Abstract
Bovine herpesvirus 1 (BHV-1) infection induces clinical symptoms in the upper respiratory tract, inhibits immune responses, and can result in life-threatening secondary bacterial infections. Following acute infection, BHV-1 establishes latency in sensory neurons within trigeminal ganglia. Periodically, reactivation from latency occurs, resulting in virus transmission. The latency-related (LR) RNA is abundantly expressed in latently infected sensory neurons, suggesting that LR gene products regulate the latency-reactivation cycle. An LR mutant virus with stop codons at the amino terminus of the first open reading frame (ORF) in the LR gene (ORF2) does not reactivate from latency, in part because it induces higher levels of apoptosis in infected neurons. ORF2 inhibits apoptosis in transiently transfected cells, suggesting that it plays an important role in the latency-reactivation cycle. ORF2 also interacts with Notch1 or Notch3 and consequently inhibits their ability to trans-activate the bICP0 early and glycoprotein C promoters. In this study, we identified ORF2 sequences that were necessary for inhibiting cold shock-induced apoptosis or Notch1-mediated trans-activation of the bICP0 early promoter and stimulation of productive infection. Relative to ORF2 sequences necessary for inhibiting apoptosis, distinct domains in ORF2 were important for interfering with Notch1-mediated trans-activation. Five consensus protein kinase A and/or protein kinase C phosphorylation sites within ORF2 regulate the steady-state levels of ORF2 in transfected cells. A nuclear localization signal in ORF2 was necessary for inhibiting Notch1-mediated trans-activation but not apoptosis. In summary, ORF2 has more than one functional domain that regulates its stability and functional properties.
INTRODUCTION
Bovine herpesvirus 1 (BHV-1) is a significant viral pathogen of cattle (72) that suppresses immune responses. Infection can result in life-threatening pneumonia due to secondary bacterial infections (reviewed in references 40, 45, and 46). Increased susceptibility to secondary infection correlates with depressed cell-mediated immunity after BHV-1 infection (6, 16–18). CD8+ T cell recognition of infected cells is impaired by repressing the expression of major histocompatibility complex class I and the transporter associated with antigen presentation (20, 24, 57). CD4+ T cell function is impaired during acute infection of calves, because BHV-1 infects CD4+ T cells and induces apoptosis (73). Three viral genes (UL49.5, bICP0, and gG) can inhibit specific immune responses in the absence of other viral genes (reviewed in references 41 and 46). The ability of bICP0 to inhibit interferon (IFN)-dependent transcription is crucial for pathogenesis, because BHV-1 does not grow in mice unless they lack IFN receptors (1). BHV-1, due to its immune-suppressive properties, is a significant risk factor for a multifactorial disease commonly referred to as bovine respiratory disease complex.
Like other Alphaherpesvirinae subfamily members, the primary site for BHV-1 latency is sensory neurons within trigeminal ganglia (TG). Lytic cycle viral gene expression (67) and infectious virus (30) are detected in TG, but latency is subsequently established. Increased corticosteroid levels, as a result of stress, can initiate BHV-1 reactivation from latency (45). Administration of the synthetic corticosteroid dexamethasone to calves or rabbits latently infected with BHV-1 reproducibly leads to reactivation from latency, virus shedding, and a secondary antibody response (30, 38, 39, 43, 44, 63). Induction of lytic cycle viral gene expression is consistently detected in TG neurons of calves latently infected with BHV-1 following dexamethasone treatment.
The BHV-1 latency-related (LR) gene expresses an abundant transcript (LR-RNA) in latently infected sensory neurons (64, 65). The LR gene contains 2 major open reading frames (ORFs), ORF2 and ORF1, and two reading frames that lack an initiating methionine, RF-C and RF-B (49). The LR gene is also antisense with respect to an important viral transcriptional regulator, bICP0, suggesting that LR-RNA reduces bICP0 levels. Small noncoding RNAs, including two microRNAs, encoded within the LR gene inhibit bICP0 protein expression in transient transfection assays (32). A mutant BHV-1 strain with 3 stop codons at the N terminus of ORF-2 (LR mutant virus) does not express detectable levels of ORF-2 or RF-C (34) but expresses reduced levels of ORF1 in cultured cells during productive infection (53). The LR mutant virus grows less efficiently in the ocular cavity or trigeminal ganglia and does not reactivate from latency following dexamethasone treatment (30, 31), suggesting that the expression of LR proteins is required for the latency-reactivation cycle in cattle. During the establishment of latency, the LR mutant virus induces higher levels of apoptosis in TG neurons of infected calves (52), and plasmids with the same stop codon mutations exhibit little or no antiapoptosis activity (7, 22).
ORF2 coding sequences, in the absence of other viral genes, inhibit apoptosis in transiently transfected cells (70), suggesting that ORF2 is a dominant function encoded by the LR gene. A yeast two-hybrid screen revealed that ORF2 interacts with Notch1 and Notch3, components of the Notch signaling pathway (74). The Notch family of cellular transcription factors regulates the differentiation and development of nearly all mammalian cell types (reviewed in references 4, 8, 11, and 47). Notch1, but not Notch3, enhances BHV-1 productive infection. Only Notch1 activates the BHV-1 immediate early transcription unit 1 (IEtu1) and bICP0 early promoters, whereas Notch1 and Notch3 both trans-activated the late BHV-1 glycoprotein C (gC) promoter. ORF2 interferes with the ability of Notch1 to trans-activate the bICP0 early promoter and Notch1 or Notch3-mediated activation of the gC promoter (74), suggesting that this function is important for establishing and/or maintaining latency.
In the current study, a panel of ORF2 mutants was analyzed for their ability to inhibit apoptosis and interfere with the trans-activation potential of Notch1. The ORF2 domains necessary for interfering with Notch1-mediated trans-activation were distinct from those important for antiapoptosis. Localization of ORF2 to the nucleus was necessary for inhibiting Notch1-mediated trans-activation of the bICP0 E promoter but not for inhibiting apoptosis. Mutating putative protein kinase A (PKA) and/or PKC phosphorylation sites stabilized ORF2 steady-state levels but had no obvious effect on inhibiting apoptosis or interfering with Notch1-mediated trans-activation of the bICP0 early promoter. In summary, distinct domains within ORF2 were necessary for inhibiting apoptosis and interfering with Notch1-mediated trans-activation of the bICP0 early promoter.
MATERIALS AND METHODS
Cells and viruses.
Murine neuroblastoma (Neuro-2A) and rabbit skin (RS) cells were grown in Earle's modified Eagle's medium (EMEM) supplemented with 5% fetal calf serum (FCS), penicillin (10 U/ml), and streptomycin (100 μg/ml).
A BHV-1 mutant containing the LacZ gene in place of the gC gene was obtained from S. Chowdury (Baton Rouge, LA) (gCblue virus) and has been previously described (13, 14, 76). Stock cultures of gCblue were prepared in established bovine kidney cells (CRIB). The virus grows to titers similar to those of the wild-type (wt) virus and expresses the LacZ gene as a late gene.
Plasmids and transient transfections.
The mammalian ORF2 construct was described previously (70). Briefly, sequences derived from cDNA at 1 day postinfection (dpi) were cloned into pCMV-Tag-2B (Stratagene, La Jolla, CA) downstream of a Flag epitope using BamHI-HindIII restriction enzymes. Sequences derived from ORF2 cDNA at 1 dpi with a nuclear localization signal (NLS) deletion (amino acids [aa] 64 to 70) (ORF2-NLS), with PKA/PKC (ORF2-P) or all (ORF2-AP) putative phosphorylation sites mutated, were synthesized by Integrated DNA Technology (IDT; Coralville, IA) and cloned into the pCMV-Tag-2B plasmid by using BamHI-HindIII restriction enzymes. A construct containing the Notch1 intracellular domain (ICD) was cloned into a human cytomegalovirus expression construct and has been previously described (74). The construction and characteristics of the bICP0 early promoter-chloramphenicol acetyltransferase (CAT) construct (EP-172) used in this study were described previously (74, 76). The number in the plasmid name refers to the length of the bICP0 E promoter fragment cloned into the promoterless vector pCAT-basic. All plasmids were transfected into Neuro-2A cells in 60-mm dishes by using TransIT Neural (MIR2145; Mirus) according to the manufacturer's instructions.
Generation of ORF2 mutants by EZ::TN in-frame linker insertion.
In-frame linker insertion mutants were prepared according to the manufacturer's instructions (Epicentre catalog number [Cat #] EZ104KN). ORF2 cDNA was released from pCMV2B-ORF2 by using BamHI-SalI endonucleases and cloned into pUC57. This plasmid was used as a target DNA in the reaction. The transposon reaction was set up as follows: 1 μl of EZ-Tn5 10× reaction buffer, 200 ng of target DNA, 1 μl of EZ-Tn5 (NotI/KAN-3) transposon, 6 μl of distilled water (dH2O), 1 μl of transposase. The reaction mixture was incubated at 37°C for 2 h. To stop the reaction, 1 μl of EZ-Tn5 10× stop solution was added, and the reaction mixture was heated at 70°C for 10 min. Top 10 DH5α electrocompetent cells (Invitrogen) were transformed with 2 μl of the reaction mixture and a 1:10 dilution was plated unto plates containing 50 μg/ml kanamycin. DNA was extracted from random colonies, and initial mapping was performed by digestion with the restriction endonucleases BamHI and HindIII. Certain clones were then sequenced to determine the precise transposon insertion. A panel of these mutants was digested with NotI to remove the kanamycin gene, religated, and then digested with BamHI and HindIII and recloned into the pCMV2B Flag-tagged vector. The Flag-tagged mutant constructs were confirmed by sequencing.
Immunofluorescence.
Neuro-2A cells cultured for 48 h after transfection were washed twice with warm EMEM without serum and fixed in 4% paraformaldehyde for 10 min at 37°C. Cells were permeabilized with cold 100% ethanol at −20°C for 5 min. After washing three times with 1× phosphate-buffered saline (PBS), slides were blocked with 3% bovine serum albumin (BSA) in PBS for 2 h and then incubated with the mouse anti-Flag antibody (Sigma F1804) (1:250) (Flag-ORF2) or rabbit anti-cleaved caspase 3 (Cell Signaling 6238S) (1:250) for 2 h at room temperature. The secondary antibody, goat anti-mouse/Alexa Fluor 633 (Invitrogen/Molecular Probes A21050) (1:100) or goat anti-rabbit/Alexa Fluor 488 (Invitrogen/Molecular Probes A11008) (1:100), was added, and cells were incubated for 1 h at room temperature in the dark. DAPI (4′,6-diamidino-2-phenylindole) (Thermo Scientific 46190) (1:1,000) was used to stain nuclear DNA. An Olympus IX 81 inverted confocal laser-scanning microscope was used to collect images (excitation/emission at 488/520 nm and 633/660 nm). For apoptosis assays, cells were incubated in EMEM with 2% FCS for 12 h. These cells were then incubated on ice (cold shock) for 4 h and subsequently processed for immunofluorescence.
Laddering assays.
Cold shock-induced apoptosis of Neuro-2A cells was performed as previously described (69, 70) with a few modifications. Briefly, 24 h after transfection, Neuro-2A cells in T25 flasks were incubated in fresh EMEM with 2% serum for 12 h. Cells were then cold shocked on ice for 1 h and allowed to recover at 37°C for 3 h. Cells were subsequently collected, and analysis of fragmented DNA was performed as described previously (69, 70).
β-Gal assay.
For apoptosis protection assays, Neuro-2A cells were grown in 60-mm plates and were cotransfected with 2 μg of pCMV-β-galactosidase (β-Gal) plasmid and 2 μg of pCMV-Tag plasmid expressing Flag-tagged ORF2 or ORF2 mutants using TransIT Neural (MIR2145; Mirus). Twenty-four hours after transfection, cells were cold shocked on ice for 2 h. Cells were then fixed and stained and the number of β-Gal+ cells counted as described previously (76). The number of β-Gal+ cells in cultures expressing the blank vector was set to 100%. The number of blue cells in cultures transfected with ORF2 or ORF2 mutants was divided by the number of β-Gal+ cells in cultures transfected with the blank vector to calculate the fold difference. The results are an average of at least three independent experiments.
For BHV-1 productive infection assays, the BHV-1 gCblue virus was used. gCblue and procedures for preparing BHV-1 genomic DNA have been previously described (76). RS cells grown in 6-well plates were cotransfected with 0.83 μg of the gCblue viral genome, 1 μg Notch1 ICD, and 1 μg ORF2 or ORF2 mutants by using Lipofectamine 2000 (11668-019; Invitrogen). Twenty-four hours after transfection, cells were fixed and stained, and the number of β-Gal+ cells were counted. The number of blue cells in cultures transfected with a blank vector was used to calculate the fold difference in cultures transfected with Notch1 and ORF2 or ORF2 mutants. The results are an average of three independent experiments.
Western blot detection of ORF2 in transfected cells.
Neuro-2A cells in 60-mm dishes were transfected with the designated plasmids, and 48 h after transfection cells were collected, washed once with 1× PBS, and suspended in hypotonic buffer (10 mM Tris [pH 7.5], 10 mM KCl, 0.5 mM EGTA, 1.5 mM MgCl2, and 0.5% Triton X-100). Cells were vortexed and then centrifuged at 5,000 × g for 2 min, and the supernatant was collected. The pellet was suspended in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1% Triton X-100) and incubated at 4°C for 30 min with rotation. Lysate was centrifuged at 14,000 rpm for 10 min and the supernatant was collected. Proteins were boiled in Laemmli sample buffer for 5 min and separated on a 12% SDS-polyacrylamide gel. Immune detection of ORF2 and its mutants was performed using a mouse anti-Flag antibody (F1804; Sigma) (1:500). A polyclonal goat anti-actin (SC-1616; Santa Cruz) antibody was used to confirm that equal protein amounts were loaded. A goat anti-histone 3 (SC-8654; Santa Cruz) was used as a control for detecting nuclear proteins in the respective cellular fractions.
CAT reporter assays.
Neuro-2A cells grown in 60-mm dishes were cotransfected with the designated plasmids. At 48 h after transfection, cell lysate was prepared by three freeze-thaw cycles in 0.25 M Tris-HCl, pH 7.4. Cell debris was pelleted by centrifugation, and protein concentration was determined by the Bradford assay. CAT activity was measured in the presence of 0.1 μCi 14C-chloramphenicol (CFA754; Amersham Biosciences) and 0.5 mM acetyl coenzyme A (acetyl-CoA) (A2181; Sigma). The reaction mixture was incubated at 37°C for 1 h. All forms of chloramphenicol were separated by thin-layer chromatography. CAT activity in 50-μg cell lysate was quantified using a Bio-Rad Molecular Imager FX (Molecular Dynamics, CA). Levels of CAT activity are expressed as fold induction relative to the vector control.
Statistical analysis of data.
The standard Student t test was used to identify statistical difference. A P value of <0.05 was considered to be significant.
RESULTS
Construction of plasmids that express ORF2 mutant proteins.
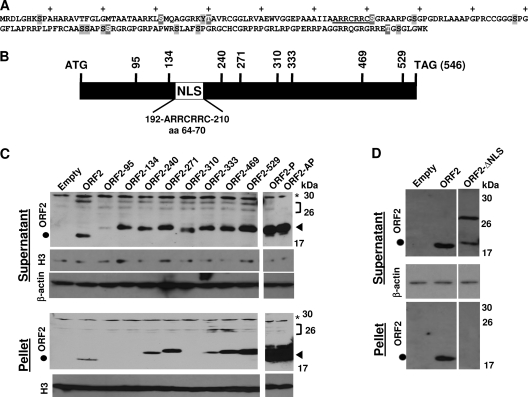
ORF2 is a 181-aa protein (Fig. 1A and B) that is expressed in a subset of trigeminal ganglionic sensory neurons of latently infected calves (26, 33). ORF2 has antiapoptosis activity (70), and a recent study demonstrated that ORF2 interacts with Notch1 and Notch3, which results in interfering with trans-activation of certain viral promoters (74). Furthermore, an alternatively spliced LR transcript encodes a protein containing part of ORF2 fused with ORF1, and this protein interacts with C/EBP-alpha (54, 74). Since ORF2 appears to have at least two distinct functions, we hypothesized that ORF2 has more than one functional domain. A BLAST analysis of ORF2 did not reveal extensive similarity to known proteins and thus did not provide insight into the mechanism by which ORF2 inhibits apoptosis or regulates transcription. To identify functional domains within ORF2, a panel of transposon insertion mutants was constructed in which a 57-nucleotide sequence was randomly inserted into the template (Fig. 1B shows the insertion sites of the respective mutants). The EZ::TN linker insertion method was previously used to successfully generate a panel of bICP0 mutants, which is also a high-GC-content template (78). This approach was used because ORF2 deletion mutants that were constructed did not encode a stable protein, whereas the transposon mutagenesis yielded a panel of mutants that spanned the protein.
Fig. 1.
Generation of BHV-1 ORF2 mutants. (A) Amino acid sequence of ORF2. NLS (underlined), 15 putative phosphorylation sites (gray shaded amino acids), and 5 consensus protein kinase A (PKA) and/or PKC phosphorylation sites (gray shaded amino acids that have white lettering) are shown. The plus signs denote every 10th amino acid in ORF2. (B) ORF2 coding sequences (BamHI-SalI) were cloned into the pUC57 vector, and transposon linker insertion reactions were performed as described in Materials and Methods. Initial mapping was performed by restriction digestion, and the precise location of the transposon insertion was confirmed by sequencing. Vertical lines with numbers indicate nucleotide positions of transposon insertions. The relative position of the consensus nuclear localization signal (NLS) is denoted by the white rectangle. (C) The transposon mutants and the two phosphorylation mutants were cloned into the pCMV-Tag-2B vector and transfected into Neuro-2A cells. At 48 h after transfection, cells were collected and lysed using hypotonic buffer as described in Materials and Methods. After centrifugation, the supernatant was removed. The nuclei and other cellular debris were suspended in RIPA buffer, and the solubilized proteins were collected after centrifuging the residual debris; this solubilized fraction was designated the pellet. Detection of Flag-tagged ORF2 mutants was performed using an Anti-Flag antibody. A β-actin antibody was used to confirm that equal amounts of protein were loaded in each lane. A histone 3 antibody was used to identify nuclear proteins in the supernatant or pellet fraction. ORF2 is predicted to migrate as a 19-kDa protein, and the black circle denotes the position of this protein. The arrow denotes the higher-molecular-weight ORF2-specific bands that migrated at approximately 30 kDa, which were detected in certain samples. The transposon mutants are predicted to migrate as a 22-kDa protein and are denoted by a closed triangle. The location of a nonspecific band in D is denoted by the asterisk. The bracket denotes the position of the ORF2-specific bands that migrated slower than expected. For each lane, 100 μg protein was loaded. (D) Neuro-2A cells were transfected with the designated plasmids and the respective samples collected at 48 h after transfection as described above. These results are representative of more than 3 independent experiments.
Each transposon mutant was cloned into a Flag-tagged cytomegalovirus (CMV) expression plasmid and transfected into Neuro-2A cells. At 48 h after transfection, proteins were extracted using hypotonic buffer. Nuclei as well as other debris were separated from the supernatant by centrifugation, and the pellet was then solubilized with RIPA buffer. Western blots were performed using an anti-Flag antibody to assess expression levels of the respective transposon mutants. ORF2 is predicted to encode a 19-kDa protein, and a prominent band migrating near the predicted molecular weight was detected (Fig. 1C, closed circle). The ORF2 transposon mutants migrated at an approximate molecular mass of 23 kDa (closed triangle) because of the additional 19 amino acids in the transposon insertion (Fig. 1C). The higher-molecular-weight bands detected by the Flag antibody (denoted by the bracket) were specific for cells transfected with wt ORF2 or the respective transposon mutants of ORF2, and were not detected in cells transfected with the empty vector. We suggest that these higher-molecular-weight bands were posttranslationally modified ORF2. β-Actin levels in the respective lanes were similar, indicating that equivalent amounts of protein were loaded. Except for ORF2-95, the other transposon mutants expressed similar levels of protein as wt ORF2 (Fig. 1C). In the pellet of cells transfected with ORF2, ORF2-240, ORF2-271, ORF2-333, ORF2-469, or ORF2-529, an ORF2-specific band was readily detected, suggesting that ORF2 and certain mutants were tightly associated with nuclear structures. This result was consistent with a previous study demonstrating that ORF2 is localized to distinct regions of the nucleus (74). Low levels of histone 3 were detected in the supernatant, indicating that this fraction contained low levels of nuclear proteins as well as cytoplasmic proteins. High levels of histone 3 were present in the pellet, as expected, confirming that tightly bound nuclear proteins were associated with the pellet. Other extraction buffers yielded variable results (data not shown), further suggesting that ORF2 was associated with cellular structures and that the extraction of ORF2 was dependent on the extraction buffers used.
Within ORF2, there are 15 potential phosphorylation sites (Fig. 1A, gray-shaded amino acids), which may explain why ORF2-specific bands migrating between 26 and 30 kDa were detected in transfected cells. Among the 15 putative phosphorylation sites, 5 consensus protein kinase A (PKA) and/or PKC phosphorylation sites were present (Fig. 1A, gray-shaded amino acids with white lettering). A mutant (ORF2-AP) containing all 15 putative phosphorylation sites and a mutant with the 5 PKA/PKC phosphorylation sites (ORF2-P) was synthesized in which the serine, threonine, or tyrosine residues in the wt sequence were replaced with alanine. Mutating the putative phosphorylation sites to alanine increased the stability of ORF2-AP (Fig. 1C, lane AP) or ORF2-P (lane P) relative to wt ORF2. Furthermore, higher levels of the ORF2 phosphorylation mutants were detected in the pellet. The phosphorylation mutants also had little or no higher-molecular-weight bands, adding support to the prediction that ORF2 may be phosphorylated by a cellular protein kinase.
A consensus nuclear localization signal (NLS) (aRRcRRc) is located between amino acids 64 and 70 of ORF2 (10) (Fig. 1A, underlined amino acids, and B). To test whether this NLS was required for nuclear localization, a mutant (ORF2-ΔNLS) containing a deletion of aa 64 to 70 was synthesized, cloned into the pCMV-FLAG vector, and transfected in Neuro-2A cells. The ORF2-ΔNLS protein levels were similar to those for wt ORF2, but ORF2-ΔNLS migrated primarily as a 29-kDa protein (Fig. 1D). Unlike wt ORF2, ORF2-ΔNLS was not detected in the nuclear pellet, suggesting that this protein was not present in the nucleus.
Subcellular localization of ORF2 mutants.
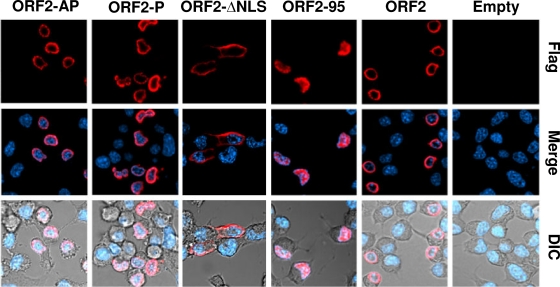
ORF2 localized to the nuclear periphery in transfected Neuro-2A cells (Fig. 2), which was consistent with a previous study (74). The ORF2-95 mutant (Fig. 2), which was expressed at lower levels in transfected cells (Fig. 1C), was more diffusely localized throughout the nucleus relative to cells expressing the wt ORF2. The other transposon mutants had a localization similar to that of wt ORF-2 (data not shown), which was somewhat surprising because certain transposon mutants appeared to be tightly associated with structures in the nucleus (Fig. 1C).
Fig. 2.
Localization of ORF2 mutants in Neuro-2A cells. Neuro-2A cells were transfected with 4 μg of the designated plasmids that express a Flag-tagged ORF2 or ORF2 mutants. Cultures were prepared for confocal microscopy at 48 h after transfection as described in Materials and Methods. ORF2+ cells were identified using the anti-Flag antibody (red), or DAPI was used to visualize the nucleus (blue). Differential interference contrast (DIC) was used to show the unstained cells. The images are representative of more than 3 experiments.
The ORF2-ΔNLS mutant was localized to the peripheral area of the cytoplasm, confirming the results in Fig. 1D, which indicated that deletion of ORF2 amino acids 64 to 70 disrupted nuclear localization. The phosphorylation mutants, ORF2-AP and ORF2-P, appeared to have a nuclear localization similar to that of wt ORF2, even though they were more difficult to extract from transfected cells. During productive infection, ORF2 is diffusely localized to the nucleus and cytoplasm (reference 53 and data not shown), suggesting that a virally encoded or induced factor regulates ORF2 localization. In summary, we have constructed and characterized a panel of ORF2 mutants that can be used to analyze the known functions of ORF2.
Analysis of antiapoptosis activity of ORF2 mutants.
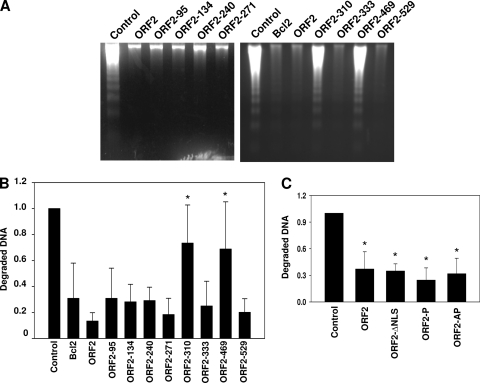
Transient transfection assays were performed to compare the antiapoptosis functions of ORF2 to the respective mutants described in Fig. 1. For these studies, plasmids expressing the respective ORF2 mutants were transfected into Neuro-2A cells, and cold shock-induced apoptosis was performed as previously described (5, 69, 70). At 24 h after transfection, cells were starved in 2% fetal calf serum for 12 h, and cultures were incubated on ice for 1 h and then returned to 37°C for 3 h. DNA was then extracted and analyzed by agarose electrophoresis. Neuro-2A cells or Neuro-2A cells placed on ice for 1 h contain no detectable DNA laddering (69, 70). However, extensive DNA laddering, indicative of apoptosis, occurs when Neuro-2A cells are returned to 37°C for 3 or 6 h (69, 70). ORF2 and Bcl-2, an inhibitor of the intrinsic apoptotic pathway (77), inhibited cold shock-induced apoptosis with similar efficiencies (Fig. 3A and B). The ORF2-310 and ORF2-469 mutants were unable to inhibit cold shock-induced apoptosis compared to wt ORF2 (Fig. 3A and B). The antiapoptosis activity of wt ORF2 was significantly different than that of ORF2-310 or ORF2-469 (P < 0.008 or 0.01, respectively). The ability of the other transposon insertion mutants to inhibit cold shock-induced apoptosis in Neuro-2A cells was not significantly different from that of wt ORF2 (P > 0.05). Although the ORF2-95 mutant was expressed at low levels relative to wt ORF2, it still retained antiapoptosis activity, suggesting that low levels of ORF2 were sufficient for inhibiting apoptosis.
Fig. 3.
Inhibition of cold shock-induced DNA laddering by ORF2 mutants. (A) Neuro-2A cells were transfected with 4 μg of the plasmid expressing N-terminally Flag-tagged ORF2 or the designated ORF2 transposon mutants. Cells were cold shocked at 4°C for 1 h and allowed to recover at 37°C for 3 h. Neuro-2A cells transfected with the empty vector were used as a negative control, while Bcl-2- or ORF2-expressing cells were used as positive controls. The agarose gel images are representative of 5 independent experiments. (B) The relative amounts of fragmented DNA in each lane in A were quantified using a Bio-Rad Molecular Imager FX. The average of 5 independent experiments is shown with standard deviation. (C) Neuro-2A cells transfected with the empty vector, ORF2, ORF2-ΔNLS, ORF2-P, or ORF2-AP were cold shocked as in A, and fragmented DNA was quantified as described in B. An asterisk denotes significant differences (P < 0.05) from the wt samples (panel B) or (empty vector) (panel C) as determined by the Student t test.
Neuro-2A cells expressing ORF2-ΔNLS (Fig. 3C) were protected against cold shock-induced apoptosis with an efficiency similar to that of wt ORF2, which was surprising because ORF2-ΔNLS was localized to the cytoplasm. Both phosphorylation mutants (ORF2-P and ORF2-AP) inhibited cold shock-induced apoptosis relative to the control (Fig. 3C, P < 0.05).
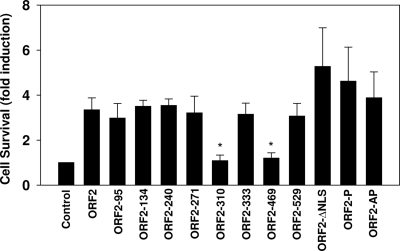
To confirm the results in Fig. 3, we used a β-Gal cotransfection assay (27, 48) that has been modified to measure the effects of various genes on apoptosis (7, 23, 28, 37, 60). When a known apoptosis stimulator is used, the number of surviving cells can be accurately measured using the β-Gal cotransfection assay. For these studies, apoptosis was induced using cold shock treatment of Neuro-2A cells. Neuro-2A cells transfected with a plasmid that expresses wt ORF2 exhibited at least a 3-fold increase in cell survival after cold shock-induced apoptosis, as judged by an increase in the number of β-Gal+ cells (Fig. 4). As expected, plasmids expressing ORF2-310 and ORF2-469 did not enhance survival. Consistent with the studies performed in Fig. 3, the other ORF2 transposon insertion mutants, ORF2-ΔNLS, and the two phosphorylation mutants (ORF2-AP and ORF2-P) inhibited apoptosis with efficiencies similar to that of wt ORF2. Cell survival was consistently high in cells expressing ORF2-ΔNLS or the phosphorylation mutants (ORF2-AP and ORF2-P) compared to wt ORF2: however, the difference was not statistically significant (Fig. 4).
Fig. 4.
Survival of Neuro-2A cells after transfection with ORF2 mutants. Neuro-2A cells were cotransfected with 2 μg of the pCMV-β-Gal plasmid and 2 μg of the designated ORF2 expression plasmids. Cells were cold shocked for 2 h, and the β-Gal assay was performed as described in Materials and Methods. The number of β-Gal+ cells in cultures expressing the blank vector was set to 100%. The number of blue cells in cultures transfected with the blank vector was divided by the number of blue cells in cultures transfected with ORF2 or ORF2 mutants to calculate the fold difference. The results are an average of three independent experiments. An asterisk denotes significant differences (P < 0.05) from the wt ORF2 results as determined by the Student t test.
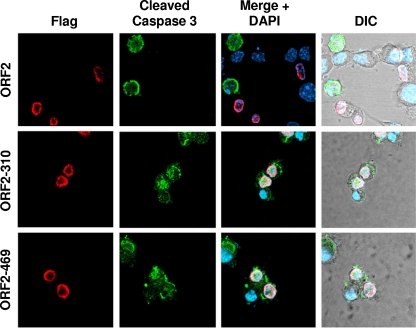
Confocal microscopy was performed to confirm that cells expressing ORF2-310 or ORF2-469 were not protected against apoptosis. For these studies, we tested whether cleaved caspase 3 was present in cells expressing either wt ORF2 or the ORF2-310 and ORF2-469 mutants. Cleavage of caspase 3 is considered to be the “point of no return” during intrinsic and extrinsic induced pathways of apoptosis and thus is an established marker for apoptosis induction (15, 58, 68). Neuro-2A cells transfected with the respective plasmids were cold shocked, incubated at 37°C, processed for immunofluorescence, and then visualized with a confocal microscope. Cleaved caspase 3 was not detected in ORF2-positive cells. However, cleaved caspase 3 was readily detected in cells not expressing ORF2, which was expected because cells that were not transfected would undergo cold shock-induced apoptosis. In contrast, cells expressing ORF2-310 or ORF2-469 were consistently positive for cleaved caspase 3, confirming that these mutants do not efficiently inhibit apoptosis (Fig. 5).
Fig. 5.
Induction of caspase 3 cleavage by ORF2 transposon mutants. Neuro-2A cells were transfected for 24 h and incubated in EMEM with 2% FCS for 12 h, and then cold shock-induced apoptosis was performed as described in Materials and Methods. Cells were then processed for immunofluorescence as described in Materials and Methods. Mouse anti-Flag antibody (red) was used to detect Flag-tagged ORF2 and the designated transposon mutants. Rabbit anti-cleaved caspase 3 antibody (green) was used to detect activated cleaved caspase 3. DAPI (blue) was used to stain the nucleus.
Identification of ORF2 mutants that interfere with trans-activation of the bICP0 E-promoter by Notch1.
As discussed above, ORF2 interacts with the cellular transcription factor Notch1, a component of the Notch signaling pathway that controls the differentiation and development of all tissues (74). Notch1 trans-activates the bICP0 early promoter, and ORF2 reduces the trans-activation potential of Notch1.
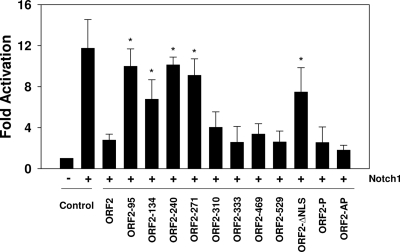
To identify domains in ORF2 that interfered with Notch1-mediated trans-activation of the bICP0 early promoter, transient transfection assays were performed with Neuro-2A cells. Notch1 overexpression activated the bICP0 early promoter approximately 12-fold, and ORF2 reduced Notch1-mediated trans-activation to 3-fold (Fig. 6) (74). Four transposon mutants, ORF2-95, ORF2-134, ORF2-240, and ORF2-271, did not significantly reduce the ability of Notch1 to stimulate bICP0 early promoter activity compared to wt ORF2. With respect to ORF2-95, we cannot distinguish whether this was due to lower protein expression levels or if an essential inhibitory domain was disrupted. The remainder of the transposon mutants inhibited trans-activation of the bICP0 early promoter by Notch1 with an efficiency similar to that of wt ORF2. The phosphorylation mutants, ORF2-P and ORF2-AP, but not ORF2-ΔNLS, also inhibited Notch1-mediated trans-activation of the bICP0 early promoter with an efficiency similar to that of wt ORF2 (P > 0.05).
Fig. 6.
Identification of ORF2 sequences that interfere with Notch1-mediated trans-activation of the bICP0 early promoter. Neuro-2A cells were cotransfected with the bICP0 E promoter construct (EP-172), a CMV promoter plasmid expressing the Notch1 intracellular domain, and the designated ORF2-expressing constructs. At 48 h posttransfection, cells were collected and processed for CAT activity as described in Materials and Methods. The CAT activity of cells transfected with the control CAT vector was set to 1. All other values are expressed as fold activation with respect to the control. These studies are the average of at least three independent experiments. An asterisk denotes significant differences (P < 0.05) from the wt ORF2 samples as determined by the Student t test.
Identification of ORF2 mutants that interfere with productive infection induced by Notch1.
Additional studies were performed to identify ORF2 mutants that interfere with the ability of Notch1 to stimulate productive infection. We hypothesized that there may be differences between wt ORF2 and the ORF2 mutants in terms of interference with Notch1 trans-activation of the bICP0 early promoter. The rationale for this prediction is summarized below. Eleven known consensus binding sites for CSL (RBP-jκ) exist (62). The Notch ICD interacts with members of the CSL family of transcriptional repressors, CBF1, Su(H), and Lag1 (also referred to as RBPjκ binding proteins) (4, 11). The BHV-1 genome contains 82 potential binding sites for CSL, of which 23 are located in noncoding regions. Thus, certain ORF2 mutants may interfere with the ability of Notch1 to trans-activate the bICP0 promoter but may not interfere with productive infection.
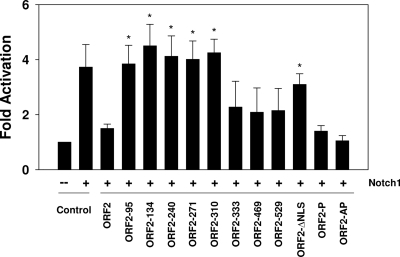
To determine whether ORF2 interfered with the ability of Notch1 to stimulate BHV-1 productive infection, rabbit skin (RS) cells were cotransfected with a plasmid expressing the Notch1 ICD or the Notch1 ICD and ORF2 plus the BHV-1 gCBlue virus genome, and the efficiency of productive infection was measured. The gCblue virus contains the LacZ gene downstream of the gC promoter, which allows measurement of the efficiency of productive infection by counting β-Gal+ cells. The number of β-Gal+ cells directly correlates with plaque formation (13, 14, 54, 75, 76). RS cells were used for these studies because they are permissive for BHV-1 and can be readily transfected. The Notch1 ICD expression plasmid consistently stimulated productive infection approximately 4-fold (Fig. 7); however, ORF2 reduced the number of β-Gal+ cells to background levels. The effects of the respective ORF2 mutants on Notch1-mediated activation of productive infection were then analyzed. Five transposon mutants (ORF2-95, ORF2-134, ORF2-240, ORF2-271, and ORF2-310) and the ORF2-ΔNLS mutant were unable to significantly interfere with the ability of Notch1 to stimulate productive infection (P < 0.05). Interestingly, ORF2-310 did not interfere with Notch1-mediated trans-activation of the bICP0 early promoter, suggesting that the ability of Notch1 to stimulate productive infection was not entirely dependent on interfering with bICP0 early promoter activity. ORF2-P and ORF-2AP, but not ORF2-ΔNLS, reduced productive infection as efficiently as wt ORF2. ORF2-333, ORF2-469, and ORF2-529, in general, had an intermediate effect on the ability of Notch1 to stimulate productive infection, which was not significantly different from that of wt ORF2 (P > 0.05). In general, the results in Fig. 6 and 7 suggested that the amino-terminal sequences in ORF2 were important for inhibiting Notch1-mediated trans-activation of the bICP0 early promoter activity and productive infection.
Fig. 7.
Identification of ORF2 sequences that interfere with Notch1-mediated trans-activation of productive infection. Neuro-2A cells were cotransfected with the gCblue virus, a CMV promoter plasmid expressing the intracellular domain of Notch1 and the designated ORF2 constructs. At 48 h after transfection, cells were fixed and a β-Gal assay was performed as described in Materials and Methods. The number of β-Gal+ cells in cultures expressing the empty vector was set to 1. The number of blue cells in cultures transfected with the empty vector was divided by the number of blue cells in cultures transfected with Notch1 or cotransfected with Notch1 and ORF2 or ORF2 mutants to calculate the fold difference. The results are an average of at least three independent experiments. An asterisk denotes significant differences (P < 0.05) from the wt ORF2 samples (empty vector) as determined by the Student t test.
DISCUSSION
LR protein expression is necessary for dexamethasone-induced reactivation from latency in cattle (30), in part because an LR mutant virus containing stop codons at the N terminus of ORF2 induces higher levels of apoptosis in sensory neurons during the establishment of latency (52). In contrast to BHV-1, other Alphaherpesvirinae subfamily members (herpes simplex virus type 1 [HSV-1] and HSV-2, for example) abundantly express several small noncoding RNAs that are proposed to mediate the latency-reactivation cycle in small-animal models (38, 39, 61). BHV-1-encoded ORF2 (7, 70) and HSV-1 LAT (2, 3, 19, 28, 60) inhibit apoptosis, which promotes the survival of infected sensory neurons. The reduced reactivation phenotype of an HSV-1 LAT-null mutant is restored to wild-type levels when an antiapoptosis gene is inserted in the LAT locus and is expressed during latency (35, 36, 56, 59), underscoring the importance of enhancing the survival of infected neurons. In addition to inhibiting apoptosis, ORF2 interacts with at least three cellular transcription factors, C/EBP-alpha, Notch1, and Notch3 (54, 74), suggesting that ORF2 regulates viral and/or cellular transcription. Thus, identifying domains in ORF2 that are important for inhibiting apoptosis relative to its other known functions may provide insight into how ORF2 regulates the latency-reactivation cycle.
A previous study identified an NLS in ORF2 that matches the NLS in the cellular transcription factor Sp1 (10). Deletion of the NLS (ORF2-ΔNLS) prevented the localization of this mutant to the nucleus. However, the ORF2-ΔNLS mutant protein was localized at or near the plasma membrane of transfected cells, suggesting that ORF2 can interact with membrane components. Surprisingly, ORF2-ΔNLS inhibited cold shock-induced apoptosis with an efficiency similar to that of ORF2, suggesting that nuclear localization of ORF2 was not required for its antiapoptosis functions or that low levels of the ORF2-ΔNLS protein were present in the nucleus. Mutating all of the putative phosphorylation sites within ORF2 or just the consensus PKA/PKC phosphorylation sites led to increased levels of ORF2 in Neuro-2A cells but had no effect on inhibiting apoptosis or Notch1 functions. At this point, it is not clear whether phosphorylation regulates an unknown function of ORF2 or if it simply destabilizes ORF2. Phosphorylation renders many proteins susceptible to degradation, as reviewed in reference 25; such susceptible proteins include the p53 tumor suppressor protein (79) and proteins that regulate cell cycle checkpoints (21). The presence of PKA phosphorylation sites in ORF2 may have biological significance, because cyclic AMP (cAMP), which activates PKA (9), stimulates HSV-1 reactivation from latency (50, 71).
ORF2-310 and ORF2-469 were unable to inhibit cold shock-induced apoptosis, but these mutants inhibited Notch1-mediated trans-activation of productive infection. These results suggested that ORF2 contains two nonoverlapping functional domains and that the amino terminus of ORF2 was important for inhibiting Notch1-mediated trans-activation of productive infection. For Notch family members to activate transcription in the canonical Notch signaling pathway, the Notch intracellular domain must interact with a CSL family member that specifically binds DNA (reviewed in references 4 and 11). At least two noncanonical Notch signaling pathways exist (66), suggesting that ORF2 interferes with one or more of these pathways. ORF2-Notch interactions may also maintain “neuronal health” during life-long latency, because activation of Notch signaling interferes with neurite formation (12, 51), which is crucial for normal neuronal functions. Conversely, ORF2 may capture Notch1 and Notch3 to regulate steps that are necessary for establishing or maintaining life-long latency in cattle. Since Notch1 and Notch3 RNA levels are increased in trigeminal ganglia after dexamethasone treatment (74), a known stimulus for reactivation from latency (74), there may be more than one reason to overcome the deleterious effects of Notch on latently infected neurons. Further studies designed to understand the effects of ORF2 on Notch signaling pathways are currently being pursued.
In summary, we predict that ORF2 promotes survival of infected neurons by at least two distinct mechanisms: (i) inhibition of apoptosis (53, 70, 75) and (ii) interference with viral transcription by sequestering cellular transcription factors such as Notch1, Notch3 (74), or C/EBP-alpha (54). C/EBP-alpha is induced during dexamethasone-induced reactivation from latency (54) and cooperates with bTIF, the HSV-1 VP16 homologue, to trans-activate the immediate early transcription unit 1 promoter (55). The ability of ORF2 or an ORF2 fusion protein to interact with cellular transcription factors that stimulate productive infection is believed to promote the establishment and/or maintenance of latency. It is unlikely that ORF2 plays a direct role in reactivation from latency, because LR promoter activity (42) and LR-RNA levels (63) are reduced dramatically during dexamethasone-induced reactivation from latency. ORF2 may not be the only important product encoded by the LR gene, because other proteins are encoded by the LR gene (29, 53) and two microRNAs encoded by the LR gene reduce bICP0 protein levels (32). The mechanism by which the additional LR gene factors support ORF2 during life-long latency is being examined.
ACKNOWLEDGMENTS
This research was supported by grants from the USDA, Agriculture and Food Research Initiative Competitive Grants Program (08-00891 and 09-01653). A grant to the Nebraska Center for Virology, in particular funds that support the microscopy core facility (1P20RR15635), have supported certain aspects of these studies. Devis Sinani was partially supported by a fellowship from Ruth L. Kirschstein National Research Service Award 1 T32 AIO60547 (National Institute of Allergy and Infectious Diseases).
Footnotes
Published ahead of print on 21 September 2011.
REFERENCES
- 1. Abril C., et al. 2004. Both viral and host factors contribute to neurovirulence of bovine herpesvirus 1 and 5 in interferon receptor-deficient mice. J. Virol. 78:3644–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmed M., Lock M., Miller C. G., Fraser N. W. 2002. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J. Virol. 76:717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Branco F. J., Fraser N. W. 2005. Herpes simplex virus type 1 latency-associated transcript expression protects trigeminal ganglion neurons from apoptosis. J. Virol. 79:9019–9025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bray S. J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678–689 [DOI] [PubMed] [Google Scholar]
- 5. Carpenter D., et al. 2007. Stable cell lines expressing high levels of the herpes simplex virus type 1 LAT are refractory to caspase 3 activation and DNA laddering following cold shock induced apoptosis. Virology 369:12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carter J. J., et al. 1989. Inhibition of T-lymphocyte mitogenic responses and effects on cell functions by bovine herpesvirus 1. J. Virol. 63:1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ciacci-Zanella J., Stone M., Henderson G., Jones C. 1999. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J. Virol. 73:9734–9740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cornell R., Eisen J. S. 2005. Notch in the pathway: the roles of Notch signalling in neural crest development. Sem. Cell Dev. Biol. 16:663–672 [DOI] [PubMed] [Google Scholar]
- 9. Das R., et al. 2007. cAMP activation of PKA defines an ancient signaling mechanism. Proc. Natl. Acad. Sci. U. S. A. 104:93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Devireddy L., Zhang Y., Jones C. 2003. Cloning and initial characterization of an alternatively spliced transcript encoded by the bovine herpes virus 1 latency related (LR) gene. J. Neurovirol. 9:612–622 [DOI] [PubMed] [Google Scholar]
- 11. Ehebauer M., Penelope P., Arias A. M. 2006. Notch, a universial arbiter of cell fate decisions. Science 314:1414–1415 [DOI] [PubMed] [Google Scholar]
- 12. Franklin J. L., et al. 1999. Autonomous and non-autonomous regulation of mammalian neurite development by Notch1 and Delta1. Curr. Biol. 9:1448–1457 [DOI] [PubMed] [Google Scholar]
- 13. Geiser V., Jones C. 2003. Stimulation of bovine herpesvirus 1 productive infection by the adneovirus E1A gene and a cell cycle regulatory gene, E2F-4. J. Gen. Virol. 84:929–938 [DOI] [PubMed] [Google Scholar]
- 14. Geiser V., Inman M., Zhang Y., Jones C. 2002. The latency related (LR) gene of bovine herpes virus 1 (BHV-1) can inhibit the ability of bICP0 to activate productive infection. J. Gen. Virol. 83:2965–2971 [DOI] [PubMed] [Google Scholar]
- 15. Granville D. J., et al. 1998. Rapid cytochrome c release, activation of caspases 3, 6, 7 and 8 followed by Bap31 cleavage in HeLa cells treated with photodynamic therapy. FEBS Lett. 437:5–10 [DOI] [PubMed] [Google Scholar]
- 16. Griebel P., Ohmann H. B., Lawman M. J., Babiuk L. A. 1990. The interaction between bovine herpesvirus type 1 and activated bovine T lymphocytes. J. Gen. Virol. 71:369–377 [DOI] [PubMed] [Google Scholar]
- 17. Griebel P., et al. 1987. T lymphocyte population dynamics and function following a primary bovine herpesvirus type-1 infection. Viral Immunol. 1:287–304 [DOI] [PubMed] [Google Scholar]
- 18. Griebel P. J., Qualtiere L., Davis W. C., Lawman M. J., Babiuk L. A. 1987. Bovine peripheral blood leukocyte subpopulation dynamics following a primary bovine herpesvirus-1 infection. Viral Immunol. 1:267–286 [DOI] [PubMed] [Google Scholar]
- 19. Hamza M. A., Higgins D. M., Feldman L. T., Ruyechan W. T. 2007. The latency-associated transcript of herpes simplex virus type 1 promotes survival and stimulates axonal regeneration in sympathetic and trigeminal ganglia. J. Neurovirol. 13:56–66 [DOI] [PubMed] [Google Scholar]
- 20. Hariharan M. J., Nataraj C., Srikumaran S. 1993. Down regulation of murine MHC class I expression by bovine herpesvirus 1. Viral Immunol. 6:273–284 [DOI] [PubMed] [Google Scholar]
- 21. Hartwell L. H., Weinert T. A. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246:6239–6234 [DOI] [PubMed] [Google Scholar]
- 22. Henderson G., Perng G.-C., Nesburn A., Wechsler S., Jones C. 2004. The latency related gene of bovine herpesvirus 1 can suppress caspase 3 and caspase 9 during productive infection. J. Neurovirol. 10:64–70 [DOI] [PubMed] [Google Scholar]
- 23. Henderson G., et al. 2002. Regulation of caspase 8- and caspase 9-induced apoptosis by the herpes simplex virus latency-associated transcript. J. Neurovirol. 8:103–111 [DOI] [PubMed] [Google Scholar]
- 24. Hinkley S., Hill A. B., Srikumaran S. 1998. Bovine herpesvirus-1 infection affects the peptide transport activity in bovine cells. Virus Res. 53:91–96 [DOI] [PubMed] [Google Scholar]
- 25. Hochstrasser M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405–439 [DOI] [PubMed] [Google Scholar]
- 26. Hossain A., Schang L. M., Jones C. 1995. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J. Virol. 69:5345–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hsu H., Xiong J., Goeddel D. V. 1995. The TNF receptor 1-associated protein TRADD signals cell death and NF- kappa B activation. Cell 81:495–504 [DOI] [PubMed] [Google Scholar]
- 28. Inman M., et al. 2001. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J. Virol. 75:3636–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Inman M., Zhou J., Webb H., Jones C. 2004. Identification of a novel transcript containing a small open reading frame that is expressed during latency, and is antisense to the latency related gene of bovine herpes virus 1 (BHV-1). J. Virol. 78:5438–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Inman M., Lovato L., Doster A., Jones C. 2002. A mutation in the latency related gene of bovine herpesvirus 1 interferes with the latency-reactivation cycle of latency in calves. J. Virol. 76:6771–6779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Inman M., Lovato L., Doster A., Jones C. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 75:8507–8515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jaber T., Workman A., Jones C. 2010. Small non-coding RNAs encoded within the bovine herpesvirus 1 latency related gene can reduce steady state levels of infected cell protein 0 (bICP0). J. Virol. 84:6297–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang Y., et al. 1998. A protein encoded by the latency-related gene of bovine herpesvirus 1 is expressed in trigeminal ganglionic neurons of latently infected cattle and interacts with cyclin-dependent kinase 2 during productive infection. J. Virol. 72:8133–8142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang Y., Inman M., Zhang Y., Posadas N. A., Jones C. 2004. A mutation in the latency related gene of bovine herpesvirus 1 (BHV-1) inhibits protein expression of a protein from open reading frame 2 (ORF-2) and an adjacent reading frame during productive infection. J. Virol. 78:3184–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jin L., et al. 2008. Cellular FLIP can substitute for the herpes simplex virus type 1 LAT gene to support a wild type virus reactivation phenotype in mice. J. Neurovirol. 14:389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin L., Perng G.-C., Nesburn A. B., Jones C., Wechsler S. L. 2005. The baculovirus inhibitor of apoptosis gene (cpIAP) can restore reactivation of latency to a herpes simplex virus type 1 that does not express the latency associated transcript (LAT). J. Virol. 12286–12295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jin L., et al. 2003. Identification of herpes simplex virus type 1 (HSV-1) latency associated transcript (LAT) sequences that both inhibit apoptosis and enhance the spontaneous reactivation phenotype. J. Virol. 77:6556–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jones C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 51:81–133 [DOI] [PubMed] [Google Scholar]
- 39. Jones C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16:79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones C. 2009. Regulation of innate immune responses by bovine herpesvirus 1 and infected cell protein 0. Viruses 1:255–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jones C., Ciacci-Zanella J. R., Zhang Y., Henderson G., Dickman M. 2001. Analysis of fumonisin B1-induced apoptosis. Environ. Health Perspect. 109(Suppl. 2):315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones C., Delhon G., Bratanich A., Kutish G., Rock D. 1990. Analysis of the transcriptional promoter which regulates the latency- related transcript of bovine herpesvirus 1. J. Virol. 64:1164–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones C., et al. 2000. Analysis of latency in cattle after inoculation with a temperature sensitive mutant of bovine herpesvirus 1 (RLB106). Vaccine 18:3185–3195 [DOI] [PubMed] [Google Scholar]
- 44. Jones C., et al. 2006. Functional analysis of bovine herpesvirus 1 (BHV-1) genes expressed during latency. Vet. Microbiol. 113:199–210 [DOI] [PubMed] [Google Scholar]
- 45. Jones C., Chowdhury S. 2010. Bovine herpesvirus type 1 (BHV-1) is an important cofactor in the bovine respiratory disease complex. Vet. Clin. N. Am. Food Anim. Pract. 26:303–321 [DOI] [PubMed] [Google Scholar]
- 46. Jones C., Chowdhury S. 2007. A review of the biology of bovine herpesvirus type 1 (BHV-1), its role as a cofactor in the bovine respiratory disease complex, and development of improved vaccines. Adv. Anim. Health 8:187–205 [DOI] [PubMed] [Google Scholar]
- 47. Justice N. J., Jan Y. N. 2002. Variations on the Notch pathway in neural development. Curr. Opin. Neurobiol. 12:64–70 [DOI] [PubMed] [Google Scholar]
- 48. Kumar S., Kinoshita M., Noda M., Copeland N. G., Jenkins N. A. 1994. Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1 beta-converting enzyme. Genes Dev. 8:1613–1626 [DOI] [PubMed] [Google Scholar]
- 49. Kutish G., Mainprize T., Rock D. 1990. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J. Virol. 64:5730–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leib D. A., Nadeau K. C., Rundle S. A., Schaffer P. A. 1991. The promoter of the latency-associated transcripts of herpes simplex virus type 1 contains a functional cAMP-response element: role of the latency-associated transcripts and cAMP in reactivation of viral latency. Proc. Natl. Acad. Sci. U. S. A. 88:48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Levy O. A., Lah J. J., Levy A. I. 2002. Notch signaling inhibits PC12 cell neurite outgrowth via RBP-J-dependent and -independent mechanisms. Dev. Neurosci. 24:79–88 [DOI] [PubMed] [Google Scholar]
- 52. Lovato L., Inman M., Henderson G., Doster A., Jones C. 2003. Infection of cattle with a bovine herpesvirus 1 (BHV-1) strain that contains a mutation in the latency related gene leads to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J. Virol. 77:4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meyer F., et al. 2007. Identification of a novel protein encoded by the latency-related gene of bovine herpesvirus 1. J. Neurovirol. 13:569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meyer F., et al. 2007. A protein encoded by the bovine herpes virus 1 (BHV-1) latency related gene interacts with specific cellular regulatory proteins, including the CCAAT enhancer binding protein alpha (C/EBP-a). J. Virol. 81:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meyer F., Jones C. 2009. The cellular transcription factor, CCAAT enhancer-binding protein alpha (C/EBP-α), has the potential to activate the bovine herpesvirus 1 immediate-early transcription unit 1 promoter. J. Neurovirol. 15:123–130 [DOI] [PubMed] [Google Scholar]
- 56. Mott K., et al. 2003. The bovine herpesvirus 1 LR ORF2 is crucial for this gene's ability to restore the high reactivation phenotype to a herpes simplex virus-1 LAT null mutant. J. Gen. Virol. 84:2975–2985 [DOI] [PubMed] [Google Scholar]
- 57. Nataraj C., et al. 1997. Bovine herpesvirus 1 downregulates the expression of bovine MHC class I molecules. Viral Immunol. 10:21–34 [DOI] [PubMed] [Google Scholar]
- 58. Nicholson D. W., Thornberry N. A. 1997. Caspases: killer proteases. Trends Biochem. Sci. 22:299–306 [DOI] [PubMed] [Google Scholar]
- 59. Perng G.-C., et al. 2002. A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels. J. Virol. 76:1224–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Perng G.-C., et al. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript (LAT). Science 287:1500–1503 [DOI] [PubMed] [Google Scholar]
- 61. Perng G.-C., Jones C. 2010. Towards an understanding of the herpes simplex virus type 1 latency-reactivation cycle. Interdiscip. Perspect. Infect. Dis. 2010:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Persson L. M., Wilson A. C. 2010. Wide-scale use of Notch signaling RTA-mediated activation of factor CSL/RBP-J kappa in Kaposi's sarcoma-associated herpesvirus lytic genes. J. Virol. 84:1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rock D., Lokensgard J., Lewis T., Kutish G. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 66:2484–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rock D. L., et al. 1987. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J. Virol. 61:3820–3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rock D. L., Beam S. L., Mayfield J. E. 1987. Mapping bovine herpesvirus type 1 latency-related RNA in trigeminal ganglia of latently infected rabbits. J. Virol. 61:3827–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Snalkumar R., Dhanesh S. B., Jackson J. 2010. Non-canonical activation of Notch signaling/target genes in vertebrates. Cell. Mol. Life Sci. 67:2957–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schang L., Jones C. 1997. Analysis of bovine herpesvirus 1 transcripts during a primary infection of trigeminal ganglia of cattle. J. Virol. 71:6786–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schmitz I., Kirchhoff S., Krammer P. H. 2000. Regulation of death receptor-mediated apoptosis pathways. Int. J. Biochem. Cell Biol. 32:1123–1136 [DOI] [PubMed] [Google Scholar]
- 69. Shen W., et al. 2009. 2009. Two small RNAs encoded within the first 1.5 kb of the herpes simplex virus type 1 (HSV-1) latency-associated transcript (LAT) can inhibit productive infection, and cooperate to inhibit apoptosis. J. Virol. 90:9131–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shen W., Jones C. 2008. Open reading frame 2 encoded by the latency related gene of bovine herpesvirus 1 has anti-apoptosis activity in transiently transfected neuroblastoma cells. J. Virol. 82:10940–10945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Smith R. L., Pizer L. I., Johnson E. M. J. R., Wilcox C. L. 1992. Activation of second-messenger pathways reactivates latent herpes simplex virus in neuronal cultures. Virology 188:311–318 [DOI] [PubMed] [Google Scholar]
- 72. Turin L., Russo S., Poli G. 1999. BHV-1: new molecular approaches to control a common and widespread infection. Mol. Med. 5:261–284 [PMC free article] [PubMed] [Google Scholar]
- 73. Winkler M. T., Doster A., Jones C. 1999. Bovine herpesvirus 1 can infect CD4(+) T lymphocytes and induce programmed cell death during acute infection of cattle. J. Virol. 73:8657–8668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Workman A., Sinani D., Pittayakhajonwut D., Jones C. 2011. A protein (ORF2) encoded by the latency related gene of bovine herpesvirus 1 interacts with Notch1 and Notch3. J. Virol. 85:2536–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Workman A., Perez S., Doster A., Jones C. 2009. Dexamethasone treatment of calves latently infected with bovine herpesvirus 1 (BHV-1) leads to activation of the bICP0 early promoter, in part by the cellular transcription factor C/EBP-alpha. J. Virol. 83:8800–8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Workman A., Jones C. 2010. Bovine herpesvirus 1 productive infection and bICP0 early promoter activity are stimulated by E2F1. J. Virol. 84:6308–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yang J., et al. 1997. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 275:1129–1132 [DOI] [PubMed] [Google Scholar]
- 78. Zhang Y., Jones C. 2005. Identification of functional domains within the bICP0 protein encoded by BHV-1. J. Gen. Virol. 86:879–886 [DOI] [PubMed] [Google Scholar]
- 79. Zhang Y., Xiong Y. 2001. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 12:175–186 [PubMed] [Google Scholar]