Abstract
Hepatitis B virus (HBV) transcription and replication are essentially restricted to hepatocytes because liver-enriched transcription factors govern viral RNA synthesis. The level of transcription from the HBV promoters depends on both the transcription factors binding to these regulatory sequence elements and their ability to recruit coactivators capable of mediating assembly of the transcription preinitiation complex containing RNA polymerase II. Nuclear receptors are a primary determinant of HBV pregenomic RNA synthesis and, hence, viral replication. Peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) enhances the activity of nuclear receptors and, consequently, HBV biosynthesis. PGC1α is also an important target of signal transduction pathways involved in hepatic glucose and lipid homeostasis, suggesting that this coactivator may have an important role in modulating HBV biosynthesis under various physiological conditions. Consistent with this suggestion, v-akt murine thymoma viral oncogene homolog/protein kinase B (AKT/PKB) is shown to modulate PGC1α activity and, hence, HBV transcription and replication in a cell line-specific manner. In addition, AKT can modulate HBV replication in some but not all cell lines at a posttranscriptional step in the viral life cycle. These observations demonstrate that growth and nutritional signals have the capacity to influence viral production, but the magnitude of these effects will depend on the precise cellular context in which they occur.
INTRODUCTION
Studies of hepatitis B virus (HBV) replication have demonstrated that viral DNA synthesis occurs by the reverse transcription of viral pregenomic 3.5-kb RNA (45, 56). Therefore, it is apparent that the level of transcription from the HBV nucleocapsid promoter is a major determinant of viral biosynthesis (48). The regulatory elements within the HBV nucleocapsid promoter have been analyzed in detail in an attempt to identify the transcription factors controlling viral RNA synthesis (18, 47). Indeed, the HBV nucleocapsid promoter binds a variety of liver-enriched and ubiquitous transcription factors that probably contribute to the level of viral pregenomic 3.5-kb RNA synthesis to different degrees depending on the physiological state of the infected hepatocyte (18, 47). Utilizing nonhepatoma cells that do not support viral pregenomic 3.5-kb RNA synthesis or HBV replication, complementation studies with individual liver-enriched transcription factors demonstrated that only nuclear receptors were capable of rescuing viral biosynthesis in the absence of additional factors (38, 48). These observations indicated that nuclear receptors may have a unique role in governing HBV biosynthesis during natural infection. Indeed, in vivo analysis using the transgenic mouse model of chronic HBV infection demonstrated that viral RNA and DNA synthesis was completely dependent on the nuclear receptor hepatocyte nuclear factor 4α (HNF4α) (22).
Nuclear receptors and their coactivators within the liver are important regulators of energy homeostasis (7, 25, 41, 58). The interactions between nuclear receptors and specific coactivators, in particular, peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α), have been shown to be critical for maintaining glucose homeostasis (58). Inhibition of gluconeogenesis by the insulin signaling pathway is particularly important for the maintenance of glucose homeostasis and involves the phosphorylation of PGC1α at serine 570 by v-akt murine thymoma viral oncogene homolog/protein kinase B (AKT/PKB), which blocks its association with nuclear receptors, reducing the expression of glucose-6-phosphatase (G6P) and phosphoenolpyruvate kinase (PEPCK) (23). Similarly, the level of transcription from the HBV nucleocapsid promoter has been shown to be modulated by the coactivator PGC1α and the corepressor small heterodimer partner (SHP) in a manner that depends on the nuclear receptor governing viral pregenomic 3.5-kb RNA synthesis (34, 35).
On the basis of these considerations, it was of interest to determine if the insulin signaling pathway and AKT, in particular, might modulate HBV pregenomic 3.5-kb RNA synthesis and viral replication in a PGC1α-dependent manner. To address this issue, the effect of activated AKT on HBV transcription and replication was evaluated in the human hepatoma cell lines Huh7 and HepG2 and the nonhepatoma cell line 293T expressing nuclear receptor liver receptor homolog 1 (LRH1). Viral biosynthesis in Huh7 cells in the absence of PGC1α was unaffected by AKT. In contrast, PGC1α-dependent HBV transcription and replication in Huh7 cells were very sensitive to phosphorylation by low levels of AKT activity. In contrast, HBV biosynthesis displayed limited sensitivity to AKT expression in HepG2 or 293T cells. These results demonstrate that HBV biosynthesis is susceptible to regulation by AKT to various degrees depending upon the precise cellular environment. Additionally, AKT serves as a model kinase demonstrating that, in principle, additional signal transduction pathways and their associated protein kinase cascades may also modulate HBV transcription and replication, in part, by regulating PGC1α activity (33, 36).
MATERIALS AND METHODS
Plasmid constructions.
The steps in the cloning of the plasmid constructs used in the transfection experiments were performed by standard techniques (42). HBV DNA sequences in these constructions were derived from the plasmid pCP10, which contains two copies of the HBV genome (subtype ayw) cloned into the EcoRI site of pBR322 (10). The HBV DNA (4.1-kbp) construct that contains 1.3 copies of the HBV genome includes the viral sequence from nucleotide coordinates 1072 to 3182 plus 1 to 1990 (48). This plasmid was constructed by cloning the NsiI/BglII HBV DNA fragment (nucleotide coordinates 1072 to 1990) into pUC13, generating pHBV(1072-1990). Subsequently, a complete copy of the 3.2-kbp viral genome linearized at the NcoI site (nucleotide coordinates 1375 to 3182 plus 1 to 1374) was cloned into the unique NcoI site (HBV nucleotide coordinate 1374) of pHBV(1072-1990), generating the HBV DNA (4.1-kbp) construct.
The pCMX-mLRH1, pcDNA-hPGC1α, pcDNA-Flag-hPGC1αS570A, pSRα-myrAKT, and pBabe-Puro-HA vectors express LRH1, PGC1α, PGC1αS570A, AKT1, and phosphatase and tensin homolog-PTEN-(PTEN) polypeptides from mouse LRH1, human PGC1α (for both PGC1α and PGC1αS570A), mouse AKT1, and human PTEN cDNAs, respectively, using the cytomegalovirus immediate-early promoter (pCMX and pcDNA), simian virus 40 early promoter (pSRα), and Moloney murine leukemia virus long terminal repeat (pBabe-Puro) (16, 23, 26, 32, 53). The PGC1αS570A polypeptide has the serine residue at position 570, which is the major phosphorylation site for AKT, changed to alanine (23). The myrAKT polypeptide, a constitutively active form of AKT, has the Src myristoylation signal fused in-frame to the AKT1 coding sequence (16).
Cells and transfections.
The human hepatoma Huh7 and HepG2 cell lines and human embryonic kidney 293T cell line were grown in RPMI 1640 medium and 10% fetal bovine serum at 37°C in 5% CO2–air. Transfections for viral RNA and DNA analysis were performed as previously described (29) using 10-cm plates containing approximately 1 × 106 cells. DNA and RNA isolation was performed at 3 days posttransfection. In 293T cells, the transfected DNA mixture was composed of 5 μg of HBV DNA (4.1 kbp) plus 1.5 μg of the nuclear receptor expression vector pCMX-mLRH1 and various amounts of the pcDNA-hPGC1α, pcDNA-Flag-hPGC1αS570A, pSRα-myrAKT, and pBabe-Puro-PTEN-HA expression vectors (16, 23, 26, 32, 53). In Huh7 and HepG2 cells, the 1.5 μg of the nuclear receptor expression vector was omitted. Controls were derived from cells transfected with HBV DNA and the expression vectors lacking a nuclear receptor, PGC1α, myrAKT, and PTEN cDNA insert (37).
Characterization of HBV transcripts and viral replication intermediates.
Transfected cells from a single plate were divided equally and used for the preparation of total cellular RNA and viral DNA replication intermediates as described previously (46), with minor modifications. For RNA isolation (6), the cells were lysed in 1.8 ml of 25 mM sodium citrate, pH 7.0, 4 M guanidinium isothiocyanate, 0.5% (vol/vol) sarcosyl, 0.1 M 2-mercaptoethanol. After addition of 0.18 ml of 2 M sodium acetate, pH 4.0, the lysate was extracted with 1.8 ml of water-saturated phenol plus 0.36 ml of chloroform-isoamyl alcohol (49:1). After centrifugation for 30 min at 3,000 rpm in a Sorval RT6000 centrifuge, the aqueous layer was precipitated with 1.8 ml of isopropanol. The precipitate was resuspended in 0.3 ml of 25 mM sodium citrate, pH 7.0, 4 M guanidinium isothiocyanate, 0.5% (vol/vol) sarcosyl, 0.1 M 2-mercaptoethanol and precipitated with 0.6 ml of ethanol. After centrifugation for 20 min at 14,000 rpm in an Eppendorf 5417C microcentrifuge, the precipitate was resuspended in 0.3 ml of 10 mM Tris hydrochloride, pH 8.0, 5 mM EDTA, 0.1% (wt/vol) sodium lauryl sulfate and precipitated with 45 μl of 2 M sodium acetate plus 0.7 ml of ethanol.
For the isolation of viral DNA replication intermediates, the cells were lysed in 0.4 ml of 100 mM Tris hydrochloride, pH 8.0, 0.2% (vol/vol) NP-40. The lysate was centrifuged for 1 min at 14,000 rpm in an Eppendorf 5417C microcentrifuge to pellet the nuclei. The supernatant was adjusted to 6.75 mM magnesium acetate plus 200 μg/ml DNase I and incubated for 1 h at 37°C to remove the transfected plasmid DNA. The supernatant was readjusted to 100 mM NaCl, 10 mM EDTA, 0.8% (wt/vol) sodium lauryl sulfate, 1.6 mg/ml pronase and incubated for an additional 1 h at 37°C. The supernatant was extracted twice with phenol, precipitated with 2 volumes of ethanol, and resuspended in 100 μl of 10 mM Tris hydrochloride, pH 8.0, 1 mM EDTA. RNA (Northern) and DNA (Southern) filter hybridization analyses were performed using 10 μg of total cellular RNA and 30 μl of viral DNA replication intermediates, respectively, as described previously (42). Filter hybridization analysis was quantified by phosphorimaging using a Packard Cyclone Storage Phosphor System.
RESULTS
PTEN/PI3K/PDK1/AKT signal transduction pathway modulates HBV biosynthesis.
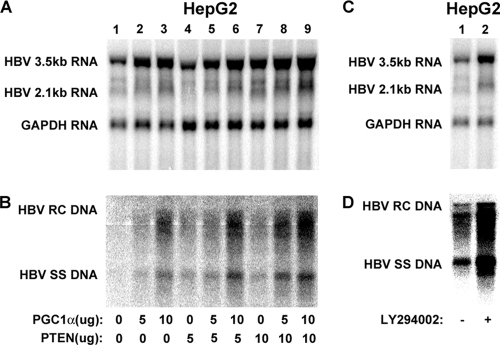
It has previously been demonstrated that activation of AKT leads to the inhibition of HBV transcription and, hence, a reduction in viral replication in HepG2 cells (12). However, mechanistic insight into this process and identification of the substrates that are phosphorylated by AKT to mediate alterations in HBV biosynthesis are lacking. Initially, the importance of the PTEN/phosphatidylinositol 3-kinase (PI3K)/phosphoinositide-dependent kinase-1 (PDK1)/AKT signal transduction pathway for the regulation of HBV biosynthesis was confirmed by expressing PTEN in HepG2 cells (Fig. 1A and B). In the presence or absence of PGC1α, expression of PTEN was associated with a modest 2-fold enhancement of HBV transcription and replication (Fig. 1A and B). These observations are consistent with the suggestion that the inhibition of AKT activity by increased PTEN expression can enhance HBV biosynthesis at the transcriptional level in HepG2 cells (12). To further confirm the role of the PTEN/PI3K/PDK1/AKT signal transduction pathway in the regulation of HBV transcription in HepG2 cells, the PI3K inhibitor LY294002 was used to reduce AKT activity (Fig. 1C and D). As observed with PTEN expression, treatment of HepG2 cells with LY294002 resulted in an approximately 3-fold increase in HBV biosynthesis at both the RNA and DNA levels, as reported previously (12). Overall, these results confirm the suggestion that the inhibition of AKT activity can increase HBV biosynthesis (12).
Fig. 1.
Effect of PTEN/PI3K/PDK1/AKT signal transduction pathway on HBV biosynthesis in the human hepatoma cell line HepG2. (A and B) Cells were transfected with the HBV DNA (4.1-kbp) construct alone (lane 1) or the HBV DNA (4.1-kbp) construct plus the PGC1α and PTEN expression vectors (lanes 2 to 9), as indicated. (C and D) Cells were treated with the PI3K inhibitor LY294002 (10 μM), as indicated. (A and C) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B and D) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA.
PGC1α-activated HBV 3.5-kb RNA synthesis and viral replication are highly sensitive to AKT inhibition in human hepatoma Huh7 cells.
Nuclear receptors, including HNF4α, retinoid X receptor α (RXRα) plus peroxisome proliferator-activated receptor α (PPARα), RXRα plus farnesoid X receptor α (FXRα), LRH1, and estrogen-related receptor (ERR), can support HBV pregenomic 3.5-kb RNA synthesis, which subsequently leads to viral DNA replication (38, 48). These nuclear receptors can recruit the coactivator PGC1α, which may result in enhanced levels of gene transcription (34, 35). PGC1α activity is regulated by a number of signal transduction pathways leading to the modulation of nuclear receptor-mediated transcription (23, 25, 33, 36, 57). Various growth factors, cytokines, and metabolically regulated hormones signal through their receptors via the PI3K/PDK1/AKT signal transduction pathway to regulate gene expression (13, 19, 58). AKT directly regulates nuclear receptor-mediated gene expression by phosphorylating PGC1α at serine 570 (23). Phosphorylation of PGC1α by AKT results in the dissociation of this coactivator from promoter regulatory sequences, with a concomitant reduction in RNA synthesis (23). As the level of HBV biosynthesis is governed by both nuclear receptors and PGC1α (34, 35, 43), it was of interest to investigate if viral transcription and replication were subject to regulation by AKT in a PGC1α-dependent manner.
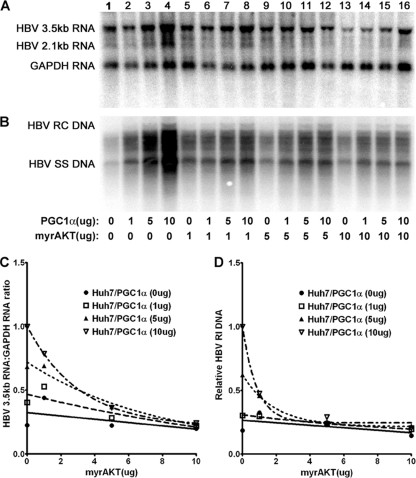
Transfection of the HBV DNA (4.1-kbp) construct into Huh7 cells supports HBV transcription and replication (Fig. 2A, lane 1). Expression of increasing levels of PGC1α activates both HBV 3.5-kb RNA synthesis and viral replication in a dose-dependent manner (Fig. 2). Expression of increasing levels of AKT inhibits both HBV 3.5-kb RNA synthesis and viral replication in a dose-dependent manner (Fig. 2). The effects of PGC1α and AKT on HBV DNA and RNA synthesis were quantitatively similar, indicating that the effects of PGC1α and AKT on transcription were reflected in the levels of observed viral replication (Fig. 2). Interestingly, PGC1α-dependent HBV RNA synthesis and DNA synthesis were highly sensitive to modest levels of AKT activity (Fig. 2C and D). However, AKT did not inhibit to any great extent viral biosynthesis that was not dependent on the expression of PGC1α (Fig. 2). These observations suggest that AKT inhibits PGC1α-dependent HBV 3.5-kb RNA synthesis by phosphorylating serine 570 and eliminating the activity of this coactivator without altering the activities of additional factors in Huh7 cells that contribute to basal levels of HBV biosynthesis.
Fig. 2.
Effect of PGC1α and AKT expression on HBV biosynthesis in the human hepatoma cell line Huh7. Cells were transfected with the HBV DNA (4.1-kbp) construct alone (lane 1) or the HBV DNA (4.1-kbp) construct plus the PGC1α and AKT expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (C) Quantitative analysis of the HBV 3.5-kb RNA from two independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software). (D) Quantitative analysis of the HBV replication intermediates from two independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software).
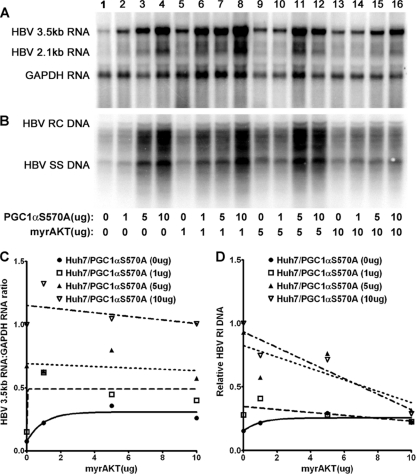
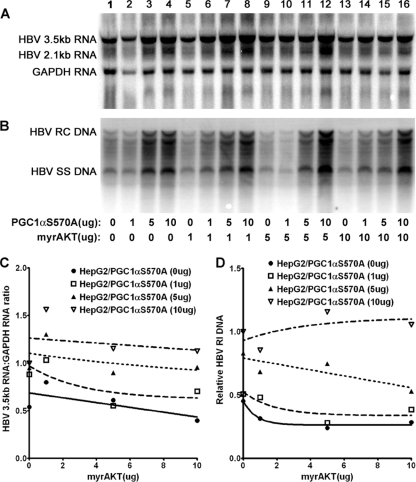
In an attempt to evaluate directly the role of phosphorylation of serine 570 by AKT on PGC1α activity, HBV biosynthesis in Huh7 cells was evaluated in cells transfected with the PGC1αS570A mutant expression vector (Fig. 3). Expression of increasing levels of PGC1αS570A activated HBV 3.5-kb RNA synthesis in a dose-dependent manner (Fig. 3A and C). However, HBV 3.5-kb RNA synthesis was resistant to AKT at all levels examined, indicating that the inhibition of HBV transcription by AKT was mediated by the phosphorylation of serine 570 in PGC1α. Interestingly, HBV replication was partially sensitive to AKT inhibition at the highest levels of AKT expression (Fig. 3B and D). This observation suggests that there is an additional target(s) for AKT phosphorylation that leads to the modulation of viral biosynthesis at a posttranscriptional step in the HBV replication cycle.
Fig. 3.
Effect of PGC1αS570A and AKT expression on HBV biosynthesis in the human hepatoma cell line Huh7. Cells were transfected with the HBV DNA (4.1-kbp) construct alone (lane 1) or the HBV DNA (4.1-kbp) construct plus the PGC1αS570A and AKT expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (C) Quantitative analysis of the HBV 3.5-kb RNA from three independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software). (D) Quantitative analysis of the HBV replication intermediates from three independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software).
PGC1α-activated HBV 3.5-kb RNA synthesis and viral replication are relatively insensitive to AKT inhibition in human hepatoma HepG2 cells.
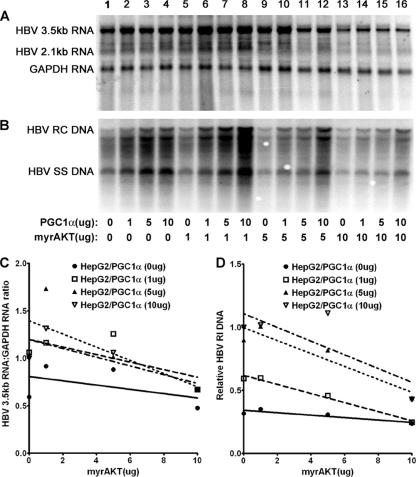
Transfection of the HBV DNA (4.1-kbp) construct into HepG2 cells supports HBV transcription and replication (Fig. 4A, lane 1). Expression of increasing levels of PGC1α modestly activates both HBV 3.5-kb RNA synthesis and viral replication (Fig. 4). Expression of increasing levels of AKT inhibits both HBV 3.5-kb RNA synthesis and viral replication in a dose-dependent manner (Fig. 4). The effects of PGC1α and AKT on HBV RNA and DNA synthesis were qualitatively similar, indicating that the effects of PGC1α and AKT on transcription were likely also reflected in the levels of observed viral replication (Fig. 4). However, unlike the results observed in Huh7 cells (Fig. 2), HBV biosynthesis in HepG2 cells was relatively insensitive to AKT-mediated inhibition, with higher levels of AKT expression being required to observe maximal reductions in HBV RNA and DNA synthesis (Fig. 4).
Fig. 4.
Effect of PGC1α and AKT expression on HBV biosynthesis in the human hepatoma cell line HepG2. Cells were transfected with the HBV DNA (4.1-kbp) construct alone (lane 1) or the HBV DNA (4.1-kbp) construct plus the PGC1α and AKT expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (C) Quantitative analysis of the HBV 3.5-kb RNA from four independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software). (D) Quantitative analysis of the HBV replication intermediates from four independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software).
The potential role of phosphorylation of PGC1α at serine 570 by AKT on HBV biosynthesis was evaluated in HepG2 cells (Fig. 4). Expression of increasing levels of PGC1αS570A activated HBV 3.5-kb RNA and DNA synthesis in a dose-dependent manner (Fig. 5A and C). However, both HBV 3.5-kb RNA synthesis and DNA synthesis were essentially resistant to AKT at all levels examined, indicating that the inhibition of HBV transcription by AKT was mediated by the phosphorylation of serine 570 in PGC1α. The observation that HBV replication was largely insensitive to AKT inhibition even at the highest levels of AKT expression suggests that the mechanism of posttranscriptional inhibition of viral replication that is apparent in Huh7 cells is absent in HepG2 cells (Fig. 5B and D). This observation suggests that there are significant differences in the regulation of HBV biosynthesis between human hepatoma cell lines, as noted previously (34, 35).
Fig. 5.
Effect of PGC1αS570A and AKT expression on HBV biosynthesis in the human hepatoma cell line HepG2. Cells were transfected with the HBV DNA (4.1-kbp) construct alone (lane 1) or the HBV DNA (4.1-kbp) construct plus the PGC1αS570A and AKT expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (C) Quantitative analysis of the HBV 3.5-kb RNA from two independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software). (D) Quantitative analysis of the HBV replication intermediates from two independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software).
PGC1α and AKT modulate LRH1-dependent HBV transcription and replication in human embryonic kidney 293T cells.
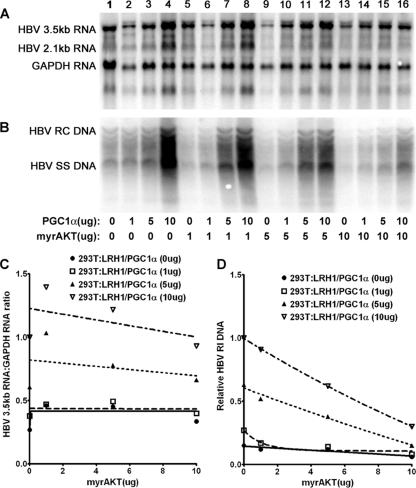
To investigate the role of PGC1α and AKT in the regulation of HBV biosynthesis in nonhepatoma cells, viral RNA and DNA synthesis was examined in 293T cells expressing LRH1 (Fig. 6 and 7). Under these circumstances, HBV 3.5-kb RNA synthesis was largely resistant to the effects of AKT activity. PGC1α and PGC1αS570A both increase viral transcription in a dose-dependent manner. However, viral transcription was not affected by AKT expression, indicating that PGC1α either was not being phosphorylated in 293T cells or was being dephosphorylated by a protein phosphatase as rapidly as AKT was phosphorylating it on serine 570. Alternatively, it is possible that phosphorylation of PGC1α on serine 570 does not affect the activity of PGC1α in 293T cells. In contrast, HBV DNA synthesis was modestly inhibited by the expression of AKT (Fig. 6 and 7). This indicates that viral replication is negatively affected by AKT activity at a posttranscriptional step in its biosynthesis, as appears to also occur in Huh7 cells (Fig. 3).
Fig. 6.
Effect of PGC1α and AKT expression on HBV biosynthesis in the human embryonic kidney cell line 293T. Cells were transfected with the HBV DNA (4.1-kbp) construct and the LRH1 expression vector (lane 1) or the HBV DNA (4.1-kbp) construct and the LRH1 expression vector plus the PGC1α and AKT expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (C) Quantitative analysis of the HBV 3.5-kb RNA from two independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad, version 5, software). (D) Quantitative analysis of the HBV replication intermediates from two independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software).
Fig. 7.
Effect of PGC1αS570A and AKT expression on HBV biosynthesis in the human embryonic kidney cell line 293T. Cells were transfected with the HBV DNA (4.1-kbp) construct and the LRH1 expression vector (lane 1) or the HBV DNA (4.1-kbp) construct and the LRH1 expression vector plus the PGC1αS570A and AKT expression vectors (lanes 2 to 16), as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (C) Quantitative analysis of the HBV 3.5-kb RNA from three independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software). (D) Quantitative analysis of the HBV replication intermediates from three independent experiments. Trend lines were calculated using one-phase decay analysis and the method of least-squares fit (GraphPad Prism, version 5, software).
DISCUSSION
HBV replicates by the reverse transcription of the viral pregenomic 3.5-kb RNA (45, 56). In nonhepatoma cells, nuclear receptors are the only transcription factors that alone have been shown to support HBV 3.5-kb RNA synthesis and viral replication (38, 48). Activation of nuclear receptor-mediated transcription by coactivators has been shown to have an important consequence for energy homeostasis in the liver (25, 39, 41). In particular, the coactivator PGC1α has been shown to be a critical determinant of both gluconeogenesis and HBV biosynthesis, suggesting that the regulation of viral transcription and key metabolic gene expression may utilize similar hepatic signal transduction pathways (34, 35, 39, 43). Insulin signaling has been extensively characterized, and a primary mode of glucose homeostasis in the liver has been shown to be mediated through the insulin/insulin receptor/PI3K/PDK1/AKT/PGC1α signal transduction pathway (13, 54). These observations have led to the suggestion that HBV biosynthesis might be similarly regulated, but to date, only the effect of PGC1α on viral biosynthesis has been examined in detail (34, 35, 43, 44).
In the current study, the effect of activated AKT on HBV biosynthesis was analyzed in two human hepatoma cell lines capable of supporting viral replication and in a nonhepatoma cell line where HBV 3.5-kb RNA synthesis was dependent on the expression of nuclear receptors (Fig. 1 to 7). Interestingly, AKT was able to inhibit HBV biosynthesis in all cell lines examined to various degrees, but the mechanisms of inhibition were quantitatively and qualitatively distinct (Fig. 8). In Huh7 cells, the phosphorylation of PGC1α by AKT on serine 570 resulted in the efficient inhibition of coactivator-dependent viral transcription and replication (Fig. 2 and 3). In addition, HBV replication was also inhibited at a posttranscriptional step by concentrations of AKT higher than those required to affect PGC1α-directed viral transcription. These observations suggest that AKT can prevent HBV transcription through PGC1α and a subsequent step in the replication cycle (Fig. 8). Although the posttranscriptional step has not been defined, it is interesting to speculate that the direct or indirect phosphorylation of the arginine-rich carboxyl-terminal domain of the HBV core polypeptide by AKT or AKT-activated downstream kinases might reduce its capacity to support viral replication (9, 11, 21, 24, 27). Indeed, the viral nucleocapsid protein is known to be phosphorylated, and these modifications can affect viral replication, making this an attractive mechanism of posttranscriptional regulation of HBV biosynthesis by AKT (11, 17, 21, 30). Additional analysis will be required to establish the significance of this suggestion for viral replication.
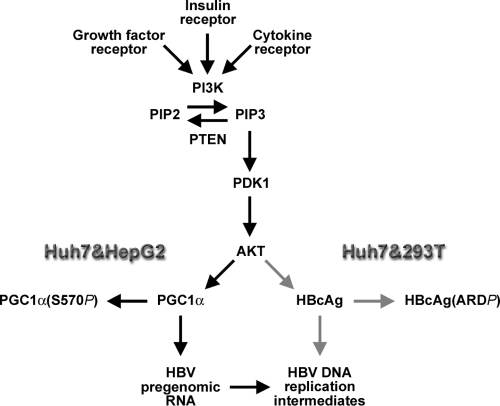
Fig. 8.
Signal transduction pathways potentially modulating HBV transcription and replication in HBV-infected hepatocytes. Growth factors, hormones, and cytokines may modulate the phosphatase and tensin homolog (PTEN)/phosphatidylinositol 3-kinase (PI3K)/phosphoinositide-dependent kinase-1 (PDK1)/AKT signal transduction pathway. PTEN converts phosphatidylinositol 3,4,5-trisphosphate (PIP3) to phosphatidylinositol 4,5-bisphosphate (PIP2). PI3K converts PIP2 to PIP3. PIP3 activates PDK1 and AKT (5, 28). AKT is known to phosphorylate PGC1α on serine 570, leading to reduced transcriptional activity on known target genes (23), including the HBV nucleocapsid gene (black arrows), as observed in Huh7 and HepG2 cells. In addition, AKT can directly or indirectly inhibit HBV replication in Huh7 and 293T cells at a posttranscriptional step in the viral replication cycle potentially involving phosphorylation of the HBV nucleocapsid polypeptide (HBcAg) in the arginine-rich carboxyl-terminal domain (ARD) (gray arrows).
The effect of AKT on HBV biosynthesis has been examined previously in HepG2 cells (12). This analysis suggested that AKT affected HBV biosynthesis at the level of viral transcription (12). Our findings are similar (Fig. 1), with the effects of AKT on viral replication being primarily a consequence of the phosphorylation of PGC1α and its effect on HBV transcription (Fig. 4 and 5). This was most readily apparent when PGC1αS570A was evaluated (Fig. 5). In this case, both HBV transcription and replication were largely resistant to inhibition by AKT, suggesting that the reduction in viral transcription was primarily due to the phosphorylation of PGC1α and its associated loss of transcriptional activity at the nucleocapsid promoter. Unlike the observations in Huh7 cells, AKT inhibited only viral transcription in HepG2 cells and failed to affect any posttranscriptional steps in HBV biosynthesis (Fig. 8).
The effect of AKT on viral biosynthesis directed by LRH1 in human embryonic kidney cells was distinct from that observed in the human hepatoma cells. Viral transcription was essentially unaffected by AKT (Fig. 6 and 7). However, HBV replication was modestly inhibited in these nonhepatoma cells independently of the potential phosphorylation status of PGC1α. Similar results were observed when HBV biosynthesis was supported by HNF4α expression in 293T cells (C. R. Ondracek and A. McLachlan, unpublished data). These observations suggest that the mechanism of action by which AKT modulated HBV biosynthesis is cell type dependent and presumably reflects the relative importance of the downstream signaling affecting HBV transcription and replication (Fig. 8). In Huh7 and HepG2 cells, AKT appeared to be able to phosphorylate PGC1α on serine 570, inhibiting its capacity to activate transcription from the HBV nucleocapsid promoter, and hence reduced viral replication, albeit to different extents, depending on the cell line (Fig. 8). In contrast, higher AKT activity can also inhibit HBV replication at a posttranscriptional step in Huh7 and 293T cells. This becomes apparent when viral transcription is activated by PGC1αS570A, which increases viral RNA synthesis because it is no longer subject to regulation by AKT (Fig. 3, 5, and 7).
In combination, these different observations suggest that there are at least two distinct signal transduction pathways downstream of AKT that regulate HBV biosynthesis. It appears that lower effective activities of AKT can regulate the contribution of PGC1α to viral transcription, whereas higher effective concentrations of AKT are necessary to facilitate the posttranscriptional effects on HBV replication. This is most obvious in Huh7 cells, where the effect on transcription is observed at lower levels of AKT expression and the effect on replication is apparent only at higher levels of AKT expression (Fig. 2 and 3). Consistent with this suggestion is the absence of any obvious posttranscriptional effect of AKT on viral biosynthesis in HepG2 cells (Fig. 5). This may reflect a lower effective AKT activity in this cell line. In contrast, only the posttranscriptional effects of AKT are readily apparent in 293T cells. This observation suggests that the effect of AKT phosphorylation on PGC1α must be mitigated by some additional activity within these cells. On the basis of the observation that PGC1α and PGC1αS570A displayed very similar effects on HBV transcription, it is simplest to postulate that 293T cells contain a protein phosphatase activity that can efficiently dephosphorylate PGC1α and, hence, prevent AKT from inhibiting PGC1α activity. Obviously, there could be alternative, more complex explanations for these observations, and additional studies will be required to resolve these issues. Regardless, it is apparent that whatever mechanism prevents AKT from inhibiting PGC1α activity in 293T cells, it does not appear to interfere with the AKT-activated signal transduction pathway inhibiting the posttranscriptional step in HBV biosynthesis, as modest reductions in viral DNA synthesis are observed in the absence of any major effects on HBV transcription (Fig. 6 and 7).
Overall, these observations indicate that two or more signal transduction pathways downstream from activated AKT can modulate either viral transcription or subsequent steps in the HBV replication cycle. Elucidating these pathways will help our understanding of the molecular processes governing HBV biosynthesis and may lead to the identification of additional targets for antiviral therapy. Importantly, further understanding of the signaling that is upstream of AKT is likely to be helpful in elucidating the relative importance of extracellular signals that govern viral production during natural infection. Glucose homeostasis mediated by insulin signaling clearly has the potential to modulate HBV biosynthesis through AKT on short- and long-term time scales, and this has led to the suggestion that viral biosynthesis may be regulated by the metabolic state of the hepatocyte (43, 44). Indeed, the observation that diabetic patients are significantly more likely to present with acute HBV infections than nondiabetic patients supports the contention that insulin signaling through AKT can influence the outcome of this viral infection (50). However, cytokines associated with acute or chronic inflammation resulting from the immune response to an HBV infection are also likely to lead to long-term alterations in the PI3K/PDK1/AKT signal transduction pathway, which may have a significant consequence for viral biosynthesis (13, 19, 58). Furthermore, chronic HBV infection is associated with the long-term development of primary hepatocellular carcinoma (PHC) (1, 2). Tumor development is often associated with activation of growth factor signal transduction pathways, leading to uncontrolled cellular proliferation (51). PHC is often associated with AKT activation resulting from either growth factor stimulation or mutations within the AKT signal transduction pathway, which may explain why viral biosynthesis is often quite limited in tumors but relatively robust in the adjacent normal liver tissue (4, 8, 14, 20, 31, 49, 52, 55). Therefore, it seems likely that the regulation of the AKT signal transduction pathway will have important roles in modulating HBV biosynthesis under a variety of physiologically relevant conditions, and hence, further understanding of the mechanisms of regulation of HBV transcription and replication by AKT may offer insight into potential targets for the development of antiviral agents against chronic HBV infection. Specifically, it may be possible to activate AKT and, hence, inhibit HBV biosynthesis while preventing the majority of the cell growth-promoting activities mediated by AKT using rapamycin to inhibit the target of rapamycin complex 1 (TORC1) (3). Additionally, AKT serves as a model kinase capable of modulating HBV pregenomic RNA transcription through the phosphorylation of PGC1α at serine 570 (23) and viral replication at the posttranscriptional level by poorly defined mechanisms (Fig. 8). Therefore, it will be of considerable interest to investigate the physiological roles of additional signal transduction pathways and their associated protein kinase cascades which are known to regulate PGC1α activity (15, 33, 36, 40) for their effects on HBV transcription and replication.
ACKNOWLEDGMENTS
We are grateful to David Mangelsdorf (Southwestern Medical Center, Dallas, TX) for the plasmid pCMX-mLRH1, Morris Birnbaum (University of Pennsylvania School of Medicine, Philadelphia, PA) for the plasmids pcDNA-hPGC1α and pcDNA-Flag-hPGC1αS570A, and Nissim Hay (University of Illinois at Chicago, Chicago, IL) for the plasmids pSRα-myrAKT and pBabe-Puro-HA.
This work was supported by Public Health Service grant AI30070 from the National Institutes of Health.
Footnotes
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Beasley R. P. 1988. Hepatitis B virus: the major etiology of hepatocellular carcinoma. Cancer 61: 1942–1956 [DOI] [PubMed] [Google Scholar]
- 2. Beasley R. P., Hwang L.-Y., Lin C.-C., Chien C.-S. 1981. Hepatocellular carcinoma and hepatitis B virus—a prospective study of 22707 men in Taiwan. Lancet ii: 1129–1133 [DOI] [PubMed] [Google Scholar]
- 3. Bhaskar P. T., Hay N. 2007. The two TORCs and Akt. Dev. Cell 12: 487–502 [DOI] [PubMed] [Google Scholar]
- 4. Boyault S., et al. 2007. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology (Baltimore, Md.) 45: 42–52 [DOI] [PubMed] [Google Scholar]
- 5. Cantley L. C., Neel B. G. 1999. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl. Acad. Sci. U. S. A. 96: 4240–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chomczynski P., Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159 [DOI] [PubMed] [Google Scholar]
- 7. Cooper M. P., et al. 2006. Defects in energy homeostasis in Leigh syndrome French Canadian variant through PGC-1α/LRP130 complex. Genes Dev. 20: 2996–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Craxi A., et al. 1984. Tissue markers of hepatitis B virus infection in hepatocellular carcinoma and cirrhosis. Hepatogastroenterology 31: 55–59 [PubMed] [Google Scholar]
- 9. Daub H., et al. 2002. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J. Virol. 76: 8124–8137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dubois M. F., Pourcel C., Rousset S., Chany C., Tiollais P. 1980. Excretion of hepatitis B surface antigen particles from mouse cells transformed with cloned viral DNA. Proc. Natl. Acad. Sci. U. S. A. 77: 4549–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gazina E. V., Fielding J. E., Lin B., Anderson D. A. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 74: 4721–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo H., et al. 2007. Regulation of hepatitis B virus replication by the phosphatidylinositol 3-kinase-Akt signal transduction pathway. J. Virol. 81: 10072–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hay N. 2011. Akt isoforms and glucose homeostasis—the leptin connection. Trends Endocrinol. Metab. 22: 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsu H. C., Lin W. S., Tsai M. J. 1983. Hepatitis-B surface antigen and hepatocellular carcinoma in Taiwan. With special reference to types and localization of HBsAg in the tumor cells. Cancer 52: 1825–1832 [DOI] [PubMed] [Google Scholar]
- 15. Jager S., Handschin C., Pierre J., Spiegelman B. M. 2007. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc. Natl. Acad. Sci. U. S. A. 104: 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kennedy S. G., et al. 1997. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 11: 701–713 [DOI] [PubMed] [Google Scholar]
- 17. Köck J., Nassal M., Deres K., Blum H. E., Von Weizsäcker F. 2004. Hepatitis B virus nucleocapsids formed by carboxy-terminally mutated core proteins contain spliced viral genomes but lack full-size DNA. J. Virol. 78: 13812–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kosovsky M. J., Qadri I., Siddiqui A. 1998. The regulation of hepatitis B virus gene expression: an overview of the cis- and trans-acting components, p. 21–50.In Koshy R., Caselmann W. H. (ed.), Hepatitis B virus: molecular mechanisms in disease and novel strategies for therapy. Imperial College Press, London, United Kingdom [Google Scholar]
- 19. Kroczynska B., Kaur S., Platanias L. C. 2010. Growth suppressive cytokines and the AKT/mTOR pathway. Cytokine 48: 138–143 [DOI] [PubMed] [Google Scholar]
- 20. Lai M. Y., et al. 1988. Status of hepatitis B virus DNA in hepatocellular carcinoma: a study based on paired tumor and nontumor liver tissues. J. Med. Virol. 25: 249–258 [DOI] [PubMed] [Google Scholar]
- 21. Lan Y. T., Li J., Liao W. Y., Ou J. H. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259: 342–348 [DOI] [PubMed] [Google Scholar]
- 22. Li L., et al. 2009. Developmental regulation of hepatitis B virus biosynthesis by hepatocyte nuclear factor 4α. PLoS One 4: e5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li X., Monks B., Ge Q., Birnbaum M. J. 2007. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature 447: 1012–1016 [DOI] [PubMed] [Google Scholar]
- 24. Liao W., Ou J.-H. 1995. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J. Virol. 69: 1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin J., Handschin C., Spiegelman B. M. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1: 361–370 [DOI] [PubMed] [Google Scholar]
- 26. Lu T. T., et al. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6: 507–515 [DOI] [PubMed] [Google Scholar]
- 27. Machida A., et al. 1991. Phosphorylation in the carboxyl-terminal domain of the capsid protein of hepatitis B virus: evaluation with a monoclonal antibody. J. Virol. 65: 6024–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manning B. D., Cantley L. C. 2007. AKT/PKB signaling: navigating downstream. Cell 129: 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McLachlan A., et al. 1987. Expression of hepatitis B virus surface and core antigens: influences of pre-S and precore sequences. J. Virol. 61: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nassal M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 66: 4107–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neuveut C., Wei Y., Buendia M. A. 2010. Mechanisms of HBV-related hepatocarcinogenesis. J. Hepatol. 52: 594–604 [DOI] [PubMed] [Google Scholar]
- 32. Nogueira V., et al. 2008. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell 14: 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Olson B. L., et al. 2008. SCFCdc4 acts antagonistically to the PGC-1α transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 22: 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ondracek C. R., Reese V. C., Rushing C. N., Oropeza C. E., McLachlan A. 2009. Distinct regulation of hepatitis B virus biosynthesis by peroxisome proliferator-activated receptor γ coactivator 1α and small heterodimer partner in human hepatoma cell lines. J. Virol. 83: 12545–12551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ondracek C. R., Rushing C. N., Reese V. C., Oropeza C. E., McLachlan A. 2009. Peroxisome proliferator-activated receptor γ coactivator 1α and small heterodimer partner differentially regulate nuclear receptor-dependent hepatitis B virus biosynthesis. J. Virol. 83: 12535–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Puigserver P., et al. 2001. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol. Cell 8: 971–982 [DOI] [PubMed] [Google Scholar]
- 37. Raney A. K., Johnson J. L., Palmer C. N. A., McLachlan A. 1997. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J. Virol. 71: 1058–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reese V. C., et al. 2011. Multiple nuclear receptors may regulate hepatitis B virus biosynthesis during development. Int. J. Biochem. Cell Biol. 43: 230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rhee J., et al. 2003. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc. Natl. Acad. Sci. U. S. A. 100: 4012–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodgers J. T., Haas W., Gygi S. P., Puigserver P. 2010. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab. 11: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rohas L. M., et al. 2007. A fundamental system of cellular energy homeostasis regulated by PGC-1α. Proc. Natl. Acad. Sci. U. S. A. 104: 7933–7938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 43. Shlomai A., Paran N., Shaul Y. 2006. PGC-1α controls hepatitis B virus through nutritional signals. Proc. Natl. Acad. Sci. U. S. A. 103: 16003–16008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shlomai A., Shaul Y. 2008. The “metabolovirus” model of hepatitis B virus suggests nutritional therapy as an effective anti-viral weapon. Med. Hypotheses 71: 53–57 [DOI] [PubMed] [Google Scholar]
- 45. Summers J., Mason W. S. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29: 403–415 [DOI] [PubMed] [Google Scholar]
- 46. Summers J., Smith P. M., Huang M., Yu M. 1991. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J. Virol. 65: 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang H., Banks K. E., Anderson A. L., McLachlan A. 2001. Hepatitis B virus transcription and replication. Drug News Perspect. 14: 325–334 [PubMed] [Google Scholar]
- 48. Tang H., McLachlan A. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. U. S. A. 98: 1841–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tsai W. L., Chung R. T. 2010. Viral hepatocarcinogenesis. Oncogene 29: 2309–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tucker M. E.Acute hepatitis B incidence double in people with diabetes. Intern. Med. News. [3 March 2011]. http://www.internalmedicinenews.com.
- 51. Walther Z., Sklar J. 2011. Molecular tumor profiling for prediction of response to anticancer therapies. Cancer J. 17: 71–79 [DOI] [PubMed] [Google Scholar]
- 52. Wang Y., et al. 2002. Different expression of hepatitis B surface antigen between hepatocellular carcinoma and its surrounding liver tissue, studied using a tissue microarray. J. Pathol. 197: 610–616 [DOI] [PubMed] [Google Scholar]
- 53. Wen S., et al. 2001. PTEN controls tumor-induced angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 98: 4622–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Whiteman E. L., Cho H., Birnbaum M. J. 2002. Role of Akt/protein kinase B in metabolism. Trends Endocrinol. Metab. 13: 444–451 [DOI] [PubMed] [Google Scholar]
- 55. Whittaker S., Marais R., Zhu A. X. 2010. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 29: 4989–5005 [DOI] [PubMed] [Google Scholar]
- 56. Will H., et al. 1987. Replication strategy of human hepatitis B virus. J. Virol. 61: 904–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xiong S., et al. 2010. PGC-1α serine 570 phosphorylation and GCN5-mediated acetylation by angiotensin II drive catalase down-regulation and vascular hypertrophy. J. Biol. Chem. 285: 2474–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yoon J. C., et al. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413: 131–138 [DOI] [PubMed] [Google Scholar]