Abstract
The double-stranded RNA (dsRNA)-dependent protein kinase (PKR) inhibits protein synthesis by phosphorylating eukaryotic translation initiation factor 2α (eIF2α). In fish species, in addition to PKR, there exists a PKR-like protein kinase containing Z-DNA binding domains (PKZ). However, the antiviral role of fish PKZ and the functional relationship between fish PKZ and PKR remain unknown. Here we confirmed the coexpression of fish PKZ and PKR proteins in Carassius auratus blastula embryonic (CAB) cells and identified them as two typical interferon (IFN)-inducible eIF2α kinases, both of which displayed an ability to inhibit virus replication. Strikingly, fish IFN or all kinds of IFN stimuli activated PKZ and PKR to phosphorylated eIF2α. Overexpression of both fish kinases together conferred much more significant inhibition of virus replication than overexpression of either protein, whereas morpholino knockdown of both made fish cells more vulnerable to virus infection than knockdown of either. The antiviral ability of fish PKZ was weaker than fish PKR, which correlated with its lower ability to phosphorylate eIF2α than PKR. Moreover, the independent association of fish PKZ or PKR reveals that each of them formed homodimers and that fish PKZ phosphorylated eIF2α independently on fish PKR and vice versa. These results suggest that fish PKZ and PKR play a nonredundant but cooperative role in IFN antiviral response.
INTRODUCTION
Compared to transcriptional control, the regulation of protein translation is more rapid and direct in the flow of genetic information, making cells adapt to diverse stresses immediately. Most translational control occurs at the initiation step, which is mediated by the eukaryotic initiation factors (eIFs), such as eukaryotic initiation factor 2 (eIF2). Under normal conditions, eIF2 interacts with GTP to deliver the initiator methionyl-tRNA to the small ribosomal subunit in the first step of translation initiation. Once translation initiation is completed, the released eIF2-GDP complex must be continuously recycled by the guanine nucleotide exchange factor eIF2B to replace GDP with GTP for another round of initiation. However, phosphorylation of the α subunit of eIF2 (eIF2α) at serine 51 blocks the recycling, thereby leading to a general shutoff of protein synthesis (19, 30).
In mammals, eIF2α is phosphorylated by a small protein family of eIF2α kinases, which consist of double-stranded RNA (dsRNA)-dependent protein kinase (PKR), PKR-like endoplasmic reticulum (ER) eIF2α kinase (PERK), general control of nitrogen metabolism kinase 2 (GCN2), and heme-regulated eIF2α kinase (HRI) (38). Of these kinases, PKR is most widely studied in the context of virus infection. The structural features of PKR include two dsRNA binding domains (dsRBDs) at its N terminus and a kinase domain (KD) at its C terminus (6, 27). In virus-infected cells, the expression of PKR is upregulated by ongoing produced interferon (IFN) (21). The latent PKR is activated by binding to dsRNA that occurs during virus replication, thus undergoing dimerization, autophosphorylation, and subsequently inhibition of viral protein synthesis via phosphorylating eIF2α (7). Consistent with its antiviral property, overexpression of PKR in many mammalian cell lines confers resistance to virus infection (17, 23). PKR-deficient cells are more permissive for several RNA viruses (28, 40) as well as DNA viruses (1), and PKR-deficient mice become highly susceptible to otherwise harmless infection of vesicular stomatitis virus (VSV) and influenza virus (2).
Therefore, PKR-mediated eIF2α phosphorylation is believed to act as a conserved antiviral pathway involved in vertebrate IFN antiviral response (9). Recent studies strengthen this notion in that the lower vertebrate fish possess many conserved IFN-stimulated genes (ISGs) and the regulatory mechanisms of IFN antiviral response (41). However, prior to characterization of fish PKR, a novel member of vertebrate eIF2α kinase, termed PKR-like or PKZ (protein kinase containing Z-DNA binding domains), was identified exclusively in fish (3, 13, 26). Fish PKZ is most homologous to mammalian PKR counterparts and exhibits expression characteristic of mammalian PKR and catalytic activity similar to mammalian PKR (3, 13, 26), leading to an ephemeral belief that it is an orthologue of mammalian PKR. However, fish PKZ protein harbors a unique N-terminal structure with two Z-DNA binding domains (Zα domains) instead of two tandem dsRBDs (3, 13, 26). Besides fish PKZ, two other cellular Z-DNA binding domain-containing proteins, ADAR (adenosine deaminase acting on RNA) (12, 16) and DAI (DNA-dependent activator of IFN-regulatory factors, also known as ZBP1 or DLM-1) (8), have been previously identified in mammals. Interestingly, these two proteins participate in IFN antiviral response (10, 35, 39). Fish PKZ is the third Z-DNA binding domain-containing protein to be identified in vertebrates, but its physiological function is not understood. Recent studies demonstrated that fish also have a conserved PKR-mediated antiviral response (25, 44). Therefore, the coexistence of PKR and PKZ in fish genomes makes it worthwhile to explore their relative contribution and functional relationship to gain insights into the molecular nature specific to fish IFN antiviral response.
In the present study, we found the coexistence of PKZ and PKR genes in crucian carp (Carassius auratus L.) and identified them as two typical IFN-stimulated genes. Further, we found that both fish PKZ and PKR displayed an ability to inhibit replication of grass carp reovirus (GCRV) in fish cells and that this antiviral effect was exerted by cooperative effects of both fish PKR and PKZ by phosphorylating eIF2α. Finally, we found that fish PKZ phosphorylated eIF2α independently of PKR and vice versa. Our results provide strong evidence that fish PKR and PKZ play a nonredundant but cooperative role in IFN-mediated antiviral response.
MATERIALS AND METHODS
Cells and virus.
Three fish cell lines, crucian carp (Carassius auratus L.) blastula embryonic cells (CAB), epithelioma papulosum cyprini cells (EPC), and ovary cells of the grass carp (Ctenopharyngodon idella) (CO), were maintained at 28°C in medium 199 (M199) supplemented with 10% fetal calf serum (FCS) and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin). Two mammalian cell lines, COS-7 and 293T, were maintained at 37°C in Dulbecco modified Eagle medium (DMEM) with 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Two cell lines, transfected stably with Stat1-ΔC or empty vector pcDNA3.1, were previously described (33). Grass carp reovirus (GCRV) was propagated in CAB cells as described previously (43).
Plasmids.
Prokaryotic expression constructs pET-PKR-N and pET-PKZ-N were generated by inserting the corresponding PCR products of PKR (amino acids [aa] 1 to 334) and PKZ (aa 1 to 162) into NcoI/XhoI sites of pET-32a (+) (Novagen). For overexpression assays, two wild-type plasmids PKR-wt and PKZ-wt were generated by insertion of the entire open reading frames (ORFs) into the EcoRI/KpnI sites of pcDNA3.1/myc-His(-) A vector (Invitrogen) or into the EcoRI/KpnI sites or NheI/KpnI sites of the pCMV-HA (CMV stands for cytomegalovirus) plasmid that was generated based on the backbone of pEGFP-N3 and by replacing the enhanced green fluorescent protein (EGFP) ORF with a three-hemagglutinin (3×HA) tag according to the previous report (11). Two catalytically inactivated mutant plasmids, PKR-K419R and PKZ-K198R, were obtained from PKR-wt and PKZ-wt plasmids by replacing lysine (K) with arginine (R). For promoter activity analysis, two promoter-driven luciferase plasmids, PKRpro-luc and PKZpro-luc, were obtained with insertion of the 5′ flanking region (+76/−460) of the PKR promoter (GenBank accession no. JN088687) or the 5′ flanking region (+47/−462) of the PKZ promoter (GenBank accession no. JN088688) into the XhoI/KpnI sites of pGL3-Basic luciferase reporter vector (Promega). All primers used in this study were given in Table S1 in the supplemental material.
Transfection and luciferase assays.
For overexpression assays, cells were seeded in 6-well plates overnight, and each well was transfected with 1.6 μg of the indicated plasmids by 4 μl of Lipofectamine 2000 (Invitrogen) in 1 ml fresh 10% FCS-containing M199 or DMEM without antibiotics. For translation inhibition assays, the cells were seeded in 24-well plates overnight, and each well was cotransfected with 200 ng of reporter plasmid pGL3 promoter and 200 ng of the indicated wild-type or mutated plasmids of PKR and PKZ. For promoter activity assays, the cells were seeded in the wells of 24-well plates overnight, and the cells in each well were cotransfected with a total amount of 0.55 μg plasmids at a ratio of 1:10 (pRL-TK and PKRpro-luc or PKZpro-luc) using Lipofectamine 2000 in 0.5 ml fresh medium. The cells were harvested according to the Dual-Luciferase reporter assay system (Promega). Luciferase activities were measured by a Junior LB9509 luminometer (Berthold) according to the protocol or the previous report (33).
Knockdown.
Two translation-blocking morpholino oligonucleotides (MOs) PKR-MO and PKZ-MO and standard control MO were synthesized and purchased from Gene Tools, LLC. MOs were resuspended in nuclease-free water at a concentration of 200 μM. The MO-mediated knockdown was performed by the scrape delivery method (20). Briefly, CAB cells were seeded in 6-well plates overnight. For each well, the culture medium was replaced with 1 ml fresh M199 containing 10% FCS, and then 53 μl of MO stock solution (200 μM) was added and swirled for 10 s to mix thoroughly. This gave a MO concentration of 10 μM. The culture plates were placed on a flat surface, and the cells were gently scraped off the surface of the well with sterile cell scrapers. The scraped cell suspensions were then gently pipetted up and down twice and transferred to new 24-well culture plates. Cells delivered with MOs were cultured for 0.5 h and then added with crucian carp recombinant IFN (rIFN) or subjected to GCRV infection. rIFN was produced by a prokaryotic expression system according to a previous study (41).
Polyclonal antiserum, Western blotting, and coimmunoprecipitation.
The prokaryotic expression and antibody production of PKR and PKZ were examined by the same protocols as in our previous studies (33, 34, 41). Human eIF2α and actin antibodies were purchased from Santa Cruz Biotech Company, and serine 51 phosphorylated human eIF2α antibody was from Sigma-Aldrich Company.
For Western blot analysis, whole-cell extracts were prepared from cells washed twice with phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay (RIPA) reagent (Beyotime)-containing protease and phosphatase inhibitor cocktails (Sigma). Other steps followed the previous protocols (33, 34). The measurement of densitometry ratio of phosphorylated eIF2α over total eIF2α followed the methods in reference 44.
For coimmunoprecipitation (Co-IP), protein samples were made by lysing cells in RIPA buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, and protease inhibitor cocktail). For anti-eIF2α Co-IP, 500 μl of a prepared cell lysate was incubated with 16 μl of eIF2α antibody (Santa Cruz) at 4°C for 2 h, which was then added to 40 μl of resuspended volume of protein A/G agarose (Santa Cruz) and incubated overnight at 4°C. Immunocomplexes were washed 5 times with PBS buffer at 4°C, collected by centrifugation at 2,000 × g for 5 min at 4°C, and then suspended in 40 μl of 2× SDS-PAGE sample buffer. The suspended immunocomplexes were further analyzed by Western blotting. For anti-HA Co-IP, HA antibody-conjugated agarose beads were washed 4 times with PBS and added to the prepared lysates at a ratio of 40 μl beads/500 μl lysate. Other steps were the same with anti-eIF2α Co-IP.
Nucleotide sequence accession numbers.
The accession numbers from GenBank for the sequences used in Fig. 1C are AB104655, NM_001048135, AY293929, JN091442, AY623897, EF685036, XM_695386, AM421526, AJ852018, AY850106, AB125660, AM850085, EF661570.1, AF193339, NM_002759, EF467667, DQ645944, NM_011163, NM_001145891, FJ179396, DQ115394, AM850088, XM_001166391, EU118259, AM850089, NM_019335, EF523422, NM_001123595, NM_214319, AM850086, AM850087, AM421523, AM421524, AM421525, AM850090, AM850091, AM850092, and AM421528.
Fig. 1.
Coexistence of fish PKR and PKZ. (A) Genomic coexistence of tandemly arranged PKR and PKZ genes in Danio rerio and Carassius auratus. The arrows indicate the 5′ to 3′ orientation of the genes. The approximate sizes of the intergenic regions are indicated below the maps. (B) Taxonomic tree of species used in the phylogenetic analysis. The taxonomic tree is constructed by NCBI taxonomy (http://www.ncbi.nlm.nih.gov/taxonomy). Arrows indicate the species in which PKZ genes have been found. (C) Phylogenetic relationship of PKR and PKZ proteins. The phylogenetic tree was constructed by maximum likelihood method from the multiple-sequence alignment of the kinase domains of PKR and PKZ without the kinase insert using the MUSCLE approach. Human and zebrafish PERK were used as outgroups for rooting the phylogenetic tree. The abbreviations used here are shown in parentheses immediately after the species name in panel B. The accession numbers of the sequences used in panel C are given in Materials and Methods.
RESULTS
Coexistence of PKR and PKZ in the fish genome.
Previously a PKZ (PKR-like) gene (GenBank accession no. AY293929) was retrieved from a subtractive cDNA library that was made with mRNA from UV-irradiated GCRV-infected CAB cells (13). Enlargement of screening the same library resulted in identification of crucian carp PKR gene (GenBank accession no. JN091442). Unlike crucian carp PKZ that contains two N-terminal Z-DNA binding domains (Zα1 and Zα2) (13), the crucian carp PKR gene encodes a 687-amino-acid protein consisting of three tandem dsRBDs at its N terminus and a kinase domain at its C terminus. Genome walking PCR amplified a DNA segment covering both genes, showing that the PKR and PKZ genes are tandemly arranged in the genome of crucian carp (Fig. 1A), further demonstrating the coexistence of the PKR and PKZ genes in the crucian carp genome. Multiple-sequence alignment reveals a series of conserved sites in fish PKR and PKZ proteins, for example, the site equivalent to lysine 296 of human PKR that is essential for ATP binding and the phosphotransfer reaction (37), and the sites equivalent to threonine 446 and 451 of human PKR in the activation segment that are crucial for PKR autophosphorylation (24) (see Fig. S1 in the supplemental material). While PKR is widely cloned in bony fish, so far PKZ is found only in members of the Cypriniformes and Salmoniformes orders (Fig. 1B and C). Phylogenetic analysis showed that fish PKZs cluster with fish PKRs, rather than form an independent clade away from all PKRs (Fig. 1C), indicating that fish PKR and PKZ originated from a common ancestor during teleost radiation.
Induction of fish PKR and PKZ by poly(I · C) and IFN.
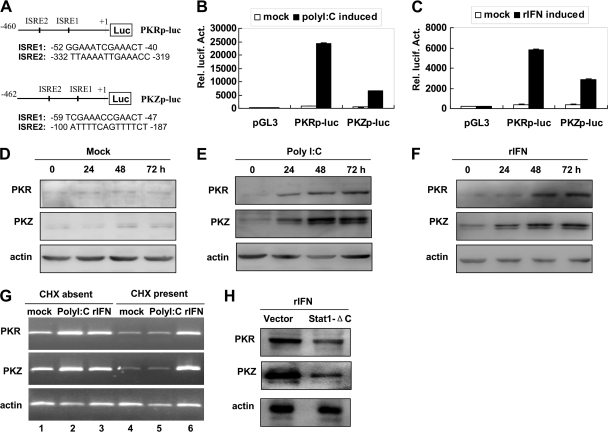
Previous studies have shown that fish PKR and PKZ are transcriptionally induced by poly(I · C) and virus infection (13, 25, 26, 44). To further characterize the expression of fish PKR and PKZ, 5′ flanking regulatory sequences of both genes were cloned, and two typical IFN-stimulated response elements (ISREs), a characteristic motif in ISG promoters (33), are found within ∼460 bp upstream from the translation start codons of both genes (see Fig. S2A and S2B in the supplemental material). Therefore, these ∼460-bp DNA fragments were identified as minimal promoters and used to generate two promoter-driven luciferase reporter constructs (Fig. 2A). Luciferase activity assays showed that the promoter activities were significantly induced by poly(I · C) (Fig. 2B) and crucian carp recombinant IFN (rIFN) (Fig. 2C).
Fig. 2.
Induction of fish PKR and PKZ by poly(I · C) and IFN. (A) Schematic illustration of promoter-driven luciferase (Luc) constructs PKRpro-luc (PKRp-luc) and PKZpro-luc (PKZp-luc). (B and C) Activation of the PKR and PKZ promoters by poly(I · C) (polyI:C) or rIFN. CAB cells in each well of 24-well plates were cotransfected with 0.05 μg pRL-TK and 0.5 μg PKRpro-luc or PKZpro-luc in 500 μl medium. At 24 h posttransfection (hpt), the cells were stimulated by transfection of poly(I · C) (0.4 μg/ml) (B) or the addition of rIFN (20 ng/ml) (C). At 48 hpt, the cells were harvested for detection of luciferase activity. Rel. lucif. Act., relative luciferase activity. (D to F) Induction of fish PKR and PKZ proteins by poly(I · C) or rIFN. CAB cells seeded in 6 well-plate were mock transfected with 1.6 μl PBS (D) or transfected with 1.6 μl of a 1-μg/μl poly(I · C) solution (E) or 20 ng rIFN was added to 1 ml medium (F). The cells were harvested at 0, 24, 48, or 72 hpt for detection of PKR and PKZ by Western blot analysis. (G) Requirement of ongoing IFN synthesis for transcriptional induction of fish PKR and PKZ genes. CAB cells seeded in 6-well plates were mock transfected with 1.6 μl PBS or transfected with 1.6 μl of a 1-μg/μl poly(I · C) solution in 1 ml medium or treated with rIFN at 20 ng/ml in the absence or presence of 8 μg/ml cycloheximide (CHX) for 24 h, and total RNA was extracted. Reverse transcription-PCR (RT-PCR) was employed to detect PKR and PKZ expression. (H) Involvement of the Stat1 pathway in PKR and PKZ induction. CAB cells stably transfected with pcDNA3.1 or Stat1-ΔC were treated with 5 ng/ml of rIFN for 10 h and then harvested for detection of PKR and PKZ protein by Western blot analysis.
To determine the expression of PKR and PKZ at the protein level, their N-terminal regions were prokaryotically expressed as His-tagged fusion proteins rPKR-N and rPKZ-N (r stands for recombinant, and N stands for N-terminal region) (see Fig. S3A and S3B in the supplemental material), and the purified proteins were used to immunize mice to prepare anti-PKR and anti-PKZ antisera. The generated antisera were able to specifically recognize the original antigen proteins PKR-N or PKZ-N overexpressed in 293T cells (Fig. S3C), and the cellular PKR and PKZ in rIFN-treated CAB cells (Fig. S3D and S3E). No cross-recognition was observed between both antisera, since such specific recognition was completely abrogated when the antisera had been preabsorbed with the original antigen but not with an unrelated one (Fig. S3D and S3E).
Using the produced antisera, Western blots revealed a basal expression of fish PKR and PKZ in normal CAB cells (Fig. 2D). Either poly(I · C) transfection or rIFN treatment significantly induced the expression of both proteins in a time-dependent manner (Fig. 2E and F). Unlike fish PKR for which one protein band was detected, two PKZ protein bands were detected in normal or treated CAB cells (Fig. 2E and 2F; see Fig. S3E in the supplemental material), likely due to alternative splicing of fish PKZ mRNA (25, 26). Further, in CAB cells that were pretreated with a canonical translation inhibitor, cycloheximide (CHX), poly(I · C) induction of PKR and PKZ mRNA was diminished (Fig. 2G, lane 5), whereas rIFN induction was maintained (Fig. 2G, lane 6), indicating that IFN is required for poly(I · C) induction of fish PKR and PKZ. Consistently, in CAB cells stably transfected with a dominant-negative mutant (Stat1-ΔC) to block the IFN-activated Stat1 pathway (33), rIFN led to a diminished expression of both proteins compared to that in control cells stably transfected with empty vector pcDNA3.1 (Fig. 2H). These results together demonstrated that fish PKR and PKZ are two IFN-inducible proteins.
Concomitant induction of eIF2α phosphorylation through the IFN-Stat1 pathway.
The level of eIF2α phosphorylation was next determined in poly(I · C)-transfected or rIFN-treated CAB cells, where PKR and PKZ proteins were confirmed to be upregulated (Fig. 2D to F). Compared to a constant level of eIF2α phosphorylation in mock-induced cells (Fig. 3A), either poly(I · C) transfection or rIFN treatment resulted in significant increase of eIF2α phosphorylation (Fig. 3B and C), in a time-dependent manner similar to the expression patterns of fish PKR and PKZ (Fig. 2E and F). Consistent with the finding that IFN induction of PKR and PKZ decreased in CAB cells stably transfected with Stat1-ΔC (Fig. 2H), the level of eIF2α phosphorylation was concomitantly attenuated (Fig. 3D). These results indicated that the phosphorylation of eIF2α is probably associated with fish PKR and PKZ in response to poly(I · C) transfection or rIFN treatment. In addition to poly(I · C), other stimuli, including poly(dA-dT) (B-DNA), poly(dG-dC) (Z-DNA), and genomic DNA, also significantly upregulated both fish proteins and concomitant eIF2α phosphorylation (Fig. 3F to H).
Fig. 3.
Concomitant induction of eIF2α phosphorylation by poly(I · C) and IFN. (A to C) Concomitant induction of eIF2α phosphorylation by poly(I · C) and rIFN. Samples loaded here are the same as those in Fig. 2D to F, respectively, but phosphorylated eIF2α (p-eIF2α) and total eIF2α were detected by Western blot analysis. (D) Involvement of the Stat1 pathway in eIF2α phosphorylation by poly(I · C) and rIFN. The same samples shown in Fig. 2H were loaded here for Western blot analysis of total eIF2α or phosphorylated eIF2α. (E) Direct interaction of eIF2α with PKR or PKZ. CAB cells seeded in 10-cm dishes were mock induced by transfection of 8 μl PBS [poly(I · C) absent] or induced by transfection of 8 μl × 1 μg/μl poly(I · C) [poly(I · C) present] in 5 ml medium for 48 h and then harvested for coimmunoprecipitation assay by anti-eIF2α antibody. PKR, PKZ, p-eIF2α, and eIF2α were analyzed by Western blotting in both immunoprecipitation (IP) samples (left blots) and input samples (right blots). (F to H) CAB cells were transfected with poly(dG · dC) which mimics Z-DNA (F), poly(dA · dT) which mimics B-DNA (G), or unsheared calf genomic DNA (g-DNA) (H), harvested at 0, 24, 48, and 72 h posttransfection (hpt), and subjected to immunoblotting with antibodies specific to PKR, PKZ, actin, p-eIF2α, and eIF2α. The relative levels of eIF2α phosphorylation as determined by quantitative densitometry are shown below the blots. The value for the sample at 0 hpt is set at 1 as a reference with which other samples are compared.
The physical interaction of the PKR and PKZ proteins was further determined by Co-IP experiments. Endogenous PKR and PKZ proteins could be coimmunoprecipitated by an anti-human eIF2α antibody in either control CAB cells or poly(I · C)-transfected cells (Fig. 3E, left blots). The specificity of these interactions was confirmed by immunoprecipitation with an irrelevant rabbit IgG antibody. Analysis of the coimmunoprecipitated complex revealed an increased proportion of phosphorylated eIF2α in the poly(I · C)-transfected cells with reference to that in control cells (Fig. 3E, left blot), which was consistent with the enhanced phosphorylation of eIF2α with total cellular lysis (Fig. 3E, right blot). These data suggest that poly(I · C) transfection leads to a consistent increase in the phosphorylation of eIF2α that is bound to PKR or PKZ and that the enhanced eIF2α phosphorylation is attributed to the activation of PKR or PKZ in response to poly(I · C). Thus, like fish PKR, fish PKZ is a functional eIF2α kinase.
Cooperative catalytic activity of fish PKZ and PKR.
In mammals, a quantitative and effective method to test eIF2α kinase activity is to determine the expression inhibition of a cotransfected reporter gene in cell lines (4, 15). Initially, we made four constructs, two wild-type constructs (PKR-wt and PKZ-wt) and two mutant constructs (PKR-K419R and PKZ-K198R) (Fig. 4, left panel). Both mutants contain a point mutation (K→R) at the site corresponding to amino acid K296 of human PKR. This point mutation abolishes the enzymatic activity of human mutant PKR-K296R but retains its binding ability to eIF2α (4, 15).
Fig. 4.
Inhibition of reporter protein synthesis by fish PKR and PKZ. CAB, CO, EPC, COS-7, and 293T cells, seeded in 24-well plates, were cotransfected with the pGL3 plasmid (200 ng) and the expression vectors (200 ng) containing the corresponding inserts indicated by the illustration shown to the left of the bar graph. The K→R mutation that abolishes kinase activity but does not affect dimerization is indicated by a black asterisk. Luciferase activity was normalized by protein contents. A control experiment by using empty vector was set at 100%. Each bar shows the average ± standard error (error bar) for three independent experiments.
Subsequently, the catalytic activity of the four constructs was analyzed in three fish cell lines (CAB, CO, and EPC) and two mammalian cell lines (293T and COS-7) (Fig. 4, right panel). Compared to the basal activity of luciferase plasmid pGL3 (100%), overexpression of the PKR-wt plasmid alone resulted in a significantly reduced luciferase activity to about 10% in all five cell lines, so did overexpression of the PKZ-wt plasmid alone to about 33%. Similarly, about 10% luciferase activity was detected by cotransfection with both wild-type constructs together. Conversely, the two mutants, PKR-K419R and PKZ-K198R, lose the catalytic activities but stimulated luciferase activity in three fish cell lines to up to 150% by PKR-K419R and about 130% by PKZ-K198R. In the two mammalian cell lines, whereas PKR-K419R transfection still stimulated luciferase activity up to about 150%, no stimulation was detected by PKZ-K198R transfection. Consistently, transfection of mammalian cells with the two mutants together yielded about 145% activity, similar to transfection with PKR-K419R alone, whereas transfection of fish cells with both mutants together resulted in 175% luciferase activity, an obvious increase relative to transfection with two mutants individually. Therefore, fish PKR and PKZ cooperatively inhibit reporter protein synthesis, and the PKZ-K198R mutant does not seem to exhibit the dominant-negative effect to endogenous PKR in fish and mammalian cells.
Cooperative antiviral roles of fish PKZ and PKR.
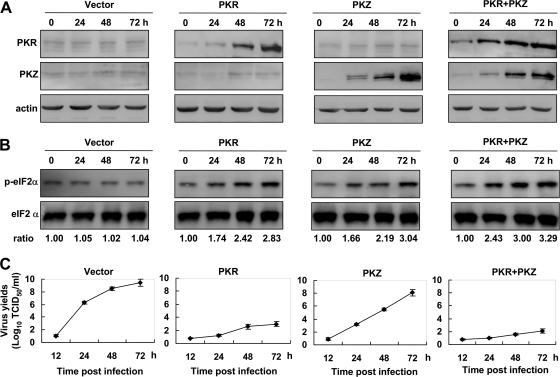
The antiviral roles of fish PKR and PKZ were investigated next. In initial assays, transfection of CAB cells with wild-type PKR and PKZ individually or collectively resulted in enhancement of eIF2α phosphorylation in a time-dependent manner (Fig. 5A and B). After infection of the transiently transfected cells with GCRV, overexpression of each fish protein severely and consistently inhibited virus replication relative to that in control cells, with the highest inhibition observed when both constructs were overexpressed together (viral titer of 101.8 50% tissue culture infective doses [TCID50]/ml versus 108.9 TCID50/ml for control cells at 72 h postinfection [p.i.]), the next highest inhibition observed when fish PKR was overexpressed (viral titer of 102.6 TCID50/ml versus 108.9 TCID50/ml of control cells at 72 h p.i.), and the third highest inhibition observed when fish PKZ was overexpressed (viral titer of 107.5 TCID50/ml versus 108.9 TCID50/ml of control cells at 72 h p.i.) (Fig. 5C), which conformed with the levels of eIF2α phosphorylation (Fig. 5B).
Fig. 5.
Inhibition of GCRV propagation by overexpression of fish PKR or PKZ. (A) CAB cells in each well of 6-well plates were cotransfected with a total amount of 1.6 μg of pcDNA3.1, PKR, PKZ, or PKR plus PKZ (1:1) plasmids in 1 ml fresh medium. At 0, 24, 48, and 72 hpt, cells were harvested for detection of PKR and PKZ proteins by Western blot analysis. (B) Phosphorylated eIF2α and total eIF2α were measured in the samples as indicated above for panel A. The relative levels of eIF2α phosphorylation as determined by quantitative densitometry are shown below the blots. The sample of 0 hpt is set at 1 as a reference with which other samples are compared. (C) The other group of transfected cells in 6-well plates was subjected to GCRV infection (each well with 1 ml of a solution with 104 TCID50/ml), and the supernatants were collected at 12, 24, 48, and 72 hpt for detection of virus titers by TCID50 assays.
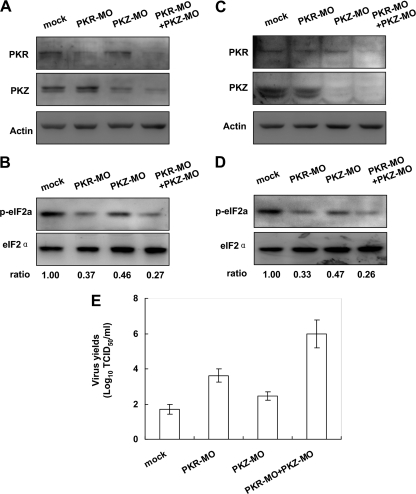
In subsequent assays, two morpholino oligonucleotides (PKR-MO and PKZ-MO) were designed to knockdown the expression of PKR and PKZ. When CAB cells were delivered with PKR-MO, the induction of PKR by rIFN was attenuated, but the induction of PKZ was not affected. The same was true for PKZ (Fig. 6A). Moreover, the level of eIF2α phosphorylation was consistently diminished in MO-treated cells relative to control cells (Fig. 6B). These results confirmed the effectiveness of two MOs to specifically knock down fish PKR and PKZ in CAB cells. The MO-treated cells were then challenged with 200 TCID50 GCRV for 48 h, and eIF2α phosphorylation was measured and the titers of the virus were determined. Western blots showed that the levels of expression of fish PKR and PKZ were diminished in MO-treated cells (Fig. 6C), and the levels of eIF2α phosphorylation were reduced accordingly (Fig. 6D). Consistently, high virus titers were measured in supernatants from MO-treated cells (Fig. 6E). Compared to the knockdown effects of PKR or PKZ individually (viral titer of 105.2 TCID50/ml for PKR knockdown cells and 103.5 TCID50/ml for PKZ knockdown cells), knockdown of both proteins together resulted in the most severe replication of GCRV (viral titer of 108.6 TCID50/ml). When the effects of knockdown between PKR and PKZ were compared, knockdown of PKR made cells more vulnerable to GCRV than knockdown of PKZ (105.2 TCID50/ml versus 103.5 TCID50/ml, respectively) (Fig. 6E), which was consistent with the relatively low level of eIF2α phosphorylation (Fig. 6D). These results together suggested that fish PKR and PKZ cooperatively inhibit replication of GCRV by phosphorylating eIF2α.
Fig. 6.
Effects on GCRV replication by knockdown of PKR or PKZ. (A and B) CAB cells seeded in 24-well plates were delivered with control MO (mock), PKR-MO, PKZ-MO, or PKR-MO plus PKZ-MO (detailed in Materials and Methods) and induced by rIFN at 20 ng/ml for 12 h and subjected to immunoblot analysis. (C to E) The other group of MO-delivered cells were subjected to GCRV infection (each well with 1 ml of a solution with 200 TCID50/ml) for 48 h. While the cells were subjected to Western blot analysis (C and D), the supernatants were collected for detection of virus titers by TCID50 assays (E). The relative levels of eIF2α phosphorylation as determined by quantitative densitometry are shown below the blots in panels B and D.
Independent dimerization of fish PKZ and PKR.
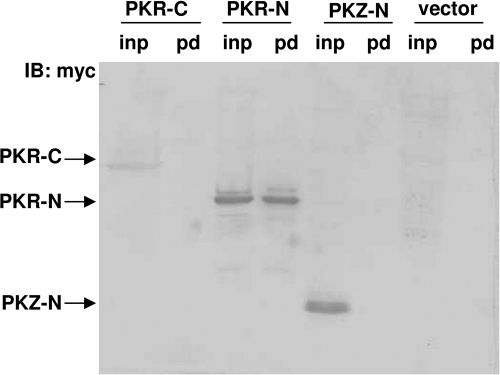
The finding that fish PKR and PKZ displayed similar abilities to inhibit virus replication prompted us to investigate whether there were reciprocal interactions for their activation. In mammals, dimerization of PKR is sufficient for its activation, and the binding of PKR N terminus to dsRNA is the first step for PKR dimerization (18). Pulldown assays showed that the three N-terminal dsRBDs of fish PKR (PKR-N) effectively bound to poly(I · C), whereas the N terminus of PKZ containing two Z-DNA binding domains (PKZ-N) did not (Fig. 7).
Fig. 7.
Interaction of poly(I · C) with the N terminus of PKR but not with the N terminus of PKZ. The N terminus of PKR containing the three dsRBDs (PKR-N) and the N terminus of PKZ containing the two Z-DNA binding domains (ZBDs) (PKZ-N) and the C terminus of PKR containing only the kinase domain (PKR-C) were constructed and inserted into pcDNA3.1(+)-myc/His (Vector) at the EcoRI/KpnI sites. The primers are shown in Table S1 in the supplemental material. COS-7 cells seeded in 10-cm dishes were transfected with 8 μg each of the constructs as well as pcDNA3.1 by 20 μl Lipofectamine 2000. Cells were harvested using the same procedures as those used in the CoIP experiment. The poly(I · C) pulldown assay was carried out by the protocol given in our previous publication (44). Equal amounts of total protein were used in each pulldown assay. Lanes: inp, input (lysate before pulldown assay); pd, complexes that are pulled down by poly(I · C). IB, immunoblotting.
The possible dimerization of fish PKR or PKZ in cells treated with poly(I · C) was further investigated. Initially, PKR and PKZ were tagged with HA or myc and transfected into 293T cells followed by transfection of poly(I · C). The successful overexpression of all constructs was confirmed by Western blotting with antibodies to the tags (Fig. 8A, top blots). Interestingly, transfection of fish PKZ constructs but not fish PKR constructs generated three protein bands that were recognized by either the anti-HA antibody or anti-myc antibody (Fig. 8A, top blots). The largest protein was the full-length one, because it was immunoblotted by anti-PKZ antiserum that was produced by immunization of the N-terminal peptide of PKZ (Fig. 8C). Analysis of the coimmunoprecipitated complexes showed that both PKZ-HA and PKR-HA were successfully immunoprecipitated by an anti-HA antibody and that PKZ-HA was associated only with PKZ-myc, not with PKR-myc, and vice versa (Fig. 8A, bottom blots). Moreover, PKR-HA interacted with not only PKR-myc but also endogenous human PKR (Fig. 8A, bottom blots). The interaction of fish PKR with endogenous human eIF2α and PKR was further confirmed by transfection of PKR-HA alone into 293T cells (Fig. 8B). However, transfection of PKZ-HA alone into CAB cells or 293T cells showed that fish PKZ was bound to endogenous eIF2α but not to fish and mammalian PKR (Fig. 8C and D). These results suggested that fish PKZ and PKR might form homodimers but not heterodimers for their activation.
Fig. 8.
Independent association of fish PKZ and PKR. (A) PKR-HA, PKZ-HA, or vector (4 μg each) was cotransfected into 293T cells, seeded in 10-cm dishes with PKZ-myc or PKR-myc (4 μg each) in 5 ml medium, and the cells were stimulated by transfection of 0.2 μg/ml poly(I · C) at 8 hpt. The cells were harvested at 24 hpt and subjected to anti-HA IP assay. HA, myc, and human PKR (hPKR) antibodies were employed to analyze the input (top blots) and IP samples (bottom blots). αHA, anti-HA antibody. (B) 293T cells seeded in 10-cm dishes were transfected with 8 μg PKR-HA alone in 5 ml medium and stimulated by transfection of 0.2 μg/ml poly(I · C) at 8 hpt. The cells were harvested at 24 hpt and subjected to anti-HA IP or anti-IgG IP as a control. HA, PKR, eIF2α, and human PKR antibodies were employed in the Western blots. (C) Parallel experiment to the experiment shown in panel B but with PKZ-HA plasmid and PKZ antibody. Other conditions were the same as in panel B. N.S, nonspecific. (D) CAB cells seeded in 10-cm dishes were transfected with 8 μg PKZ-HA alone in 5 ml medium and stimulated by transfection of 0.2 μg/ml poly(I · C) at 8 hpt. The cells were harvested at 48 hpt and subjected to anti-HA IP or anti-IgG IP as a control. HA, PKZ, eIF2α, and crucian carp PKR antibodies were employed in Western blotting (immunoblotting [IB]).
DISCUSSION
Previous attempts to characterize fish IFN antiviral response resulted in identification of the PKR-like/PKZ gene in crucian carp (13) and later in other fish species (3, 26). The unique N-terminal structure and catalytic activity to the eIF2α substrate have suggested that PKZ is a novel member of the vertebrate eIF2α kinase family (3, 13, 26). However, the biological roles of PKZ remained to be established. In the present study, we extend these findings by confirming that fish PKZ is associated with cellular eIF2α under normal conditions and in response to IFN or poly(I · C) treatment, it is able to phosphorylate eIF2α, directly and firmly identifying fish PKZ as a functional eIF2α kinase. The results of overexpression and knockdown experiments showed that PKZ-mediated inhibition of viral replication is concomitant with the phosphorylation level of eIF2α bound to PKZ, suggesting that PKZ exerts antiviral function by phosphorylating eIF2α.
There are four eIF2α kinases in mammals. The feature of induction by IFNs distinguishes PKR from the other three kinases in mammals (38). The fact that fish PKZ is a typical IFN-inducible kinase indicates that it functions like PKR participating in an IFN antiviral response. Considering the fact that PKZ exists exclusively in fish, we have undertaken to characterize the biophysical and functional properties of PKZ in fish cells to compare them with the role of fish PKR in the IFN antiviral response. First, both proteins displayed similar expression patterns in response to IFN or all kinds of IFN stimuli, including poly(I · C), poly(dA-dT), poly(dG-dC), and genomic DNA (Fig. 2 and 3). Second, under these conditions, they were activated phosphorylating a common substrate, eIF2α (Fig. 3). Third, overexpression of both proteins together conferred much more significant inhibition of GCRV replication in fish cells than overexpression of either protein alone, whereas knockdown of both proteins together made fish cells more vulnerable to virus infection than knockdown of either protein alone (Fig. 5). Similarly, overexpression of both dominant-negative mutants (PKR-K419R and PKZ-K198R) in fish cells stimulated reporter gene expression more than that of either protein alone (Fig. 4). Finally, PKR and PKZ are arranged in a head-to-tail orientation in the genome from crucian carp (Fig. 1); this structure is thought to be very important for similar transcriptional activation after immunostimulation (25). Likewise, zebrafish PKZ mRNA and PKR mRNA are ubiquitously expressed in all kinds of investigated tissues and induced by poly(I · C) in the common tissues at almost equivalent expression levels (25). These results together suggest that fish PKR and PKZ play overlapping and cooperative roles in IFN-mediated antiviral response.
Whereas mixed antiviral effects of fish PKR and PKZ were observed in rIFN- and poly(I · C)-treated fish cells, overexpression and knockdown assays showed that fish PKR displayed a higher ability to block virus replication than PKZ did (Fig. 5 and 6). First, their promoters respond to poly(I · C) at different levels, since luciferase assays confirmed that fish PKR exhibited about 3-fold-stronger inhibition of reporter gene translation than PKZ did (Fig. 4). Second, based on the variable phosphorylation levels of eIF2α, which correlated with the different susceptibilities of fish cells to GCRV infection in the context of overexpression or knockdown of fish PKR and PKZ individually or collectively (Fig. 5 and 6), the higher antiviral ability of fish PKR over PKZ might be attributed to the stronger eIF2α phosphorylation ability of PKR over PKZ. Moreover, there might be other possible mechanisms resulting in this difference, for example, different sensitivities of fish PKR and PKZ to viral inhibitors.
It is well-known that the latent PKR is activated by binding to dsRNA prior to phosphorylating the eIF2α substrate. Given eIF2α phosphorylation as an index of fish PKR and PKZ activation, interesting questions of how the PKZ and PKR proteins are activated during transfection assays and in response to IFN or IFN stimuli are raised. In mammals, PKR is activated during transfection, since asymmetric transcription of transfected plasmid DNA might generate mRNA containing dsRNA features (15, 22). However, even if the activation of fish PKR during transfection assays is dependent on the mRNA with dsRNA features generated, the activation of fish PKZ remains unknown, since the two unique Z-DNA binding domains of PKZ specifically recognize Z-DNA or Z-RNA (26). GCRV is a member of the Aquareovirus genus of the family Reoviridae and contains a conserved structure involved in viral dsRNA interaction and transcription to other members of Reoviridae (5). The genomic dsRNA of GCRV is speculated to be recognized by host sensors for induction of IFN in UV-inactivated GCRV-infected CABs (42). In mammals, RIG-I and MDA5 recognize reovirus genome-derived dsRNA that trigger the IFN response, although reovirus dsRNA is rarely seen to be exposed during natural infection (29). Given that there is leaked dsRNA during GCRV replication and that the exposed dsRNA is responsible for induction of IFN and activation of fish PKR, it is hard to understand how PKZ is activated in GCRV-infected cells. Of course, we cannot rule out the possibility that PKZ is activated indirectly by activated PKR though binding to dsRNA or directly by Z-DNA produced by unknown mechanisms in virus-infected cells.
In the present study, poly(I · C) was bound to the N terminus of fish PKR but not to the N terminus of PKZ (Fig. 7), indicating that dsRNA is not able to activate PKZ. Poly(I · C) is consistently not able to activate recombinant Atlantic salmon PKZ (3). However, we observed that fish IFN treatment resulted in upregulation of both PKR and PKZ and a concomitant increase of eIF2α phosphorylation (Fig. 2 and 3). Therefore, one interpretation is that IFN might activate both fish proteins, at least partly, even in GCRV-infected cells. This speculation is supported by a recent finding that PKR is indeed a dual-specificity kinase (31, 32). Although PKR has been characterized as a serine/threonine kinase, IFNs induce tyrosine phosphorylation of human PKR by activating JAK1 and Tyk2, which are essential for kinase activation and eIF2α phosphorylation of PKR (32). Autophosphorylation of human PKR at Thr 446, a site essential for the optimal activation of PKR and eIF2α phosphorylation (7), is also observed and correlated with tyrosine phosphorylation in IFN-treated cells (32). Interestingly, we recently found that fish IRF3 is activated by IFN treatment, which is different from mammalian IRF3 (33). Although the details of the mechanism are not known and remain to be elucidated, this may highlight the physiological relevance of IFN-mediated activation of fish PKR and PKZ. That is, at an early phase of virus infection, both PKR and PKZ are activated and inhibit protein synthesis, while IFN is induced by virus infection. At the later phase of infection, the IFN produced acts in an autocrine and paracrine manner to induce and activate both fish kinases, which in turn cooperatively shut down protein synthesis.
In mammals, eIF2α kinase activation involves dimerization and autophosphorylation (6). The results of crystal structural analysis of a complex of the PKR kinase domain with an N-terminal fragment of eIF2α strengthen the importance of the catalytic-domain dimerization for activation of PKR and specific eIF2α substrate recognition (6), which is supported by the results of mutagenesis analysis (7). In this model, the binding of dsRBD to dsRNAs brings two PKR monomers into close proximity, promoting the dimerization of catalytic domains; such dimerization facilitates activation segment autophosphorylation, which in turn stabilizes the dimerization and promotes the specific recognition of the eIF2α substrate (7). In the present study, we observed the association of fish PKZ molecules or PKR molecules with either mammalian eIF2α (Fig. 8) or fish eIF2α (Fig. 3E). Further, fish PKR is associated even with human PKR molecules, but not with fish PKZ molecules (Fig. 8), suggesting that fish PKR and PKZ form homodimers, but not heterodimers, to phosphorylate eIF2α.
The independent dimerization of fish PKR or PKZ can explain why a dominant-negative effect was found in transfection of mammalian cells with fish PKR-K419R rather than with fish PKZ-K198R, although overexpression of fish PKR or PKZ individually is able to block reporter gene translation (Fig. 4). The reason is that mammalian cells have only PKR and not PKZ (25). In these luciferase assays, whereas fish PKR-K419R stimulates 1.5-fold reporter gene expression compared with control vector in mammalian and fish cells, fish PKZ-K198R stimulates the reporter to 1.3-fold only in fish cells and not in mammalian cells. In human cells, the corresponding enzymatically inactive mutant PKR-K296R stimulates reporter gene expression about 2- to 6-fold, probably by interacting with endogenous PKR leading to a dominant-negative effect (15, 36). Cotransfection of fish PKR-K419R and PKZ-K198R together exhibited a cooperatively stimulating effect in fish cells but not in mammalian cells. Consistent with our results, the zebrafish PKZ-K199R mutant does not stimulate reporter gene expression in human 293T cells and rodent CHO cells (26). These results together also suggest that dimerization of fish PKR or PKZ is required for them to arrest protein synthesis. Actually, dimerization of human PKR in the absence of dsRNA is sufficient to activate PKR (7, 18). The data described in this study reveal that fish PKR and PKZ play cooperative roles in the IFN antiviral immune response.
In conclusion, the data described in this study reveal the antiviral functions of both fish PKR and PKZ that are coexpressed in crucian carp. The common property of IFN induction or virus induction enables the proteins to function cooperatively during the IFN antiviral response. However, the independent association of PKR and PKZ suggests that each of these proteins is activated and subsequently phosphorylates eIF2α independently. In mammals, in addition to being an antiviral effector, PKR also acts as a dsRNA sensor detecting virus infection in the cytoplasm. It is reasonable to infer that fish PKZ is a new cytosolic sensor to detect Z-DNA or Z-RNA due to the unique N-terminal Zα domains. Interestingly, two Z-DNA binding domain-containing proteins identified in mammals, DAI (ZBP1/DLM-1) and ADAR1, have been confirmed to be involved in IFN signaling (10, 35, 39). So far, ADAR1 has been found either in mammals or in fish (14), while PKZ homologues are not found in mammals (13, 25) and fish seem not to contain DAI (unpublished data). Therefore, the coexistence of PKR and PKZ in the fish genome might be especially important to taxonomically lower fish species, since harboring different N-terminal structures enable fish PKR and PKZ to recognize different viral nucleic acids. This feature makes the fish innate immune system more effective by broadening their ability to sense viruses and to inhibit viral replication cooperatively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from 973 National Basic Research Program of China (2010CB126301 and 2010CB126303), the National Natural Science Foundation of China (30871922), and the national transgenic project (2009ZX08010-021B).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 21 September 2011.
REFERENCES
- 1. Al-khatib K., Williams B. R. G., Silverman R. H., Halford W., Carr D. J. J. 2003. The murine double-stranded RNA-dependent protein kinase PKR and the murine 2′,5′-oligoadenylate synthetase-dependent RNase L are required for IFN-beta-mediated resistance against herpes simplex virus type 1 in primary trigeminal ganglion culture. Virology 313:126–135 [DOI] [PubMed] [Google Scholar]
- 2. Balachandran S., et al. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129–141 [DOI] [PubMed] [Google Scholar]
- 3. Bergan V., Jagus R., Lauksund S., Kileng Ø., Robertsen B. 2008. The Atlantic salmon Z-DNA binding protein kinase phosphorylates translation initiation factor 2 alpha and constitutes a unique orthologue to the mammalian dsRNA-activated protein kinase R. FEBS J. 275:184–197 [DOI] [PubMed] [Google Scholar]
- 4. Cai R., Williams B. R. G. 1998. Mutations in the double-stranded RNA-activated protein kinase insert region that uncouple catalysis from eIF2α binding. J. Biol. Chem. 273:11274–11280 [DOI] [PubMed] [Google Scholar]
- 5. Cheng L., Fang Q., Shah S., Atanasov I. C., Zhou Z. H. 2008. Subnanometer-resolution structures of the grass carp reovirus core and virion. J. Mol. Biol. 382:213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dar A. C., Dever T. E., Sicheri F. 2005. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell 122:887–900 [DOI] [PubMed] [Google Scholar]
- 7. Dey M., et al. 2005. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell 122:901–913 [DOI] [PubMed] [Google Scholar]
- 8. Fu Y., et al. 1999. Cloning of DLM-1, a novel gene that is up-regulated in activated macrophages, using RNA differential display. Gene 240:157–163 [DOI] [PubMed] [Google Scholar]
- 9. Garner J. N., Joshi B., Jagus R. 2003. Characterization of rainbow trout and zebrafish eukaryotic initiation factor 2alpha and its response to endoplasmic reticulum stress and IPNV infection. Dev. Comp. Immunol. 27:217–231 [DOI] [PubMed] [Google Scholar]
- 10. George C. X., Gan Z., Liu Y., Samuel C. E. 2011. Adenosine deaminases acting on RNA, RNA editing, and interferon action. J. Interferon Cytokine Res. 31:99–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Herbst A., Tansey W. P. 2000. HAM: a new epitope-tag for in vivo protein labeling. Mol. Biol. Rep. 27:203–208 [DOI] [PubMed] [Google Scholar]
- 12. Hough R. F., Bass B. L. 1994. Purification of the Xenopus laevis double-stranded RNA adenosine deaminase. J. Biol. Chem. 269:9933–9939 [PubMed] [Google Scholar]
- 13. Hu C. Y., Zhang Y. B., Huang G. P., Zhang Q. Y., Gui J. F. 2004. Molecular cloning and characterisation of a fish PKR-like gene from cultured CAB cells induced by UV-inactivated virus. Fish Shellfish Immunol. 17:353–366 [DOI] [PubMed] [Google Scholar]
- 14. Jin Y., Zhang W., Li Q. 2009. Origins and evolution of ADAR-mediated RNA editing. IUBMB Life 61:572–578 [DOI] [PubMed] [Google Scholar]
- 15. Kaufman R. J., Davies M. V., Pathak V. K., Hershey J. W. 1989. The phosphorylation state of eucaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol. Cell. Biol. 9:946–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim U., Wang Y., Sanford T., Zeng Y., Nishikura K. 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. U. S. A. 91:11457–11461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee S. B., Esteban M. 1993. The interferon-induced double-stranded RNA-activated human p68 protein kinase inhibits the replication of vaccinia virus. Virology 193:1037–1041 [DOI] [PubMed] [Google Scholar]
- 18. Lemaire P. A., Lary J., Cole J. L. 2005. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J. Mol. Biol. 345:81–90 [DOI] [PubMed] [Google Scholar]
- 19. Mathews M. B., Sonenberg N., Hershey J. W. B. (ed.). 2007. Translational control in biology and medicine. Cold Spring Harbor Monograph Series 48. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20. Partridge M., et al. 1996. A simple method for delivering morpholino antisense oligos into the cytoplasm of cells. Antisense Nucleic Acid Drug Dev. 6:169–175 [DOI] [PubMed] [Google Scholar]
- 21. Patel R. C., Sen G. C. 1992. Identification of the double-stranded RNA-binding domain of the human interferon-inducible protein kinase. J. Biol. Chem. 267:7671–7676 [PubMed] [Google Scholar]
- 22. Patel R. C., Stanton P., Sen G. C. 1996. Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J. Biol. Chem. 271:25657–25663 [DOI] [PubMed] [Google Scholar]
- 23. Rojas M., Arias C. F., Lopez S. 2010. PKR is the kinase responsible for the phosphorylation of eIF2alpha in rotavirus infection. J. Virol. 84:10457–10466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Romano P. R., et al. 1998. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol. Cell. Biol. 18:2282–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rothenburg S., Deigendesch N., Dey M., Dever T., Tazi L. 2008. Double-stranded RNA-activated protein kinase PKR of fishes and amphibians: varying the number of double-stranded RNA binding domains and lineage-specific duplications. BMC Biol. 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rothenburg S., et al. 2005. A PKR-like eukaryotic initiation factor 2α kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl. Acad. Sci. U. S. A. 102:1602–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ryter J. M., Schultz S. C. 1998. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17:7505–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Samuel M. A., et al. 2006. PKR and RNase L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80:7009–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sherry B. 2009. Rotavirus and reovirus modulation of the interferon response. J. Interferon Cytokine Res. 29:559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sonenberg N., Hinnebusch A. G. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Su Q., et al. 2006. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2α RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 103:63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su Q., et al. 2007. Interferons induce tyrosine phosphorylation of the eIF2alpha kinase PKR through activation of Jak1 and Tyk2. EMBO Rep. 8:265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun F., et al. 2010. Characterization of fish IRF3 as an IFN-inducible protein reveals evolving regulation of IFN response in vertebrates. J. Immunol. 185:7573–7582 [DOI] [PubMed] [Google Scholar]
- 34. Sun F., et al. 2011. Fish MITA serves as a mediator for distinct fish IFN gene activation dependent on IRF3 or IRF7. J. Immunol. 187:2531–2539 [DOI] [PubMed] [Google Scholar]
- 35. Takaoka A., et al. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501–505 [DOI] [PubMed] [Google Scholar]
- 36. Terenzi F., et al. 1999. The antiviral enzymes PKR and RNase L suppress gene expression from viral and non-viral based vectors. Nucleic Acids Res. 27:4369–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thomis D. C., Samuel C. E. 1992. Mechanism of interferon action: autoregulation of RNA-dependent P1/eIF-2 alpha protein kinase (PKR) expression in transfected mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 89:10837–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wek R. C., Jiang H. Y., Anthony T. G. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34:7–11 [DOI] [PubMed] [Google Scholar]
- 39. Wulff B. E., Sakurai M., Nishikura K. 2011. Elucidating the inosinome: global approaches to adenosine-to-inosine RNA editing. Nat. Rev. Genet. 12:81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yeung M. C., Chang D. L., Camantigue R. E., Lau A. S. 1999. Inhibitory role of the host apoptogenic gene PKR in the establishment of persistent infection by encephalomyocarditis virus in U937 cells. Proc. Natl. Acad. Sci. U. S. A. 96:11860–11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu F. F., et al. 2010. Fish virus-induced interferon exerts antiviral function through Stat1 pathway. Mol. Immunol. 47:2330–2341 [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y. B., et al. 2007. The innate immune response to grass carp hemorrhagic virus (GCHV) in cultured Carassius auratus blastulae (CAB) cells. Dev. Comp. Immunol. 31:232–243 [DOI] [PubMed] [Google Scholar]
- 43. Zhang Y. B., Zhang Q. Y., Xu D. Q., Hu C. Y., Gui J. F. 2003. Identification of antiviral-relevant genes in the cultured fish cells induced by UV-inactivated virus. Chin. Sci. Bull. 48:581–588 [Google Scholar]
- 44. Zhu R., Zhang Y. B., Zhang Q. Y., Gui J. F. 2008. Func-tional domains and the antiviral effect of the double-stranded RNA-dependent protein kinase PKR from Paralichthys olivaceus. J. Virol. 82:6889–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.