Abstract
Immature retrovirus particles are assembled from the multidomain Gag protein. In these particles, the Gag proteins are arranged radially as elongated rods. We have previously characterized the properties of HIV-1 Gag in solution. In the absence of nucleic acid, HIV-1 Gag displays moderately weak interprotein interactions, existing in monomer-dimer equilibrium. Neutron scattering and hydrodynamic studies suggest that the protein is compact, and biochemical studies indicate that the two ends can approach close in three-dimensional space, implying the need for a significant conformational change during assembly. We now describe the properties of the Gag protein of Moloney murine leukemia virus (MLV), a gammaretrovirus. We found that this protein is very different from HIV-1 Gag: it has much weaker protein-protein interaction and is predominantly monomeric in solution. This has allowed us to study the protein by small-angle X-ray scattering and to build a low-resolution molecular envelope for the protein. We found that MLV Gag is extended in solution, with an axial ratio of ∼7, comparable to its dimensions in immature particles. Mutational analysis suggests that runs of prolines in its matrix and p12 domains and the highly charged stretch at the C terminus of its capsid domain all contribute to this extended conformation. These differences between MLV Gag and HIV-1 Gag and their implications for retroviral assembly are discussed.
INTRODUCTION
Expression of a single virus-coded protein, termed Gag, in permissive host cells is sufficient for efficient assembly of immature retrovirus-like particles (VLPs) (52). In most retroviruses, Gag molecules are initially targeted to the plasma membrane where they accumulate and organize into a protein lattice, ultimately leading to the formation and budding of immature VLPs. After the immature particle is released from the cell, the Gag proteins within it are cleaved by the viral protease into their constituent domains to form a mature virus. Immature VLPs are roughly spherical, pleomorphic membrane-enclosed structures, with a diameter of ∼120 nm. Under the membrane exterior, several thousand Gag molecules are arranged radially, with their matrix (MA) domains in contact with the lipid bilayer and their nucleocapsid (NC) domains facing the interior of the particle, presumably interacting with RNA. The Gag proteins in the immature particle are highly extended rods, with a length of ∼20 nm and a width of only ∼2 to 3 nm (2, 57, 58).
Each Gag protein molecule that participates in the structural lattice of these nearly regular VLPs must interact in a highly specific way with several other Gag molecules. In turn, these interactions must depend upon the overall conformation of the participating monomers. A structural feature of the immature protein lattice, conserved in all retroviruses studied, is the hexagonal arrangement of Gag molecules, mediated largely by interactions between their CA domains (13). The presence of irregular defects, which allows for curvature of the hexagonal lattice to form an enclosed sphere, has been observed in several retroviruses (13, 57).
The similarity in organizational principles of immature retroviral VLPs across genera might suggest similarities in the overall structural properties of Gag proteins. Indeed, there are conserved features between Gag proteins from different genera, despite a low degree of sequence conservation. All retroviral Gag proteins are multidomain proteins, always containing (from the N terminus to the C terminus) the MA, capsid (CA), and NC domains (52), though Gag proteins often contain one or more genus-specific domains in addition to the three canonical domains. Structural studies on isolated domains of Gag reveal conservation in three-dimensional shape: the MA domains of HIV-1, equine infectious anemia virus, Rous sarcoma virus, and Moloney murine leukemia virus (MLV) Gag show a remarkable similarity in three-dimensional shape (8, 21, 22, 32, 34, 45), as do the N-terminal domains (NTDs) of CA from HIV-1 and MLV (18, 35). In contrast to studies on isolated domains, detailed structural information on intact Gag polyproteins has not been forthcoming, presumably due to their larger size and conformational heterogeneity.
We have previously analyzed the properties of purified, recombinant unmyristoylated HIV-1 Gag protein. The free protein is soluble but assembles spontaneously into VLPs upon addition of nucleic acid (3, 4). In the absence of nucleic acid, the protein is in monomer-dimer equilibrium in solution; the dimerization interface was localized to the C-terminal portion (CTD) of the CA domain (12). This interface in mature CA protein was previously shown to participate in the assembly of the conical core structure within mature HIV-1 particles (30, 54, 55) and is probably involved in the assembly of Gag proteins into immature HIV-1 particles as well.
Monomeric HIV-1 Gag protein appears to adopt relatively compact conformation(s) in solution (9), or when bound to lipid membranes (10), and biochemical experiments suggest that its N and C termini can be quite near each other in three-dimensional space (12). It is known to contain several unstructured and/or flexible regions, including the C-terminal regions of the MA (32, 53) and CA (19, 55) domains, the SP1 linker (38), and the NC domain (28). Since it is compact in solution but extended in VLPs, assembly must involve a reorganization of the relative positions of its major globular domains.
In the present report, we describe the properties of the recombinant unmyristoylated Gag protein from MLV, a prototypical gammaretrovirus. While immature MLV particles are very similar in overall structure and organization to immature HIV-1 particles (17, 58), we found that monomeric MLV Gag molecules are different in several respects from their HIV-1 counterparts. MLV Gag has a much weaker propensity for interprotein interactions and is monomeric under dilute conditions. Moreover, despite being a multidomain protein, MLV Gag has a rigid and extended structure, with dimensions in solution similar to those in VLPs. Its rodlike shape implies that the linkers between its domains are all relatively rigid. The implications of these observations are discussed.
MATERIALS AND METHODS
Recombinant protein expression and purification.
Beginning with an infectious molecular clone of MLV, expression plasmids for MLV Gag, mutants, and chimeras (see Fig. 1A) were generated in pET3xc vectors by standard molecular cloning protocols and expressed in BL21-CodonPlus-(DE3) RPIL Escherichia coli (Stratagene). HIV-1 Gag protein was purified using phosphocellulose chromatography as described earlier (11, 12). Plasmids containing MLV/HIV-1 chimeras were kindly provided by Michael Emerman, Fred Hutchinson Cancer Research Center. The mutant MLV 18PA has 18 of its proline codons (at positions 92 to 94, 105 to 107, 109, 110, 124, 125, 161 to 163, 169 to 171, 203, and 204) replaced with alanine codons. Cells expressing proteins were grown to an optical density of ∼0.8 and induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 5 h at 37°C. The purifications of the MLV Gag proteins and chimeras were similar to that of the HIV-1 Gag protein (11, 12). Briefly, MLV Gag was fractionated from the cell lysate by the addition of saturated ammonium sulfate to 40% saturation. The precipitated proteins were resolubilized in 20 mM Tris-HCl (pH 7.4)–0.5 M NaCl–5 mM dithiothreitol (DTT) and purified by phosphocellulose (PC) affinity chromatography as described for HIV-1 Gag (11, 12). The PC-purified MLV Gag proteins were further purified by size exclusion chromatography (SEC) on a Superose 12 column (GE Healthcare), equilibrated in 20 mM Tris-HCl (pH 7.4)–0.5 M NaCl–5 mM DTT, and stored at −80°C after the addition of glycerol to 10% (vol/vol). For sedimentation equilibrium (SE) experiments and small-angle X-ray scattering (SAXS) measurements, the protein was further purified on an SP-Sepharose column (GE Healthcare) and dialyzed into 20 mM Tris-HCl (pH 8.0) with 0.5 M NaCl and 1 mM tris(2-carboxyethyl)phosphine (TCEP).
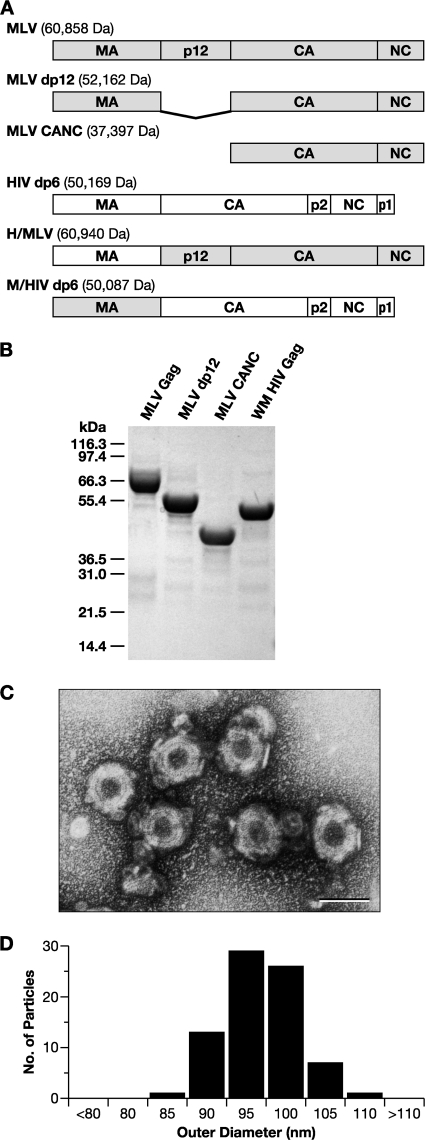
Fig. 1.
(A) Schematic representation and molecular masses of the Gag proteins in the present study, showing domain organization. (B) SDS-PAGE gel of purified Gag proteins, stained with Coomassie brilliant blue. WM HIV-1 Gag, a monomeric mutant of HIV-1 Gag, is included. (C) Negative stain EM of VLPs assembled from MLV Gag in vitro. Scale bar, 100 nm. (D) Histogram of the size distribution of in vitro assembled MLV Gag VLPs.
In vitro assembly.
MLV Gag at 1 mg/ml in 20 mM Tris-HCl (pH 7.4)–0.5 M NaCl–5 mM DTT was incubated with 10% (wt/wt) yeast tRNA and subjected to overnight dialysis at 4°C against 20 mM Tris-HCl (pH 8.0)–0.1 M NaCl–5 mM β-mercaptoethanol (assembly buffer). Assembled VLPs in the dialysate were visualized by electron microscopy (EM) after negative staining with 2% uranyl acetate.
Analytical ultracentrifugation.
All sedimentation equilibrium (SE) experiments were carried out at 4°C in an Optima XL-A analytical centrifuge (Beckman-Coulter Instruments). Protein samples were in 20 mM Tris-HCl (pH 7.4)–0.5 M NaCl–1 mM TCEP. For SE, 12-mm cells were loaded with 180 μl of protein, while 3-mm cells were loaded with 50 μl and centrifuged to equilibrium at 8,000, 10,000, and 15,000 rpm. The data were acquired at 280 and 254 nm at radial increments of 0.001 cm and with 10 repeats. The software Sedenterp (http://www.bbri.org/RASMB/rasmb.html) (26) was used to estimate the partial specific volumes of proteins and buffer parameters, viscosity (η), and density (ρ). Data analysis was performed using SEDPHAT (46; reviewed in reference 27). The goodness of fit in SEDPHAT is the “reduced chi-square” value (χ2).
Boundary sedimentation velocity (SV) experiments were performed at 45,000 rpm and 20°C or 4°C, using either 400 μl (12-mm cells) or 100 μl (3-mm cells) of sample. The data acquired at 280-nm and 0.003-cm radial increments were analyzed with SEDFIT (http://www.analyticalultracentrifugation.com/), using previously described computational methodology (47). Briefly, the SV data were subjected to maximum entropy regularization with the confidence level set to P = 0.95 for the most parsimonious distribution of sedimenting species. The results of this computational analysis give c(s) versus s plots, where c(s) is the concentration of protein divided by the sedimentation coefficient at the respective s position.
SEC and light scattering.
A Rainin HPXL solvent delivery system connected to a Rainin Dynamax UV-1 detector and a Wyatt systems Dawn EOS static and quasi-elastic light scattering (SLS and QELS, respectively) detector was used to study the hydrodynamic properties of the proteins on a Superose 12 (GE Healthcare) column. A solvent viscosity of 0.00936 centipoise and a temperature dependence of −1.95E−4 g/cm s K were assumed. The data collected simultaneously from light detectors 5 through 18, with the exception of detector 13, were used for SLS, while detector 13 was modified for QELS measurements.
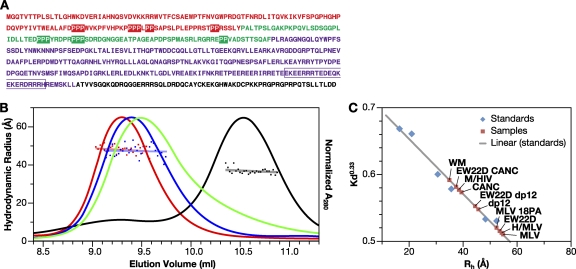
The column was calibrated with standards (GE Healthcare), including RNase A (Rh 16.4 Å), chymotrypsin (Rh 20.9 Å), ovalbumin (Rh 30.5 Å), bovine serum albumin (BSA; Rh 35.5 Å), aldolase (Rh 48.1 Å), and catalase (Rh 52.2 Å), for the determination of Rh from retention time. Retention time was used to calculate the parameter (KD)1/3, which has a linear relationship to the Rh of the eluting species (48). Sample proteins, except for MLV 18PA and M/HIV, were injected onto the column at concentrations of 2 to 3 mg/ml, such that the concentrations in the eluting peaks ranged from 0.22 to 0.35 mg/ml. M/HIV and MLV 18PA were injected at ∼0.8 mg/ml, resulting in concentrations of ∼ 0.06 mg/ml in the eluting peaks.
SAXS and modeling.
Both SAXS and wide-angle X-ray scattering (WAXS) were performed at beam lines 12-ID of Advanced Photon Sources (APS) at Argonne National Laboratory (Chicago site). The wavelength (λ) of X-ray radiation was set as 0.689 Å, and the scattered X-ray photons were recorded with a charge-coupled device X-ray detector. A cylindrical quartz capillary X-ray flow cell with a diameter of 1.5 mm and a wall of 10 μm was used. The X-ray beam, with size of 0.1 by 0.2 mm2, was adjusted to pass through the center of the cell. The exposure time was set to 0.4 to 0.6 s to avoid detector saturation and radiation damage. Potential radiation damage was further reduced by maintaining a constant flow of the samples. The range of momentum transfer q (= 4π sinθ/λ, where 2θ is the scattering angle) of SAXS experiments was 0.006 to 0.260 Å−1 and that of WAXS was 0.1 to 2.6 Å−1. Twenty images were acquired for each sample or buffer solution. The two-dimensional scattering images of buffers and samples, azimuthally averaged after solid angle correction, were normalized with the intensity of the incident X-ray beam. The resulting one-dimensional scattering data sets were averaged before buffer background subtraction. The background subtractions were performed as follows. First, the WAXS profile was obtained by using equation 1:
| (1) |
The value of α, an adjustable parameter, was tuned to eliminate the scattering from buffer, indicated by the disappearance of the solvent peak at ∼2.0 Å −1. The resulting WAXS profile was then used as a guide for the SAXS background subtraction, by tuning the value of α in SAXS subtraction and overlaying the resulting SAXS profile with the WAXS profile at the overlapping q range, i.e., 0.1 to 0.26 Å −1 in our experiments.
All samples were measured at three concentrations, ranging from 0.6 to 3.2 mg/ml. The final SAXS profiles were obtained from linear extrapolation of data from these concentrations to that of zero concentration. The total scattering profiles were obtained by piecing the resulting SAXS and WAXS data together in the range of 0.006 to 2.5 Å−1.
The radius of gyration (Rg) was calculated from data at low q values in the range of qRg < 1.2, using the Guinier approximation (equation 2):
| (2) |
The scattering intensities at and near q = 0 were extrapolated with the Guinier equation. The Kratky plots, q2 · I(q) versus q (see Fig. 4A), indicate that all molecules under study are in folded form.
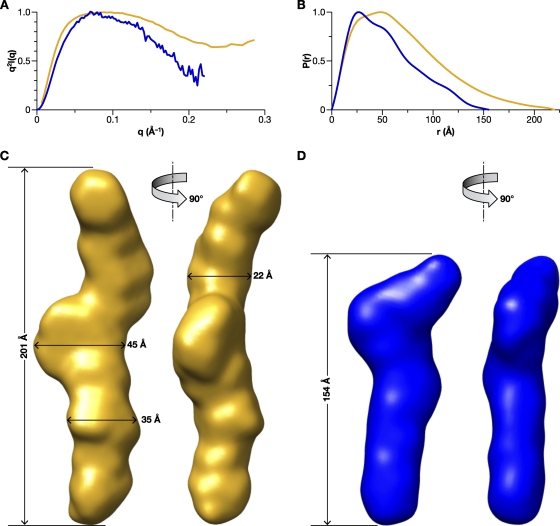
Fig. 4.
SAXS analysis of MLV Gag and CANC proteins. (A) Kratky plots for MLV Gag (yellow) and CANC (blue). (B) p(r) analysis of SAXS data for MLV Gag (yellow) and CANC (blue). (C) Bead model, shown in molecular surface mode, generated from SAXS data using DAMMIF, as described in detail in Materials and Methods, for MLV Gag. (D) Bead model for CANC. The data indicate that both proteins are rod-shaped, with maximum dimensions of ∼200 and 150 Å, respectively. The normalized spatial discrepancy (NSD) scores of both the models are <0.7, suggesting a good consistency among all models and an excellent fit to the experimental SAXS data.
The pairwise distance distribution function (PDDF), or p(r), in real space was calculated using GNOM (50). To avoid underestimation of the molecular dimension and consequent distortion in low resolution structural reconstruction, the parameter Rmax, the upper end of distance r, was manually scanned in the range of 2Rg to 6Rg, and chosen such that the resulting PDDFs gradually approach zero at high r.
Molecular envelopes (also known as bead models) were obtained using the program DAMMIF (16), a fast version of DAMMIN (51). In DAMMIF, a spherical space with a radius of Rmax, read from the PDDF result, is initially filled with multiple-phase dummy atoms. To avoid distortion caused by possible underestimation of Dmax, DAMMIF can automatically adjust the value of Rmax during the calculation. At each step, the envelope evolves by randomly phasing a dummy-atom in (as a part of the molecule) or out (as a part of the solvent). A simulated annealing algorithm drives the envelope evolution, by reducing the discrepancy between the experimental and calculated scattering curves during the annealing process. Thirty-two DAMMIF calculations were performed for each molecule, running in the “slow” mode with default setting of DAMMIF. The resulting structural models were subjected to averaging and the normalized spatial discrepancy (NSD) values between each pair of models were computed, using DAMAVER (41). The model with lowest average NSD with respect to the rest of models was chosen as the reference model. The remaining models were superimposed onto the reference model using SUPCOMB (24), except that possible outliers identified by NSD criteria were discarded. The dummy atoms of these superimposed models were remapped onto a densely packed grid of atoms, with each grid point marked with its occupancy factor. The grids with non-zero occupancies were chosen to generate a final consensus model with the volume equal to the average excluded volume of all of the models. Scattering data in a q range of 0 to 0.22 Å −1, which reflect the global shape without significant undesired influence from the internal structure, were used in DAMMIF calculations. The goodness-of-fit Rf values (51; http://www.embl-hamburg.de/biosaxs/manual_dammin.html) were defined as follows:
| (3) |
and were in the range of 0.004 to 0.006, indicating good match between the experimental scattering data and the calculated ones for individual models. In equation 3, sc is a scaling factor which makes the experimental data and a given back-calculated profile have the best match in intensities. The average NSDs for these bead models were 0.70 to 0.95, which indicate good convergences in both individual DAMMIF fits and overall bead model ensembles for each sample.
The averaged molecular envelope/bead models were further smoothed by using the program Situs (56), version 2.4, and the smoothed molecular envelope images shown in Fig. 4 were produced by using the UCSF program Chimera (42) (version 1, build 2540).
RESULTS
In vitro assembly of MLV Gag.
The proteins used in our studies, including MLV Gag, WM HIV-1 Gag (a monomeric mutant of HIV-1 Gag [12]), and a series of related proteins, are depicted in Fig. 1A. They were purified from E. coli lysates as described in Materials and Methods. Their purity is indicated by the Coomassie brilliant blue-stained SDS-PAGE gel of several of the proteins (Fig. 1B).
MLV Gag was tested for its ability to assemble into VLPs in a defined in vitro system. As in prior studies with HIV-1 Gag, the protein was dialyzed at 1 mg/ml into assembly buffer in the presence of 100 μg of yeast tRNA/ml at 4°C. Under these assembly conditions, roughly spherical VLPs were formed, as visualized by negative stain EM (Fig. 1C) and transmission EM of thin sections (data not shown). These VLPs had a median diameter of 95 nm with a standard deviation of 4.5 nm (Fig. 1D), similar to that of immature VLPs assembled in cells (58). The VLPs have a somewhat irregular appearance, and thin sections of these VLPs made after high-speed centrifugation often show “collapsed” structures (not shown), suggesting that the VLPs are fragile.
Oligomerization of MLV Gag in solution.
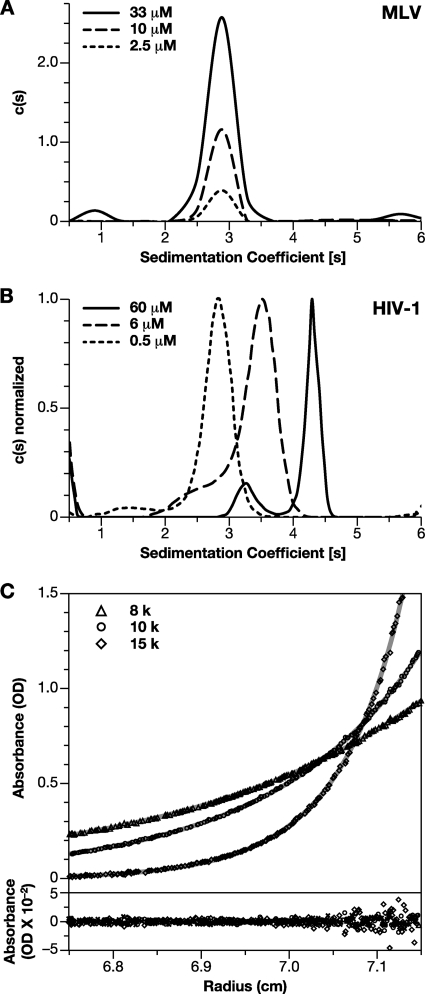
Recombinant HIV-1 Gag protein is in monomer-dimer equilibrium in solution, with a Kd of ∼0.5 × 10−5 M (12). We tested for oligomerization of MLV Gag in several ways. MLV Gag at concentrations ranging from 0.15 mg/ml (i.e., 2.5 μM) to 2 mg/ml (33 μM) was subjected to SV analysis. Boundary sedimentation measurements (data not shown) were performed to determine the weight-average sedimentation coefficient of MLV Gag and the data analyzed by the Sedfit program. Figure 2A shows the c(s) versus S resulting from this analysis. The profiles show a predominant single peak with an S value of 2.85 at all three concentrations at 20°C. In contrast, the peak of the weight-average S value for HIV-1 dp6, which dimerizes with a Kd of ∼5 μM, shifts from 2.8 to 4.2 as the concentration is increased from 0.5 to 60 μM (Fig. 2B). The weight-average S values for HIV-1 Gag at intermediate concentrations of 2 and 20 μM were 3.2 and 3.8, respectively (data not shown). The invariance of the MLV Gag S value across the concentration range tested suggests that the oligomeric composition of MLV Gag is not significantly altered under these conditions. The S value of 2.83 (S20,W 3.15) for MLV Gag, whose molecular mass is 6.0 × 104 Da, represents an f/f0 of 1.85 and an Rh of ∼47 Å (Table 1), suggesting that the molecule is highly asymmetric or extended. MLV dp12 and CANC were also subjected to SV analysis (data not shown). MLV dp12 had an S value of 2.73 (S20,W 3.04) and an f/f0 of 1.62, while CANC had an S value of 2.31 (S20,W 2.57) and an f/f0 1.64, indicating that these portions of MLV Gag also are extended and/or asymmetric.
Fig. 2.
Oligomeric properties of MLV Gag protein. (A) c(s) analysis of boundary sedimentation data for MLV Gag protein at concentrations between 2.5 and 33 μM. (B) c(s) analysis for boundary sedimentation data for HIV-1 dp6 protein at concentrations ranging from 0.5 to 60 μM. (C) SE analysis of Gag oligomerization. Solutions of Gag at 2, 6, and 20 μM were centrifuged to equilibrium at 8,000 (▵), 10,000 (○), and 15,000 (♢) rpm. The data points were globally fit to a model of dimeric association with a Ka of 2.45 × 103 M−1 and a global reduced chi-square value (χ2) of 1.32. For clarity, the figure shows only the three A280 profiles and residuals for the 20 μM protein sample. The best fit root-mean-square error is 0.0055.
Table 1.
Rh values for proteins in this study
| Method | Rh (Å)a |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MLV | H/MLV | MLV 18PA | EW22D | dp12 | EW22D dp12 | CANC | EW22D CANC | WM | M/HIV | BSA | |
| SEC | 54.6 | 54 | 52.1 | 53.3 | 45.5 | 44.3 | 39.2 | 37.2 | 36 | 38.3 | 35.5 |
| QELS | 48 | 47 | ND | 47 | 42 | 39 | 37 | 34 | 38* | ND | 35 |
| SV | 47 | ND | ND | ND | 41.3 | ND | 36.4 | ND | 41* | ND | ND |
ND, not determined. *, Data from Datta et al. (9).
As a more rigorous test for weak self-association, SE analysis of MLV Gag in solution was performed, and the data were fitted to different models using SEDPHAT (46). MLV Gag at concentrations of 2, 6, and 20 μM was centrifuged to equilibrium at 8,000, 10,000, and 15,000 rpm. The SE profiles obtained were first modeled assuming the presence of a single noninteracting species, with a mass of 60 kDa. This resulted in a fit with a global reduced chi-square value (χ2) of 1.823. However, allowing the molecular mass to float while assuming the presence of a single species resulted in a predicted mass of 68 kDa and a better fit to the data (χ2 = 1.346). This indicated that the MLV Gag protein might have a propensity for weak association. Since HIV-1 Gag protein has previously been shown to dimerize (12), the self-association of MLV Gag was initially modeled as a monomer-dimer equilibrium. All nine profiles were analyzed to obtain a global fit with a monomer-dimer association model, assuming the monomer mass to be 60,000 Da. The top panel in Fig. 2C shows only three profiles (20 μM sample) of nine (for clarity). The continuous line through each data set is the nonlinear regression fit to the data, yielding an association constant (Ka) of 2.45 × 103 M−1 for dimeric association of the monomers, with a global reduced chi-square value (χ2) of 1.32 for the fit. The bottom panel shows a plot of the respective residuals for each fit shown above. It is evident that the residuals are all close to zero and mostly randomly distributed. The determined association constant corresponds to a Kd of ∼400 μM. The same data were also subjected to a monomer-trimer model for self-association (data not shown). Here, the best fit obtained was for a Ka of 3.63 × 107 M−2, with a global reduced chi-square value (χ2) of 1.37. Such analysis was performed with SE data obtained using three separately purified batches of protein, and we have obtained Kd values for monomer-dimer equilibrium ranging from ca. 200 to 500 μM (data not shown). Although the very weak interactions observed with MLV Gag do not allow us to determine the strength or mode of interaction unambiguously, these results demonstrate that MLV Gag does self-associate, though far more weakly than HIV-1 Gag.
Conformation of MLV Gag in solution.
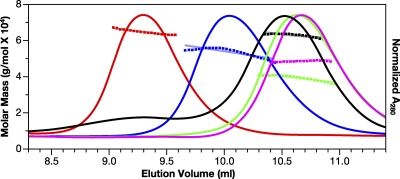
We characterized MLV Gag (∼60 kDa) by SEC. It elutes (red profile) from Superose 12 more rapidly than WM HIV-1 Gag (∼50 kDa, pink profile in Fig. 3). In fact, it elutes at a position comparable to that of a dimer of BSA (∼130 kDa, black profile) (a shallow peak before the BSA monomer). The elution position of MLV Gag was constant over injection concentrations ranging from 4 to 25 μM (data not shown). MLV dp12 (∼52 kDa, blue profile) also elutes well before BSA (∼67 kDa), while MLV CANC (∼37 kDa, green profile) elutes slightly before monomeric WM HIV-1 Gag (∼50 kDa). The high ionic strength of the buffer precludes nonspecific interactions with the column matrix. The eluate was simultaneously monitored by both SLS, to determine the molecular mass of the eluting material, and QELS, as a measure of the Rh of the eluting species. The molar mass for MLV Gag measured by SLS (Fig. 3) was 6.07 × 104 Da, while those for MLV dp12 and MLV CANC were 5.4 × 104 Da and 4.0 × 104 Da, respectively, confirming that the proteins were predominantly monomeric under these conditions. Under identical conditions, the molar mass of BSA was estimated to be 6.47 × 104 Da, while that of WM HIV-1 Gag was 4.89 × 104 Da. The unusual elution behaviors of MLV Gag and derived proteins on the size exclusion column were therefore a result of their conformations and not their oligomeric status. The QELS data gave Rh values of 48, 42, and 37 Å for the MLV, dp12, and CANC proteins, respectively, while that for WM HIV-1 Gag was 38 Å (Table 1).
Fig. 3.
SEC and SLS of MLV and HIV-1 Gag proteins. MLV (red), dp12 (blue), CANC (green), and WM HIV-1 Gag (pink) Gag solutions, as well as BSA (black) at 1.5 mg/ml, were chromatographed on a Superose 12 column. Elution of protein was simultaneously monitored by determining the A280, giving rise to the elution profiles, and by SLS, yielding the data points above the elution profiles. The points indicate the molar mass of the protein in each fraction. These masses, in turn, were averaged over the breadth of each peak, producing the roughly horizontal lines above the elution profiles.
As a completely independent approach to determining the conformation of these proteins, we performed SAXS analysis. SAXS data for MLV Gag and CANC were first analyzed to generate Kratky plots, q2I(q) versus q (Fig. 4A), as described in Materials and Methods. The shape of the Kratky plot is an indicator of the “unfoldedness” of a protein. In the case of multidomain proteins, the plot is also indicative of how domains are extended or packed and has been used as an indicator of global protein conformation. Specifically, folded proteins have a characteristic peak at low q, followed by a drop in the curve at high q, while unfolded proteins do not have the characteristic peak and increase monotonically with q (43). Since Gag is a multidomain protein, the Kratky plots are well suited for characterization of the global domain organization. Figure 4A indicates that while both MLV Gag and CANC are folded, MLV Gag has a more extended conformation with some disorder.
The pairwise distance distribution function [PDDF or p(r)] plots, i.e., histograms of distances between scattering atoms in the protein, give an indication of the shape and a measure of the dimensions of the protein. The p(r) functions for MLV Gag and CANC proteins calculated using GNOM (Fig. 4B) are asymmetric, with a tail at high r values. This is a feature of elongated molecules; the p(r) of globular proteins is symmetrical about the peak. The Dmax, derived from the distance r at which p(r) drops to zero, is indicative of the longest dimension in the molecule. This was estimated to be ∼210 ± 10 Å for MLV Gag and 150 Å ± 10 Å for MLV CANC. The radius of gyration (Rg), determined by SAXS for MLV Gag (57.8 Å), and that of MLV CANC (41.1 Å) were both higher than that of WM HIV-1 Gag (38 Å [data not shown]) or for WM HIV-1 Gag previously determined by small-angle neutron scattering as ∼34 Å (9, 10).
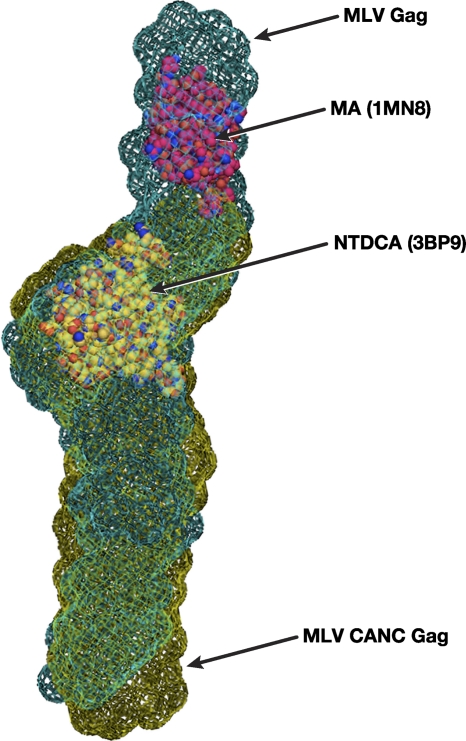
Bead models of the proteins, derived from the SAXS data using DAMMIF (as described in detail in Materials and Methods), are shown in Fig. 4C and D. It is striking to see that ab initio modeling of SAXS data yields a rod-shaped model for MLV Gag, with a long axis of the molecular envelope estimated at ∼201 Å. Similarly, the SAXS data show that CANC is evidently elongated as a rod ∼150 Å long. The width of the bead model at different regions for MLV Gag ranges from 35 to 45 Å, with a narrow dimension of 22 Å. These dimensions are common to both models, as are the presence of “kinks” or bends along the length. The tentative superimposition of molecular envelopes of MLV Gag and CANC with the known crystallographic structures of MLV MA (45) and the NTD of MLV CA (35) is displayed in Fig. 5.
Fig. 5.
Molecular envelope in mesh of MLV Gag generated by molecular modeling of SAXS data as described in Materials and Methods. A tentative superposition of the derived molecular envelopes for MLV Gag (green) and MLV CANC (gold), with crystallographic structures for MA (1MN8) and the NTD of CA (3BP9), is shown.
Structural features of MLV Gag.
We examined the sequence of MLV Gag in an effort to identify possible features contributing to its extended conformation. We noted that both the C-terminal half of the MA domain and the N-terminal half of the p12 domain are very proline-rich and in fact contain several short runs of prolines (Fig. 6A). It seemed possible that these runs might form short polyproline helices or other such extended structures and thus contribute to the rigidity of the protein. To test this possibility, we mutated 18 proline codons (boxed in Fig. 6A) to alanine codons. The resulting mutant Gag protein, MLV 18PA, has a molecular mass of 60,389 Da, which is very similar to that of MLV Gag (60,858 Da). However, its elution from an SEC column is clearly delayed (Fig. 6B), indicating a decrease in Rh from 54.6 to 52.1 Å (Fig. 6B and Table 1). These data suggest that the runs of prolines in the C-terminal half of MA and the N-terminal half of p12 are at least in part responsible for the rigidity of the MLV Gag protein.
Fig. 6.
Hydrodynamic characterization of mutant MLV Gag proteins. (A) Primary amino acid sequence of MLV Gag. The MA (red), p12 (green), CA (purple), and NC (black) domains are indicated. The proline residues in the MA and p12 domains changed to alanine in MLV P18A Gag are boxed. The 22 amino acid residues deleted in the “electric wire” region of CA to generate E22D are outlined. (B) SEC and DLS of mutant MLV Gag proteins. MLV Gag (red), EW22D (blue), MLV 18PA (green), and BSA (black) were chromatographed on Superose 12, and the Rh was estimated by in-line DLS. (C) Retention volume used to estimate Rh for proteins, using proteins with known Rh values as calibrants (48).
Another striking feature of the MLV Gag sequence is an extraordinary run of charged residues near the C terminus of the CA domain, termed the “electric wire” or “charged assembly helix motif” (6). Cheslock et al. (6) presented genetic evidence suggesting that this region forms an α-helix during assembly. These researchers also showed that the deletion of 22 residues (outlined in Fig. 6A) is compatible with the proper assembly, and indeed with the infectivity, of the virus. We determined the Rh of MLV proteins lacking these 22 residues (designated “EW22D” by Cheslock et al.). EW22D MLV Gag elutes slightly after the wild type but before MLV 18PA (Fig. 6B). We also tested the effect of the EW22D deletion in the context of dp12 and CANC proteins (QELS data not shown). As indicated in Table 1, both of these changes significantly decrease the Rh of these smaller proteins, as well as that of full-length Gag. Therefore, the charged stretch in the C-terminal domain of CA also contributes to the extended structure of MLV Gag.
In light of the large differences in molecular dimensions between MLV Gag and HIV-1 Gag (Fig. 3, Table 1), it was of interest to determine the contribution of individual domains to these differences. Deminie and Emerman (14, 15) have previously characterized viruses in which the MA domains of HIV-1 and MLV Gags have been exchanged. These chimeric Gag proteins were purified and analyzed by SEC. As shown in Table 1, replacement of the MA domain of either Gag with the heterologous MA domain did not significantly affect the Rh of either parent protein.
DISCUSSION
Gag proteins of different retroviruses assemble into very similar VLPs in vivo (2, 13). Since the information content for the assembly of VLPs exists within the Gag polyprotein (5), a comparison of the properties of Gag from viruses from different genera might give insight into common underlying mechanisms of assembly.
We have previously characterized the solution properties of unmyristoylated HIV-1 Gag protein. Briefly, it contains a dimer interface in the CTD of its CA domain; the monomer-dimer equilibrium in solution has a Kd of ∼5 × 10−6 M (12). Further, monomeric HIV-1 Gag appears to adopt relatively compact conformation(s) in solution (9). It assembles into small, thin-walled VLPs (4), while presumably still in a compact conformation. An extension in length to ∼20 nm can be observed in vitro when both ends are supplied with their preferred ligands, i.e., inositol phosphates or lipids, and nucleic acid (3, 10).
We now describe the properties of the unmyristoylated MLV Gag protein. Remarkably, it is different from HIV-1 Gag protein in each of these respects. Thus, its tendency to oligomerize in solution is barely detectable: we estimate that the affinity of monomers for each other is at least 40-fold weaker than in the case of HIV-1 (Fig. 2). Further, it exhibits a rod-like conformation in solution, apparently similar to that of the Gag monomers in immature VLPs; this conformation was evident from the SV (Fig. 2A), SEC (Fig. 3), and SAXS data (Fig. 4). This extended conformation presumably enables it to assemble into full-size VLPs upon addition of nucleic acid alone (Fig. 1C and D) and without a major change in its overall dimensions.
MLV and HIV-1 Gag are similar to each other with respect to the secondary and tertiary structures of the MA, NTD, and CTD regions (18, 20, 22, 32, 35, 45); however, the linkers between these regions are apparently unstructured and/or flexible in HIV-1 Gag but rigid in MLV Gag. The fact that exchanging MA domains between the two proteins has a negligible effect on Rh (Table 1) is consistent with this assumption.
What might be responsible for the extended conformation of MLV Gag? Examination of its sequence reveals two notable features not present in HIV-1 Gag (Fig. 6A). First, MLV Gag is quite proline-rich: 11.9% of its residues are prolines, compared to 5% in HIV-1 dp6 and 5% in the protein database (33). Further, 28% of the amino acids between positions 90 and 214 (i.e., the C-terminal region of MA and p12) are proline residues. They are often found as short runs of two to four consecutive prolines; such runs frequently have an extended conformation in known protein structures (49). In fact, we found that a change of 18 prolines in MA and p12 to alanines, which in themselves favor helical structures, results in a detectable decrease in the Rh of the protein (Fig. 6B and Table 1).
It has previously been reported that the p12 domain of MLV Gag (which has no obvious counterpart in HIV-1 Gag) lacks a unique structure (25); it seems likely that the short runs of prolines can assume an ensemble of structures, but that these are predominantly extended. We also noted that deletion of p12 (84 amino acids) causes a drop of Rh from ∼50 to 43 Å, while a further deletion of MA (130 amino acids) to generate CANC reduces the Rh from 43 to 39 Å (Table 1). The larger change in Rh due to a deletion of p12 suggests that this domain, even if disordered, might adopt relatively extended conformation(s) keeping the globular domains of MA and CA far apart.
Second, the C-terminal end of the CA domain contains a remarkable run of ∼33 charged residues. This region, called the “charged assembly helix motif” or “electric wire,” carries no net charge; it is critical for proper VLP assembly (6). Like MLV Gag, MLV CANC displays unusual mobility in SEC: although its mass is only 37.4 kDa, it elutes slightly before monomeric HIV-1 Gag (50.2 kDa) (Fig. 3). The p(r) plot from the SAXS data (Fig. 4B), as well as the Rg of ∼41 Å from SAXS and the Rh of ∼39 Å from SEC and QELS (Table 1) for CANC, all suggest that it also has an extended structure. Since retroviral NC domains are very small and largely unstructured (28), it seemed possible that the “electric wire” region might be responsible in part for the extended structure of MLV CANC and Gag proteins. We found that deletion of 22 of these residues in either MLV Gag, dp12, or CANC caused a detectable decrease in the Rh of the proteins (Fig. 6A and B, Table 1). This was particularly apparent in the context of the smaller CANC construct.
What is the biological significance of these features? Interestingly, several mutants in which individual proline tracts in p12 are replaced with alanine tracts retain infectivity (29, 59), as does the electric wire deletion that we studied (6). It thus appears that the rigidity and/or length of the monomer can be compromised somewhat without loss of function in vivo. In addition, several observations suggest that VLP assembly may be more dependent upon Gag-RNA interaction in MLV than in HIV-1. First, deletion of the NC domain, the primary RNA-binding domain of Gag, modestly reduces VLP assembly in vivo in HIV-1 (39) but virtually eliminates it in MLV (36). Second, digestion of detergent-stripped VLPs with RNase disrupts MLV VLPs (37) but not HIV-1 VLPs (3). It seems plausible that the stronger interaction between free HIV-1 Gag molecules, relative to MLV Gag molecules, renders its lattice structure less dependent upon nucleic acid binding for stability.
On the other hand, HIV-1 MA is known to bind RNA, and it has been suggested that this is responsible for the ability of HIV-1 Gag to assemble with no NC domain (1, 31, 40, 44). Several reports show that in HIV-1 Gag, the MA domain participates along with the NC domain, in interactions with RNA (7, 23). It will be of interest to learn whether similar phenomena are observed in MLV Gag, in which the MA and NC domains are probably always physically distant from each other.
In conclusion, the “building blocks” for HIV-1 and MLV virus particles are surprisingly different from each other. Unlike HIV-1 Gag, MLV Gag in solution has dimensions comparable to that in VLPs; the protein would undergo very little, if any, change in global conformation during the process of assembly. This is a striking example of the contrasts in fundamental properties between members of different retroviral genera.
ACKNOWLEDGMENTS
We thank Demetria Harvin for technical assistance and Richard Frederickson for help with illustrations. S.A.K.D. gratefully thanks Ritu Kanwar for many fruitful scientific discussions.
This study was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research, and in part with federal funds from the National Cancer Institute, NIH, under contract HHSN26120080001E.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Alfadhli A., Still A., Barklis E. 2009. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 83:12196–12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Briggs J. A., et al. 2009. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U. S. A. 106:11090–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campbell S., et al. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. U. S. A. 98:10875–10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campbell S., Rein A. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campbell S., Vogt V. M. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487–6497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheslock S. R., et al. 2003. Charged assembly helix motif in murine leukemia virus capsid: an important region for virus assembly and particle size determination. J. Virol. 77:7058–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chukkapalli V., Oh S. J., Ono A. 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. U. S. A. 107:1600–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conte M. R., Matthews S. 1998. Retroviral matrix proteins: a structural perspective. Virology 246:191–198 [DOI] [PubMed] [Google Scholar]
- 9. Datta S. A., et al. 2007. Conformation of the HIV-1 Gag protein in solution. J. Mol. Biol. 365:812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Datta S. A., et al. 2011. HIV-1 Gag extension: conformational changes require simultaneous interaction with membrane and nucleic acid. J. Mol. Biol. 406:205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Datta S. A., Rein A. 2009. Preparation of recombinant HIV-1 gag protein and assembly of virus-like particles in vitro. Methods Mol. Biol. 485:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Datta S. A., et al. 2007. Interactions between HIV-1 Gag molecules in solution: an inositol phosphate-mediated switch. J. Mol. Biol. 365:799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Marco A., et al. 2010. Conserved and variable features of Gag structure and arrangement in immature retrovirus particles. J. Virol. 84:11729–11736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deminie C. A., Emerman M. 1994. Functional exchange of an oncoretrovirus and a lentivirus matrix protein. J. Virol. 68:4442–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deminie C. A., Emerman M. 1993. Incorporation of human immunodeficiency virus type 1 Gag proteins into murine leukemia virus virions. J. Virol. 67:6499–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Franke D., Svergun D. I. 2009. DAMMIF, a program for rapid ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42:342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuller S. D., Wilk T., Gowen B. E., Krausslich H. G., Vogt V. M. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729–738 [DOI] [PubMed] [Google Scholar]
- 18. Gamble T. R., et al. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285–1294 [DOI] [PubMed] [Google Scholar]
- 19. Gamble T. R., et al. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849–853 [DOI] [PubMed] [Google Scholar]
- 20. Ganser B. K., Cheng A., Sundquist W. I., Yeager M. 2003. Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. EMBO J. 22:2886–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hatanaka H., et al. 2002. Structure of equine infectious anemia virus matrix protein. J. Virol. 76:1876–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hill C. P., Worthylake D., Bancroft D. P., Christensen A. M., Sundquist W. I. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. U. S. A. 93:3099–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones C. P., Datta S. A., Rein A., Rouzina I., Musier-Forsyth K. 2011. Matrix domain modulates HIV-1 Gag's nucleic acid chaperone activity via inositol phosphate binding. J. Virol. 85:1594–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kozin M. B., Svergun D. I. 2001. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34:33–41 [Google Scholar]
- 25. Kyere S. K., Joseph P. R., Summers M. F. 2008. The p12 domain is unstructured in a murine leukemia virus p12-CA(N) Gag construct. PLoS One 3:e1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laue T. M., Shah B., Ridgeway T., Pelletier S. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90–125 In Harding S., Rowe A., Horton J. (ed.), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, Cambridge, United Kingdom [Google Scholar]
- 27. Lebowitz J., Lewis M. S., Schuck P. 2002. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11:2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee B. M., De Guzman R. N., Turner B. G., Tjandra N., Summers M. F. 1998. Dynamic behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 279:633–649 [DOI] [PubMed] [Google Scholar]
- 29. Leung J., et al. 2006. Interaction of Moloney murine leukemia virus matrix protein with IQGAP. EMBO J. 25:2155–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li S., Hill C. P., Sundquist W. I., Finch J. T. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409–413 [DOI] [PubMed] [Google Scholar]
- 31. Lochrie M. A., et al. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type 1 Gag polyprotein. Nucleic Acids Res. 25:2902–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Massiah M. A., et al. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198–223 [DOI] [PubMed] [Google Scholar]
- 33. McCaldon P., Argos P. 1988. Oligopeptide biases in protein sequences and their use in predicting protein coding regions in nucleotide sequences. Proteins 4:99–122 [DOI] [PubMed] [Google Scholar]
- 34. McDonnell J. M., et al. 1998. Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus. J. Mol. Biol. 279:921–928 [DOI] [PubMed] [Google Scholar]
- 35. Mortuza G. B., et al. 2004. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431:481–485 [DOI] [PubMed] [Google Scholar]
- 36. Muriaux D., et al. 2004. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 78:12378–12385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muriaux D., Mirro J., Harvin D., Rein A. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U. S. A. 98:5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Newman J. L., Butcher E. W., Patel D. T., Mikhaylenko Y., Summers M. F. 2004. Flexibility in the P2 domain of the HIV-1 Gag polyprotein. Protein Sci. 13:2101–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ott D. E., et al. 2003. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J. Virol. 77:5547–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ott D. E., Coren L. V., Gagliardi T. D. 2005. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 79:13839–13847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Petoukhov M. V., Konarev P. V., Kikhney A. G., Svergun D. I. 2007. ATSAS 2.1: towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 40:s223–s228 [Google Scholar]
- 42. Pettersen E. F., et al. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 43. Pollack L., et al. 1999. Compactness of the denatured state of a fast-folding protein measured by submillisecond small-angle x-ray scattering. Proc. Natl. Acad. Sci. 96:10115–10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Purohit P., Dupont S., Stevenson M., Green M. R. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7:576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Riffel N., et al. 2002. Atomic resolution structure of Moloney murine leukemia virus matrix protein and its relationship to other retroviral matrix proteins. Structure (Cambridge) 10:1627–1636 [DOI] [PubMed] [Google Scholar]
- 46. Schuck P. 2003. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320:104–124 [DOI] [PubMed] [Google Scholar]
- 47. Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78:1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Siegel L. M., Monty K. J. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation: application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta 112:346–362 [DOI] [PubMed] [Google Scholar]
- 49. Stapley B. J., Creamer T. P. 1999. A survey of left-handed polyproline II helices. Protein Sci. 8:587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Svergun D. I. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25:492–503 [Google Scholar]
- 51. Svergun D. I. 1999. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76:2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Swanstrom R., Wills J. W. 1997. Synthesis, assembly, and processing of viral proteins, p. 263–334 In Coffin J. M., Hughes S. H., Varmus H. E. (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY: [PubMed] [Google Scholar]
- 53. Tang C., Ndassa Y., Summers M. F. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537–543 [DOI] [PubMed] [Google Scholar]
- 54. von Schwedler U. K., Stray K. M., Garrus J. E., Sundquist W. I. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Worthylake D. K., Wang H., Yoo S., Sundquist W. I., Hill C. P. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 55(Pt. 1):85–92 [DOI] [PubMed] [Google Scholar]
- 56. Wriggers W., Chacon P. 2001. Using Situs for the registration of protein structures with low-resolution bead models from X-ray solution scattering. J. Appl. Crystallogr. 34:773–776 [Google Scholar]
- 57. Wright E. R., et al. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26:2218–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yeager M., Wilson-Kubalek E. M., Weiner S. G., Brown P. O., Rein A. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. U. S. A. 95:7299–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yuan B., Li X., Goff S. P. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]