases that are activated during conditions of aerobiosis and anaerobiosis, respectively. These enzymes catalyze the reduction of ribonucleoside di- or triphosphates, thereby providing the building blocks required for DNA replication and repair. During anaerobic growth, L. lactis switches on the nrdDG operon and relies on the ability of NrdG to generate the glycyl radical crucial to the reducing activity of NrdD (25, 60). Repression of the aerobic ribonucleotide reductase (nrdEF) and induction of the anaerobic enzyme (nrdDG) would therefore suggest a switch to growth conditions resembling anaerobic fermentation in L. lactis IL1403.
c2 phage infection induces stress response genes in L. lactis IL1403.
The L. lactis IL1403 response to phage infection also included a strong upregulation of genes (ythB, ythC, yneH, kinD, and llrD; Tables 1 and 2) that have been associated with the cell wall stress response in various organisms. YthB (67 aa) is identical to the membrane-anchored phage shock protein C (PspC; Protein Family PF04024) acting as a stress-responsive transcriptional regulator in E. coli and Yersinia enterocolitica. The Psp system is thought to protect the cell from a variety of extracytoplasmic stressors affecting membrane integrity, causing dissipation of the PMF and a likely change in the cell's redox state (14, 24). Topology analysis predicts YthB to be an integral membrane protein with the N terminus inserted into the lipid layer, the C terminus forming a transmembrane helix, and the last 7 aa residues projecting out of the membrane, which suggest that it could be involved in stress signal transduction (see Fig. S1c in the supplemental material). Conversely, YthC (371 aa) is probably a cytoplasmic protein and shares homology to a family of conserved proteins of unknown function (Conserve Domain COG3595). Sequence analysis of the regions surrounding ythB and ythC revealed a three-gene operon-like structure where ythC precedes the genes ythB and ythA. Similarly to YthB, YthA (154 aa) contains a PspC-like domain and is predicted to be located inside the membrane (see Fig. S1b in the supplemental material). The ythCBA genes are probably transcribed as a single polycistronic unit, as promoter sequences for efficient gene expression were retrieved upstream of only ythC, whereas a strong rho-independent terminator signal (ΔG = −7.1) is located downstream of ythA (see Fig. S1a in the supplemental material). Also induced at high levels was the gene spxB (formerly yneH), encoding one of the seven lactococcal paralogs of the conserved Spx proteins acting as global transcriptional regulators in many Gram-positive species (44). Spx homologues play an important role in growth, general stress protection, and biofilm formation in S. aureus (48), regulate the global response to oxidative stress in Bacillus subtilis (43), and mediate the cell wall stress response in L. lactis (29, 41, 65). Moreover, L. lactis IL1403 highly induced the cesSR genes (Tables 1 and 2), encoding a two-component system thought to coordinate the cell wall stress response and to control the expression of both spxB and psp genes in L. lactis (41, 65).
DISCUSSION
In this study, a transcriptomic approach was used to analyze the holistic response of L. lactis IL1403 at the onset of a phage challenge in order to identify any putative strategy adopted by the host to avoid phage infection. We observed a host response strongly targeted to the cell wall, which indicates that phage infection is sensed as an assault on the cell membrane affecting its integrity. To our knowledge, our results describe for the first time the molecular response of L. lactis counteracting the phage-mediated perturbations to its physiology.
An MOI of 800 was experimentally determined to be required to synchronize cell infection, thus obtaining a homogeneous gene expression profile of the L. lactis IL1403 response to phage c2 attack. The requirement of such a high MOI is probably associated with the L. lactis IL1403 R/M system (54), which seems to delay and alter the physiology of phage c2 infection compared to that on the more sensitive host L. lactis MG1363. In L. lactis IL1403, phage c2 propagates with 3-log-unit less efficiency, plaque size is reduced by 2- to 3-fold, and the lytic cycle is extended to 40 min (unpublished observations). Although L. lactis MG1363 encodes an R/M system too, the two systems are likely to have different restriction efficiencies, as low similarities exist at the amino acid level between the respective subunits (HsdR, 34% identity; HsdM, 41% identity; HsdS, 37% identity). Consequently, the L. lactis IL1403 R/M system is most likely to account for the high MOI required in our experiments, but also, a possible interference from other unknown resistance mechanisms cannot be excluded. A potentially high number of abortive adsorptions or DNA injections by phage c2 could also contribute to the high MOI. These events occur at high rates (50 to 75%) even in highly efficient systems such as the coliphages T4 and T5 (1, 20) and could therefore affect the infection of phage c2 (42, 64). High MOIs (>100) can be associated with lysis of the cell population (68), but in both E. coli and Lactobacillus lactis, the most common effect appears to be inhibition of host lysis and extension of the latent period (2, 68). In addition, T4- and T2-infected E. coli cells are known to be resistant to passive lysis due to high MOIs and are also able to exclude superinfecting phages (3, 12).
In our study, a number of observations support the idea that the MOI of 800 is not altering cell viability and, consequently, not introducing any artifact biasing the final results. First, flow cytometric analysis of membrane potential changes in L. lactis IL1403 25 min after phage c2 infection showed a metabolically active bacterium capable of restoring an energized membrane and gradually reuptaking the rhodamine 123 lost from the cell at 10 min postinfection (Fig. 2b). Additionally, production of phage progeny after 40 min implies that the infection process proceeds regularly as a result of a metabolically functional host machinery. Finally, a number of responses observed in L. lactis IL1403, such as the induction of regulatory elements associated with the cell wall stress response, the downregulation of energy-consuming functions, and the putative switch to anaerobic respiration, have also been reported in systems where much lower MOIs have been used (26, 50).
The identity of the regulated genes suggests that L. lactis IL1403 activates a four-prong response to phage infection involving induction of membrane stress proteins, d-alanylation of the cell wall, maintenance of the PMF, and energy conservation. Upregulation of the dltABCD operon indicates that L. lactis IL1403 is increasing the incorporation of d-alanine esters into WTAs and LTAs, and this could be associated with an attempt to resist phage infection. WTAs and LTAs are phosphate-rich glycopolymers containing d-alanine esters as main backbone substitutions (66). The extent of this modification exerts profound modulatory effects on cell physiology and host-mediated responses, including a protective function against UV and acid stress (7, 15), autolysins (58), cationic antimicrobials (30), and, interestingly, phage attack (51). Indeed, the degree of LTA substitutions appears to affect phage binding to bacterial cells when the receptor is either LTA or a different molecule. Phage-resistant strains of Lactobacillus delbrueckii subsp. lactis were found to contain higher levels of d-alanine in their LTAs than phage-sensitive ones (51), whereas galactosyl-modified LTAs were suggested to prevent phage adsorption to a L. lactis subsp. cremoris strain by steric shielding of the actual receptor site (56).
As regards phage c2, the nature of the primary receptor used to infect lactococcal cells is unclear. It was reported to adsorb to rhamnose moieties on the cell wall of L. lactis C2 (42), and a similar receptor may be used to infect related strains such as L. lactis IL1403. Rhamnose is also a component of LTAs that have recently been proposed to be receptor candidates for 936-type lactococcal phages (57, 61). On the basis of these observations, the upregulation of the dlt operon observed in L. lactis IL1403 adds more weight to the suggestion that phage c2 could use LTAs as a primary receptor. Also, it can be speculated that an increased d-alanylation of LTAs could correspond to the host attempt to evade phage infection by sterically impeding the already adsorbed phage from accessing and irreversibly binding the secondary receptor Pip protein. This strategy would resemble an adsorption-blocking mechanism and could be very effective at controlling phage infection by rendering the cell impervious to the phage DNA and leaving the cell and its biochemical activities unaltered (16). Alternatively, d-alanylation of LTAs could act as a superinfection exclusion mechanism affecting the initial (and reversible) adsorption of secondary phages via modification of the primary receptor. Although the outcome is still a successful phage infection, such a mechanism could allow the survival of a small percentage of the cell population when facing in vivo low-titer phage attacks.
Flow cytometry analysis of ΔΨ revealed that L. lactis IL1403 was able to gradually restore the ΔΨ physiological value that had been altered during the early stage of phage c2 infection. These results are consistent with the depolarization-repolarization phenomenon observed in E. coli (27, 31) and, to the best of our knowledge, have never been described so far in L. lactis. The partial and transient depolarization of the E. coli membrane following adsorption of phages T4 and T5 results from the opening of a membrane channel, mediating the transfer of phage DNA into the host and consequent efflux of potassium and calcium ions. Infected bacteria respond by a repolarization that leads to a new steady state of reduced ΔΨ (6, 27, 31). Our results suggest that, regardless of the phage-host system, a strong depolarization of the host membrane is a common effect induced by phage infection. Moreover, our transcriptional data provide insights into the strategy adopted by the cell to obtain a rapid repolarization of the membrane. By downregulating the expression of genes (yshC and citEF) central to the F0F1-ATPase activity and to the citrolactic fermentation pathway, L. lactis IL1403 can simultaneously modulate both ΔΨ and ΔpH components of the PMF and thus restore the physiological preinfectious condition in the shortest time frame possible. Observations that increased respiratory rate and proton influx are also associated with membrane depolarization in E. coli cells (35) suggest that this strategy is commonly used by cells to reverse the phage-induced effect on ΔΨ.
Concomitantly, L. lactis IL1403 was found to engage in the global repression of growth- and development-promoting functions as well as to switch metabolism to anaerobic growth. Energy-consuming activities such as biogenesis of lipid membrane or DNA replication are postponed by the bacterium probably as a means of better utilization of energy resources, which are redirected toward the activities necessary to cope with the phage-induced stress. In tune with this, the switch to anaerobic growth would be part of a genetic reprogramming of the host cell aiming at saving energy and achieving an efficient reallocation of cellular resources in order to sustain the PMF and the overall response to phage.
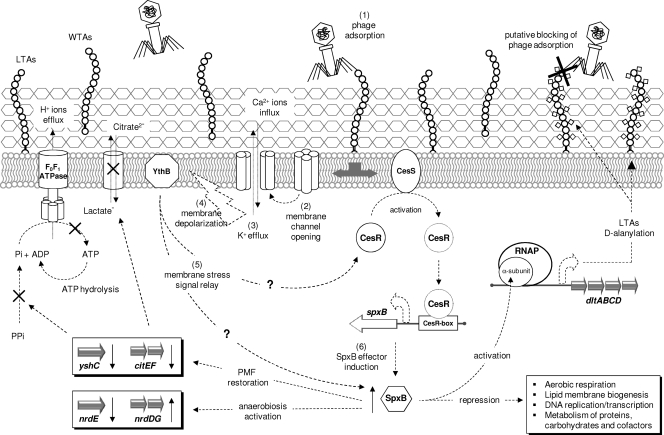
The overall response of L. lactis IL1403 to phage infection is possibly orchestrated by a complex regulatory network, including Psp homologues, SpxB, and the two-component system CesSR. In E. coli, phage-induced stress induces a response similar to that obtained in our study and suggested to be regulated by the psp system (26, 50). Attack by the lytic phage PRD1 triggers upregulation of genes involved in anaerobic respiration early (i.e., 5 to 15 min) in the infection, whereas Psp proteins are induced throughout the infection process (50). A homologue of protein IV from filamentous phage also induces a psp system response in E. coli that switches cell metabolism to anaerobic fermentation and maintains the PMF by sustaining ΔΨ and ΔpH (26). In both Gram-negative and Gram-positive bacteria, the psp system appears to be devoted to counteracting any stress affecting cell envelope integrity and the associated PMF. During infection by the coliphage PRD1, a peak of expression of the psp system is observed in coincidence with the production of PRD1 holin protein (50), whereas in L. lactis, psp homologues are strongly induced in a nisin-resistant strain (29) and following exposure to Lcn972, a bacteriocin that inhibits cell wall synthesis (41). Thus, the psp system is likely to play a key role in L. lactis IL1403 genetic reprogramming directed to restore a physiological PMF and to adjust energy use to favor maintenance of the new metabolic steady state. Four genes (pspFABC) are suggested to constitute the minimal functional psp system in Gram-negative bacteria, and a basic working model implies that the stress signal is sensed by the membrane proteins PspB and/or PspC and then relayed to the effector complex PspAF (14). In L. lactis IL1403, the operon ythCBA encodes the membrane-anchored PspC-like proteins YthB and YthA, but no homologues of PspAF can be found in the surrounding regions. This suggests that L. lactis could have evolved a novel organization of this responsive system, where cell envelope stress is still sensed by a PspC-like protein (YthB) but then relayed to an effector system different from PspAF. This effector could correspond to the highly induced regulator SpxB that, in L. lactis, has been reported to mediate a multistep response increasing peptidoglycan resistance to lysozyme (65) and to be strongly upregulated in response to the cell wall-targeting antimicrobials nisin (29) and Lcn972 (41). Spx proteins play pleiotropic functions in various organisms (41, 43, 44, 48, 65), and their bivalent regulatory activity appears to be essential for protecting and maintaining the general well-being of bacterial cells under stressful conditions. They would act as negative regulators to postpone energy-consuming functions related to growth while using their activator capacities to mobilize the operations necessary to reverse the stress-induced effects (43, 44). Therefore, SpxB could play a leading effector role in the overall response of L. lactis IL1403 to the phage stimulus, leading to cell wall d-alanylation, PMF restoration, and energy conservation. The stress signal could be relayed to SpxB by YthB via direct interaction or, alternatively, via the two-component system CesSR that was also highly induced in L. lactis IL1403. This system has been associated with the cell envelope stress response in L. lactis and seems to control the expression of both spxB and psp genes via the regulator CesR (41, 65). On the basis of these findings, we suggest that YthB, SpxB, and CesSR could all be part of the same signal transduction pathway or cell wall stressosome that in L. lactis IL1403 would react to phage infection, according to the putative working model proposed in Fig. 3. The phage-mediated stress would be sensed by the membrane-anchored sensor YthB, which would then relay the stress signal to the effector SpxB presumably via the two-component system CesSR or via direct interaction. Upon induction, SpxB would use its global regulator capacities to increase the d-alanine content of cell wall TAs, to restore a physiological PMF, and to activate an energy-saving program by postponing growth-related functions and switching cell metabolism to anaerobiosis. Another option is that the Psp and SpxB proteins could exert a regulatory control over specific responses, such as PMF restoration by Psp proteins, d-alanylation of LTAs, and energy conservation by SpxB. Determining the putative crossover or separation of the regulatory activities of these proteins is of great interest and will be the subject of future investigations. Results of this study and our knowledge to date (41, 65) suggest that SpxB, Psp proteins, and the CesSR system most probably interact and are part of a common strategy (or stressosome) aiming at protecting L. lactis from any cell envelope stress. Considering the nature of the different stressors (lysozyme, antimicrobial peptides, phages) stimulating its activation in L. lactis, this stressosome probably represents a general defense mechanism committed to preserve not only cell wall integrity but also, ultimately, cell viability. While much is known about this system in Gram-negative bacteria, very few data are available for Gram-positive species. As such, we anticipate further investigations to improve our understanding of this vital defense system in L. lactis and in Gram-positive bacteria in general.
Fig. 3.
Putative working model of the cell wall stressosome of L. lactis IL1403 in response to phage infection.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the Irish Dairy Levy. Vincenzo Fallico received a grant from the Irish Research Council for Science, Engineering and Technology (IRCSET) under the Embark initiative.
We thank Aldert Zomer for helpful discussions on the optimization of the microarray protocol.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Abedon S. T. 1994. Lysis and the interaction between free phages and infected cells, p. 398 In Karam J. D. (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, DC [Google Scholar]
- 2. Alatossava T., Jutte H., Seiler H. 1987. Transmembrane cation movements during infection of Lactobacillus lactis by bacteriophage Ll-H. J. Gen. Virol. 68: 1525–1532 [DOI] [PubMed] [Google Scholar]
- 3. Anderson C. W., Eigner J. 1971. Breakdown and exclusion of superinfecting T-even bacteriophage in Escherichia coli. J. Virol. 8: 869–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernsel A., Viklund H., Hennerdal A., Elofsson A. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37: W465–W468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bolotin A., et al. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp lactis IL1403. Genome Res. 11: 731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boulanger P., Letellier L. 1988. Characterization of ion channels involved in the penetration of phage T4 DNA into Escherichia coli cells. J. Biol. Chem. 263: 9767–9775 [PubMed] [Google Scholar]
- 7. Boyd D. A., et al. 2000. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 182: 6055–6065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Britton R. A. 2009. Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 63: 155–176 [DOI] [PubMed] [Google Scholar]
- 9. Brunskill E. W., Bayles K. W. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J., et al. 1990. Pyrophosphatase is essential for growth of Escherichia coli. J. Bacteriol. 172: 5686–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conradt H., Hohmannberger M., Hohmann H. P., Blaschkowski H. P., Knappe J. 1984. Pyruvate formate-lyase (inactive form) and pyruvate formate-lyase activating enzyme of Escherichia coli: isolation and structural properties. Arch. Biochem. Biophys. 228: 133–142 [DOI] [PubMed] [Google Scholar]
- 12. Cornett J. B. 1974. Spackle and immunity functions of bacteriophage T4. J. Virol. 13: 312–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cox K. H., et al. 2009. Inactivation of DltA modulates virulence factor expression in Streptococcus pyogenes. PLoS One 4: e5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Darwin A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57: 621–628 [DOI] [PubMed] [Google Scholar]
- 15. Duwat P., Cochu A., Ehrlich S. D., Gruss A. 1997. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 179: 4473–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emond E., Moineau S. 2007. Bacteriophages and food fermentations, p. 93–123 In McGrath S., van Sinderen D. (ed.), Bacteriophage: genetics and molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 17. Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garvey P., Fitzgerald G. F., Hill C. 1995. Cloning and DNA-sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl. Environ. Microbiol. 61: 4321–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garvey P., Hill C., Fitzgerald G. F. 1996. The lactococcal plasmid pNP40 encodes a third bacteriophage resistance mechanism, one which affects phage DNA penetration. Appl. Environ. Microbiol. 62: 676–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldberg E., Grinius L. L., Letellier L. 1994. Recognition, attachment, and injection, p. 353 In Karam J. D. (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, DC [Google Scholar]
- 21. Hahn F. M., Hurlburt A. P., Poulter C. D. 1999. Escherichia coli open reading frame 696 is idi, a nonessential gene encoding isopentenyl diphosphate isomerase. J. Bacteriol. 181: 4499–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hugenholtz J., Perdon L., Abee T. 1993. Growth and energy generation by Lactococcus lactis subsp. lactis biovar diacetylactis during citrate metabolism. Appl. Environ. Microbiol. 59: 4216–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jarvis A. W. 1984. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl. Environ. Microbiol. 47: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joly N., et al. 2010. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34: 797–827 [DOI] [PubMed] [Google Scholar]
- 25. Jordan A., et al. 1996. The ribonucleotide reductase system of Lactococcus lactis: characterization of an NrdEF enzyme and a new electron transport protein. J. Biol. Chem. 271: 8779–8785 [DOI] [PubMed] [Google Scholar]
- 26. Jovanovic G., Lloyd L. J., Stumpf M. P. H., Mayhew A. J., Buck M. 2006. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J. Biol. Chem. 281: 21147–21161 [DOI] [PubMed] [Google Scholar]
- 27. Kalasauskaite E. V., Kadisaite D. L., Daugelavicius R. J., Grinius L. L., Jasaitis A. A. 1983. Requirement for membrane potential in Escherichia coli infection by phage T4. Eur. J. Biochem. 130: 123–130 [PubMed] [Google Scholar]
- 28. Koprivnjak T., et al. 2006. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J. Bacteriol. 188: 3622–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kramer N. E., Van Hijum S. A. F. T., Knol J., Kok J., Kuipers O. P. 2006. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob. Agents Chemother. 50: 1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kristian S. A., et al. 2005. d-Alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 187: 6719–6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Labedan B., Letellier L. 1981. Membrane-potential changes during the first steps of coliphage infection. Proc. Natl. Acad. Sci. U. S. A. 78: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larsen R., Buist G., Kuipers O. P., Kok J. 2004. ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. J. Bacteriol. 186: 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larsen R., van Hijum S. A. F. T., Martinussen J., Kuipers O. P., Kok J. 2008. Transcriptome analysis of the Lactococcus lactis ArgR and AhrC regulons. Appl. Environ. Microbiol. 74: 4768–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lerner A., et al. 2009. The Azospirillum brasilense Sp7 noeJ and noeL genes are involved in extracellular polysaccharide biosynthesis. Microbiology 155: 4058–4068 [DOI] [PubMed] [Google Scholar]
- 35. Letellier L., Labedan B. 1985. Release of respiratory control in Escherichia coli after bacteriophage adsorption: process independent of DNA injection. J. Bacteriol. 161: 179–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levander F., Andersson U., Radstrom P. 2001. Physiological role of beta-phosphoglucomutase in Lactococcus lactis. Appl. Environ. Microbiol. 67: 4546–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lolkema J. S., Poolman B., Konings W. N. 1995. Role of scalar protons in metabolic energy generation in lactic-acid bacteria. J. Bioenerg. Biomembr. 27: 467–473 [DOI] [PubMed] [Google Scholar]
- 38. Lubbers M. W., Schofield K., Waterfield N. R., Polzin K. M. 1998. Transcription analysis of the prolate-headed lactococcal bacteriophage c2. J. Bacteriol. 180: 4487–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lubbers M. W., Waterfield N. R., Beresford T. P. J., Lepage R. W. F., Jarvis A. W. 1995. Sequencing and analysis of the prolate-headed lactococcal bacteriophage-c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61: 4348–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marchler-Bauer A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37: D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martinez B., Zomer A. L., Rodriguez A., Kok J., Kuipers O. P. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64: 473–486 [DOI] [PubMed] [Google Scholar]
- 42. Monteville M. R., Ardestani B., Geller B. L. 1994. Lactococcal bacteriophages require a host-cell wall carbohydrate and a plasma-membrane protein for adsorption and ejection of DNA. Appl. Environ. Microbiol. 60: 3204–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakano S., Kuster-Schock E., Grossman A. D., Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 100: 13603–13608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakano S., Nakano M. M., Zhang Y., Leelakriangsak M., Zuber P. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. U. S. A. 100: 4233–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Neuhaus F. C., Baddiley J. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67: 686–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Novo D., Perlmutter N. G., Hunt R. H., Shapiro H. M. 1999. Accurate flow cytometric membrane potential measurement in bacteria using diethyloxacarbocyanine and a ratiometric technique. Cytometry 35: 55–63 [DOI] [PubMed] [Google Scholar]
- 47. O'Driscoll J., Glynn F., Fitzgerald G. F., van Sinderen D. 2006. Sequence analysis of the lactococcal plasmid pNP40: a mobile replicon for coping with environmental hazards. J. Bacteriol. 188: 6629–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pamp S. J., Frees D., Engelmann S., Hecker M., Ingmer H. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188: 4861–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Poranen M. M., et al. 2006. Global changes in cellular gene expression during bacteriophage PRD1 infection. J. Virol. 80: 8081–8088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raisanen L., et al. 2007. Molecular interaction between lipoteichoic acids and Lactobacillus delbrueckii phages depends on d-alanyl and alpha-glucose substitution of poly(glycerophosphate) backbones. J. Bacteriol. 189: 4135–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ravantti J. J., Ruokoranta T. M., Alapuranen A. M., Bamford D. H. 2008. Global transcriptional responses of Pseudomonas aeruginosa to phage PRR1 infection. J. Virol. 82: 2324–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rey S., et al. 2005. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 33: D164–D168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schouler C., Clier F., Lerayer A. L., Ehrlich S. D., Chopin M. C. 1998. A type IC restriction-modification system in Lactococcus lactis. J. Bacteriol. 180: 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schouler C., Gautier M., Ehrlich S. D., Chopin M. C. 1998. Combinational variation of restriction modification specificities in Lactococcus lactis. Mol. Microbiol. 28: 169–178 [DOI] [PubMed] [Google Scholar]
- 56. Sijtsma L., Wouters J. T. M., Hellingwerf K. J. 1990. Isolation and characterization of lipoteichoic acid, a cell-envelope component involved in preventing phage adsorption, from Lactococcus lactis subsp. cremoris SK110. J. Bacteriol. 172: 7126–7130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Spinelli S., et al. 2006. Modular structure of the receptor binding proteins of Lactococcus lactis phages-The RBP structure of the temperate phage TP901-1. J. Biol. Chem. 281: 14256–14262 [DOI] [PubMed] [Google Scholar]
- 58. Steen A., et al. 2005. Autolysis of Lactococcus lactis is increased upon d-alanine depletion of peptidoglycan and lipoteichoic acids. J. Bacteriol. 187: 114–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Terzaghi B. E., Sandine W. E. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29: 807–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Torrents E., et al. 2000. The anaerobic (class III) ribonucleotide reductase from Lactococcus lactis: catalytic properties and allosteric regulation of the pure enzyme system. J. Biol. Chem. 275: 2463–2471 [DOI] [PubMed] [Google Scholar]
- 61. Tremblay D. M., et al. 2006. Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J. Bacteriol. 188: 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tusher V. G., Tibshirani R., Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Utaida S., et al. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149: 2719–2732 [DOI] [PubMed] [Google Scholar]
- 64. Valyasevi R., Sandine W. E., Geller B. L. 1991. A membrane-protein is required for bacteriophage-c2 infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 173: 6095–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Veiga P., et al. 2007. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 282: 19342–19354 [DOI] [PubMed] [Google Scholar]
- 66. Weidenmaier C., Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6: 276–287 [DOI] [PubMed] [Google Scholar]
- 67. Yang Y. H., et al. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Young R. Y. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56: 430–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zdobnov E. M., Apweiler R. 2001. InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]
- 70. Zhang Y., Cronan J. E. 1998. Transcriptional analysis of essential genes of the Escherichia coli fatty acid biosynthesis gene cluster by functional replacement with the analogous Salmonella typhimurium gene cluster. J. Bacteriol. 180: 3295–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.